ABSTRACT
Recently studies reported that long non-coding RNAs (lncRNAs) may take part in a lot of congenital diseases, meanwhile, Hirschsprung's disease (HSCR) is a major congenital digestive tract malformation. Nevertheless whether lncRNAs participate in the occurrence of HSCR and how it contributes to this disease are still unknown. LOC100507600 was selected from our gene expression microarray data obtained from bowel tissues from HSCR patients and negative controls. Subsequently, we used qRT-PCR to prove the result in 64 pairs of HSCR disease bowel stenosis tissues and negative controls. Transwell assay, CCK-8 assay and flow cytometry were employed to explore whether cellular functions change after knocking down the LOC100507600 in SH-SY5Y cell and human 293T cell. Dual-luciferase reporter assay was used to confirm the competitive relationship between BMI1 and LOC100507600 through their association with hsa-miR128–1-3p. Protein extraction and Western blotting were used to further confirm the relationship between LOC100507600 and BMI1. We found that LOC100507600 was obvious reduced in tissues from HSCR patients with noteworthy correlation with BMI1. Furthermore, Downregulation of LOC100507600 repressed cell migration and proliferation and didn't affect cell apoptosis or cycle. Dual-luciferase reporter assay, qRT-PCR and Western blotting assay verified that LOC100507600 serves as a competitive endogenous RNA of miR128–1-3p and down-regulates BMI1 expression by sponging miR128–1-3p in HSCR. In sum, our study researches the potential diagnostic value of LOC100507600 in HSCR and deduces that LOC100507600 can contributes to HSCR as a competitive endogenous RNA to regulate BMI1 expression by sponging miR128–1-3p.
Introduction
Hirschsprung's disease (HSCR), which is also known as congenital megacolon or intestinal aganglionosis, is distinguished by an absence of enteric (intrinsic) neurons from variable lengths of the most distal bowel [Citation1]. The incidence of HSCR disease in newborns is about 1/5000 and the male: female ratio is 4:1 [Citation2]. By now EDNRB and RET are still the most important genes proved to contribute to this disease [Citation3]. According to our previous studies LOC101926975, Circular RNA ZNF609, miR-206, and even HN12 participate in the occurrence of HSCR [Citation4–7]. However, the exact underlying mechanism is unknown. So the pathogenesis of HSCR needs further exploration.
Long noncoding RNAs (lncRNAs), once thought to be transcriptional noise [Citation8], have been clearly demonstrated to regulate a variety of biochemical processes in some levels such as pre-transcriptional, post-transcriptional and translational level [Citation9–11]. However, more and more evidences suggest that this type of RNAs contributes to the occurrence of HSCR [Citation4]. Weather some more lncRNAs participate in this disease needs further research.
MicroRNAs are known to have significant functions in many growth and development processes [Citation12]. Recently, some lncRNAs have been proved to work as ceRNA to influence gene expression [Citation13]. It needs to be explored whether lncRNAs can serve as competing endogenous RNAs in the occurrence of HSCR disease.
Our prevenient work has proved the expression profile of long non-coding RNA in HSCR [Citation14]. One of them is LOC100507600, which is obviously down regulated among HSCR cases. LOC100507600 is lied in chromosome 2 (135820191..135823087) with a neighbor gene called miR128–1. miR128–1 suppresses prostate cancer by inhibiting BMI1 to inhibit the proliferation and other capacity of tumor-initiating cells [Citation15]. So, we planned to explore the potential function of LOC100507600 in HSCR. We hypothesized whether LOC100507600 is crucial for the development of enteric nervous system as a competitive endogenous RNA to regulate BMI1 expression. Experiments were performed to uncover the biological functions of LOC100507600, which may be a potential factor of HSCR.
Results
The expression of LOC100507600 in HSCR
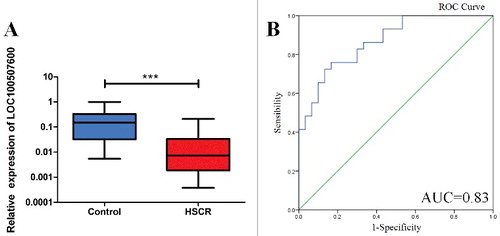
We collected 64 pairs of colon tissues containing 64 HSCR cases and 64 matched negative controls for this scientific research. The age, sex and body weight of patients were recorded when we were collecting samples, with parents approached for consent as soon as possible. There is no obviously difference between HSCR cases and normal controls in days, body weight and sex as shown in . According to the qRT-PCR assays, the expression of LOC100507600 was obviously down-regulated in HSCR cases compared with the negative controls (). As is known, lncRNAs can also have the potential diagnostic value in some diseases. So, we used ROC curve to evaluate whether LOC100507600 can distinguish HSCR disease from normal controls (). We found the value of AUC was 0.83. Maybe LOC100507600 serve as biomarker of HSCR.
Table 1. Clinical features of study population.
Figure 1. LOC100507600 is down-regulated in HSCR. (A) The expression of LOC100507600 in HSCR tissues (n = 64) and control tissues (n = 64). LOC100507600 was significantly reduced in patient tissues compared with control tissues. (B): Receiver Operating Characteristic (ROC) curve for the LOC100507600 to distinguish HSCR cases from controls.
The knockdown of LOC100507600 inhibits cell proliferation and migration without influence on cell apoptosis or cycle
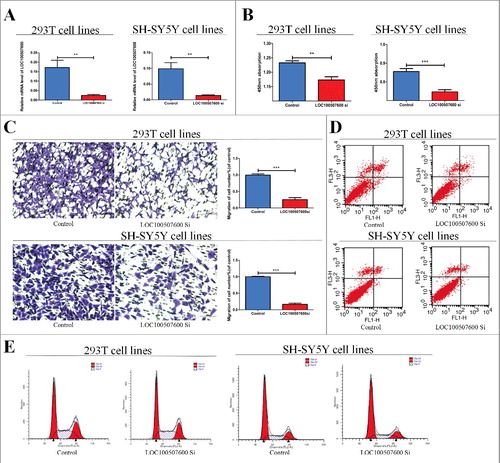
To learn more about LOC100507600, We purchased short interfering RNAs (siRNAs) to down-regulate the expression of LOC100507600 in SH-SY5Y cells and human 293T cells. After transfection we confirmed that the expression of LOC100507600 could be obviously down-regulated by siRNA (). The CCK8 assays and Transwell assays were conducted to verify whether proliferation and migration change with suppression of LOC100507600. Experimental results showed that the ability of cell proliferation and migration was obviously inhibited by down-regulation of LOC100507600 ( and ). Flow cytometry verified that the suppression of LOC100507600 can not obvious affect cell cycle progression and apoptosis ( and ).
Figure 2. Cytobiology change after treating cells with LOC100507600 siRNA. (A) Human 293T and SH-SY5Y cell lines were transfected with LOC100507600 siRNA and then qRT-PCR is repeated three times to determine the efficiency of transfection. (B) Human 293T and SH-SY5Y cell lines were transfected with LOC100507600 siRNA to regulate its expression levels and cell proliferation was detected using the CCK8 assay. Knockdown of LOC100507600 suppressed cell proliferation. (C) Transwell assay was performed as described in method and indicated that down-regulation of LOC100507600 delayed cell migration. Pictures were captured under a light microscope with the magnification, × 10. (D and E)Flow cytometry demonstrated that the down-regulation of LOC100507600 had no effect on cell cycle progression and apoptosis.
Subcellular localization of LOC100507600
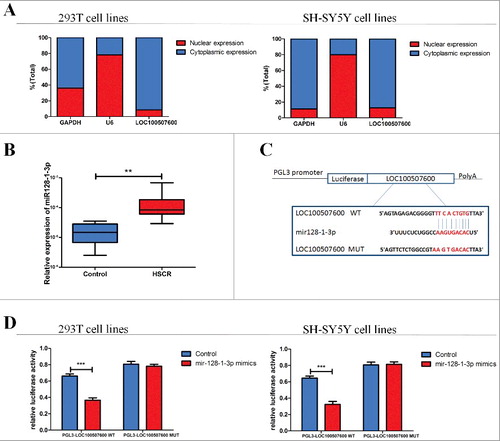
As is known, the subcellular localization of lncRNAs determines its type of action. We separated the total RNA of cells into nuclear and cytoplasmic fractions. We used the U6 and the GAPDH as the control because the U6 lied mostly in the nuclear fraction, meanwhile the GAPDH distributed mainly in the cytoplasmic fraction. The results of qRT-PCR showed the LOC100507600 was detected 87.5% and 91.5% in the cytoplasm fraction of SH-SY5Y cells and human 293T cells respectively (). So the LOC100507600 located mainly in the cytoplasm fractions, which indicates it may play a part in the post-transcriptional regulation of gene.
Figure 3. LOC100507600 serves as a sponge for miR128–1-3p. (A) The levels of nuclear control transcript (U6), cytoplasmic control transcript (GAPDH), and LOC100507600 were assessed by qRT-PCR in nuclear and cytoplasmic fractions. (B) The expression of miR128–1-3p in HSCR tissues and normal tissues. miR128–1-3p was significantly rose in patient tissues compared with normal tissues. (C) Superstratum:sequence alignment of human miR128–1-3p with LOC100507600. Bottom: mutations in the LOC100507600 sequence to create the mutant luciferase reporter constructs. (D) Luciferase reporter assay in 293T and SH-SY5Y cells after transfected with negative control or miR128–1-3p mimics, renilla luciferase vector pRL-SV40 and the reporter constructs. Both frefly and renilla luciferase activities are measured in the same sample. Firefly luciferase signals were normalized with renilla luciferase signals.
LOC100507600 serves as a sponge for miR128–1-3p
Now we have proved that LOC100507600 was obvious suppression in HSCR tissues and could down-regulate cell migration and proliferation. But how it contributes to the occurrence of HSCR needs further prove. Lately, more and more evdience indicated that lncRNAs could function as sponges of miRNA. We predicted that LOC100507600 have binding sites with several miRNAs by RegRNA (http://regrna.mbc.nctu.edu.tw/html/prediction.html), and miR128–1-3p whose gene position is near to the gene of LOC100507600 has very high score. The relative miRNA level of miR128–1-3p was detected in HSCR tissues and negative controls, as shown in . To further confirm the relationship between miR128–1-3p and LOC100507600, an episode of LOC100507600 containing the mutant target site or the predicted target site was added to the downstream of the firefly luciferase gene (named as pGL3-LOC100507600-Wild and pGL3-LOC100507600-Mut) (). The plasmid was co-transfected with miR128–1-3p negative control or mimics into SH-SY5Y cells or 293T cells. As shown in miR128–1-3p mimics mediated a down regulation of the luciferase activity in pGL3-LOC100507600-Wild compared with the negative control. On the contrary, there was no obvious discrepancy in relative luciferase expression of pGL3-LOC100507600-Mut between miR128–1-3p mimics and the negative control (). These results indicated that LOC100507600 directly targets miR128–1-3p in vitro.
LOC100507600 regulates the miR128–1-3p target, BMI1
To investigate the molecular mechanisms through which miR128–1-3p exerts its effects, we used several bioinformatics tools, such as Regrna TargetScan, Microcosm V5, and miRanda to screen its potential targets. Based on the sequence complementarity to miR128–1-3p seed sequence, we chose BMI1 which has been notarized to be the target of miR128–1-3p in prostate cancer [Citation15]. To measure this conclusion, the mRNA expression level and protein expression level of BMI1 were detected in HSCR tissues and negative controls. BMI1 was significant suppressed in HSCR patients compared with negative controls (,). We applied Bivariate correlation analysis to explore the relationship between LOC100507600 and BMI1 in normal and HSCR tissues (). The consequences showed that the relative expression of BMI1 positively correlated with that of LOC100507600 in control and HSCR tissues. To verify whether miR128–1-3p interacts with BMI1, We transfected the SH-SY5Y cells and human 293T cells with plasmid pGL3-BMI1-Wild and pGL3-BMI1-Mut, which include the mutant binding site and predicted binding site (). The miR128–1-3p mimics in cells which were transfected with the pGL3-BMI1-Wild obviously suppress the level of luciferase activity compared with the negative controls. On the contrary, there is no difference in cells transfected with pGL3-BMI1-Mut between miR128–1-3p mimics and negative controls (). All in all, the findings further proved that BMI1 is a biological target of miR128–1-3p. Meanwhile we found that miR128–1-3p inhibitor reversed the relative mRNA levels of BMI1 which was downregulated by siRNA of LOC100507600 (). Western blotting analysis of BMI1 overall corroborated the qRT-PCR results. In conclusion, these results uphold that LOC100507600 regulates BMI1 by competitive binding miR128–1-3p.
Figure 4. LOC100507600 regulates the miR-128–1-3p target, BMI1. (A) Relative expression of BMI1 in HSCR tissues in comparison with control tissues. BMI1 was significantly reduced in patient tissues. (B) Protein level of BMI1 in HSCR tissues and normal control samples were detected by Western Blot assay. (C) Bivariate correlation analysis of the relationship between LOC100507600 and BMI1 expression level. (D) Superstratum:the putative miRNA binding sites in the BMI1 sequence. The putative miRNA recognition sites were cloned downstream of the luciferase gene and named pGL3-BMI1-Wild. Bottom: mutations in the BMI1 sequence to create the mutant luciferase reporter constructs named pGL3-BMI1-Mut. (E) Left: The luciferase reporter in 293T cell lines. Right: the luciferase reporter in SH-SY5Y cells. Luciferase activity was determined using the dual luciferase assay and shown as the relative luciferase activity normalized to renilla activity.
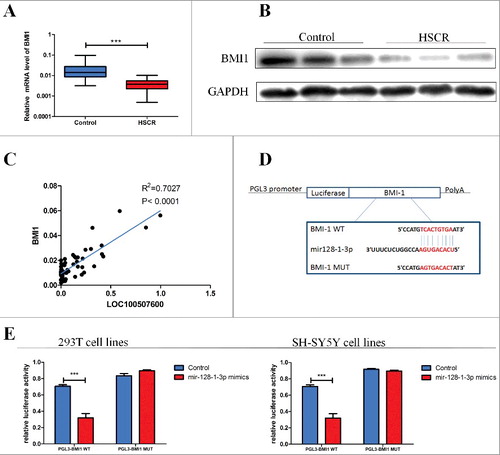
Figure 5. LOC100507600-miR128–1-3p regulatory loop is critical for cell function. (A) LOC100507600 siRNA with or without miR128–1-3p inhibitor was transfected into 293T cells and the relative expression of BMI1 was evaluated by qRT-PCR, × 20 (B) Western blot analysis of BMI1 protein level following treatment of 293T and SH-SY5Y cells with LOC100507600 siRNA or with LOC100507600 siRNA plus miR128–1-3p inhibitor. GAPDH was used as control. (C and D) CCK8 assay and Transwell assays were performed to determine the proliferation and migration of miR128–1-3p transfected cells and treated with LOC100507600 siRNA plus miR128–3p inhibitor.
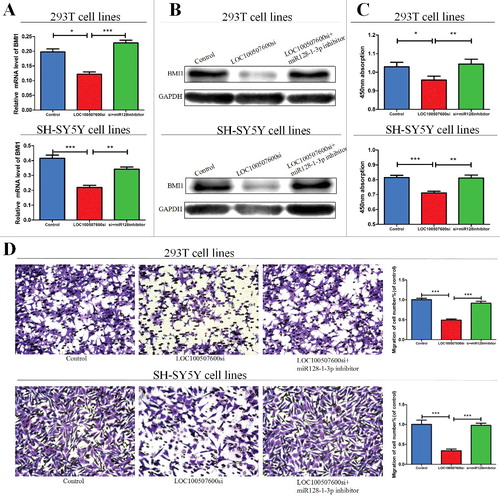
LOC100507600-miR128–1-3p regulatory loop is important for cellular functions
We next explored whether miR128–1-3p can participate in cell migration and proliferation. The expression of miR128–1-3p was decreased by miR128–1-3p inhibitor. The migration and proliferation ability of cells deal with LOC100507600 siRNA were obviously reversed by miR128–1-3p inhibitor ( and ). Above all, these results reflected the interaction among LOC100507600, miR128–1-3p, and BMI1.
Discussion
HSCR characterized by the absence of enteric neurons in the distal gut is the second most common digestive malformation in the newborns [Citation16]. Abdominal distension and constipation are the main clinical symptoms. Uncured HSCR is a deadly disease especially with Hirschsprung's disease associated enterocolitis [Citation17]. So, it is necessary to investigate the nosogenesis of HSCR. Recently long non-coding RNA have been proved to play important roles in a lot of diseases [Citation18–20]. According to the Microarray expression results of lncRNAs between HSCR and negative controls [Citation14]. LOC100507600 is significantly down-regulated in HSCR tissues. We verified this phenomenon by qRT-PCR assay between colon tissues from HSCR patients and control patients. Assays in vitro showed that LOC100507600 influences cell migration and cell proliferation without impacts on cell cycle or apoptosis.
LncRNAs can serve as biomarkers in cancers [Citation21]. For example, lncRNA-n336928 is related to bladder cancer [Citation22]. Traditional biomarkers are mainly based on the blood, which may affect the stability and sensitivities of outcomes [Citation23]. So we tried to evaluate the potential diagnostic value of LOC100507600 among tissue samples. We found LOC100507600 is significantly down-regulated in the tissues of HSCR patients with the AUC of 0.83, which means that LOC100507600 may efficiently distinguish HSCR cases from negative controls. Generally speaking, we attest that down-regulation of LOC100507600 can impact cell migration and proliferation and LOC100507600 can act as biomarker for HSCR disease. But as is known to us all, tissues are much more difficult to obtain, we planned to further explore the potential diagnostic value of faeces.
However, the molecular mechanisms of the phenomenon need further study. To further explore the mechanism of LOC100507600, miR128 which is in the vicinity of LOC100507600 on chromosome came into our notice. LncRNAs positively or negatively contribute to the relative expression of neighboring genes namely cis regulation [Citation24]. Some well-known lncRNAs such as Air and Xist can regulate nearby even distant genes by acting on histone modification complexes [Citation25,26]. In consequence, we wonder whether LOC100507600 regulates miR128–1-3p at the level of pre-transcription. Then we explored the subcellular localization of LOC100507600 in cells and discovered the LOC100507600 mainly lied in the cytoplasm fraction, which indicates the probability that it functions at the level of post-transcription. Anyhow whether LOC100507600 paticipates in the process from Pre-miR128 to mature miR128–1-3p needs further study.
As is known, lncRNAs can also function as miRNA sponge in thyroid cancer [Citation27], cervical cancer [Citation28] and gastric cancer [Citation29]. We speculated that LOC100507600 might competitive bind to the miR128–1-3p, thereby stopping its target RNA from suppression. To demonstrate this viewpoint, we verified the miRNA by sequence analysis and operated dual-luciferase reporter gene assay. As expected, miR128–1-3p has a high score according to bioinformatic prediction and could diminish the fluorescence of pGL3-LOC100507600-Wild. These results verified the combination between the miR128–1-3p and LOC100507600. Moreover, lower expression of miR128–1-3p by inhibitor reversed the cell migration and proliferation, which was repressed by the knockdown of LOC100507600. To sum up, these results proved the conjecture that LOC100507600 influenced cellular functions by interacting with miR128–1-3p.
Then we took miR128–1-3p for further study. Coincidentally, the site of binding between LOC100507600 and miR128–1-3p is extremely similar to this between miR128–1-3p and BMI. In our assays, BMI1 is conformed to be a authentic target gene of miR128–1-3p by luciferase assay. Meanwhile, qRT-PCR and western blot demonstrated BMI1 was down-regulated in HSCR. So upregulation of miR128–1-3p is related to cell proliferation, migration, and so on by binding to BMI. What's more, miR128–1 suppresses BMI1 to inhibit the proliferation and other capacity of tumor-initiating cells has been proved in prostate cancer [Citation15]. In a word, our study attest the hypothesis that LOC100507600 serves as a miR128–1-3p sponge and influence the expression of the miR128–1-3p target gene BMI1. In the meantime LOC100507600 may contribute to the diagnosis of HSCR.
Material and methods
Tissue collection
We got permitted by the Institutional Ethics Committee of Nanjing Medical University. All the tissue samples were gathered after a consent form. A total of 64 pairs of HSCR and negative controls were gathered from Children's Hospital of Nanjing Medical University, Nanjing China from 2011 to 2016 and immediately stored at −80 °C. The diagnosis was finally conformed via postoperative pathological analysis of bowel tissues. The normal colon tissues which were regarded as negative controls were from patients diagnosed to be without congenital malformation. The clinical information of the 64 pairs of patients are listed in .
RNA extract and qRT-PCR
We extracted total RNA from each cell line and tissue on the basis of the product instruction of Trizol reagent (Life Technologies, CA, US) and separate it into cytoplasmic and nuclear part according to the Manufacturer Product Description of RNAqueous™ Total RNA Isolation Kit (Thermo Scientifc, Wilmington, DE, USA). To assure the purity and concentration of RNA, we used NanoDrop 2000 Spectrophotometer (Thermo Scientifc, Wilmington, DE, USA). The total RNAs (500 ng) were reversed transcription by Reverse Transcription Kit 36A (Takara, Tokyo, Japan) for the qRT-PCR assays on the ABI 7900HT (Applied Biosystems) to measure the relative expression of lncRNA, miRNA and mRNA. The reaction conditions of qRT-PCR were performed according to the universal method. All the assays were in triplicate respectively. Statistical analysis in relative expression were calculated by 2−ΔCt. The primers involved in this experiment are summarized in .
Table 2. Sequences of primers for qRT-PCR and siRNA related sequence.
Protein extraction and Western blotting
We used RIPA buffer containing protease inhibitors (cOmplete, ULTRA, mini, EDTA-free,EASY pack Roche) to extract total proteins from cultured cells and tissues for western blot. To assure the concentration of RNA bicinchoninic acid (BCA) solution Beyotime (Nantong, China) was used. The Western Blot assays were performed with the generic method. The control primary antibodies (antiGAPDH) and the secondary antibodies were purchased from Beyotime (Nantong, China). In addition, another primary antibodies (anti-BMI1) was purchased from ProteinTech (Chicago, IL).
Cell culture and SiRNA transfection
SH-SY5Y and human 293T cell were purchased from American Type Culture Collection (ATCC, Manassas VA, USA) which were cultured in DMEM (Hyclone, UT, USA), mixed with 10% fetal bovine serum (10% FBS), penicillin (100 U/ml), and streptomycin (100 µg/mL) at 37 °C, 5% CO2. The small interfering RNA against LOC100507600, miR128–1-3p mimics and controls () were obtained from GenePharma (Shanghai,China). All cells, which were cultured to about 50%–60% view of cell-culture dish, were transfected with 50nM negative control or 100nM siRNA by Lipofectamine 2000 Reagent (Invitrogen, CA, USA).
Migration assay
Transwell migration chambers (8 μm pore size, Millipore Corporation, Billerica, MA) were utilized to measure the ability of cell migration. Every single-cell suspension of 1 × 105 transfected or control cells in 100 µl of serum-free culture medium was injected to the upper chamber meanwhile 600ul DMEM/F12 medium with 10% FBS was added to the bottom well. After incubating for 24–48 h, the cells were fasten by methanol for 20 min, dyed by crystal violet staining solution for 30 min (Beyotime, Nantong, China) and washed by PBS for three times. The quantity and shape of cells were observed with Image-pro Plus 6.0. All the assays were performed for triple times respectively.
Proliferation assay
Cells were cultivated on 96-well plates and incubated for 24 h then kept 1 h with CCK8 (Beyotime, Nantong, China). Then we used the TECAN infnite M200 Multimode microplate reader (Tecan, Mechelen, Belgium) to measure the absorbance at 450nm of the treated cell. All assays were performed triple times independently.
Dual-luciferase reporter assay
The binding sites of LOC100507600 and miR128–1-3p called LOC100507600-Wild, LOC100507600-Mut were stepped in the KpnI and SacI sites of pGL3 promoter vector (Realgene, Nanjing, China) for Dual-luciferase reporter assay. The binding sites of BMI1 and miR128–1-3p called BMI1-Wild, BMI1-Mut were also stepped in the KpnI and SacI sites of pGL3 promoter vector (Realgene, Nanjing,China) for Dual-luciferase reporter assay. The first, Cells were plated on 24-well plates. The second 5 ng renilla luciferase vector pRL-SV40, 80 ng plasmid, 50 nM miR128–1-3p mimics and controls mixed with lipofectamine 2000 (Invitrogen, Shanghai, China) were transfected into cells. Last but not the least cells were gathered and measured according to the standard steps by utilizing the DualLuciferase Assay (Promega, Madison, WI, USA) after transfecting for 36–48 h. All assays were repeated in triplicate.
Statistical analysis
SPSS20.0 and GraphPad prism software were used for statistical analysis. Chisquare tests and Student's t-test were employed to detect statistical discrepancy in demographic data. Receiver operating characteristic (ROC) curves analysis was executed to assess the diagnostic value of LOC100507600 expression levels in HSCR. Pearson correlation analysis were employed to analyze the relationship between BMI1 and LOC100507600. And other results of cells or tissues experiments were evaluated via double-sided Student's t-test. Results of Statistical analysis were considered different at P< 0.05 level.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Xiaoqun Xu, Qiming Gen, Jie Zhang, Huan Chen, Changgui Lu, Weiwei Jiang and Wei Li (Nanjing Children's Hospital Affiliated to Nanjing Medical University) for sample collection. This work was supported by Natural Science Foundation of China (NSFC 81370473, NSFC 81570467), and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Additional information
Funding
References
- McKeown SJ. HSCR disease: a developmental disorder of the enteric nervous system. WIREs Dev Biol. 2013;2:113–129. doi:10.1002/wdev.57.
- Goldberg EL. An epidemiological study of HSCR disease. Int J Epidemiol. 1984;13:479–485. doi:10.1093/ije/13.4.479. PMID:6240474
- Wallace AS, Anderson RB. Genetic interactions and modifier genes in HSCR's disease. World J Gastroenterol. 2011;17:4937–4944. doi:10.3748/wjg.v17.i45.4937. PMID:22174542
- Shen Z. Downregulated expression of long non-coding RNA LOC101926975 impairs both cell proliferation and cell cycle and its clinical implication in HSCR disease patients. Int J Med Sci. 2016;13(4):292–297. doi:10.7150/ijms.14187. PMID:27076786
- Peng L. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150–5p in HSCR disease. Oncotarget. 2017 Jan 3;8(1):808–818. doi: 10.18632/oncotarget.13656.
- Sharan A. Corrigendum: down-regulation of miR-206 is associated with HSCR disease and suppresses cell migration and proliferation in cell models. Sci Rep. 2015 Mar 20;5:9302. doi: 10.1038/srep09302. doi:10.1038/srep09302.
- Du C. Apoptotic neuron-secreted hN12 inhibits cell apoptosis in HSCR disease. Int J Nanomedicine. 2016;11:5871–5881. doi:10.2147/IJN.S114838. PMID:27853370
- Brosius J. Waste not, want not–transcript excess in multicellular eukaryotes. Trends Genet. 2005;21:287–288. doi:10.1016/j.tig.2005.02.014. PMID:15851065
- Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90:430–440. doi:10.1093/cvr/cvr097. PMID:21558279
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi:10.1016/j.cell.2013.02.012. PMID:23498938
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi:10.1038/nrm3679. PMID:24105322
- Ghildiyal, M, Zamore, PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi:10.1038/nrg2504. PMID:19148191
- Liu XH. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331–3p in gastric cancer. Mol Cancer. 2014 Apr 28;13:92. doi: 10.1186/1476-4598-13-92. doi:10.1186/1476-4598-13-92.
- Shen Z. Microarray expression profiling of dysregulated long non-coding RNAs in HSCR's disease reveals their potential role in molecular diagnosis. Neurogastroenterol Motil. 2016 Feb;28(2):266–273. doi: 10.1111/nmo.12722. Epub 2015 Nov 17. doi:10.1111/nmo.12722.
- Jin M. miR-128 suppresses prostate cancer by inhibiting BMI1 to inhibit tumor-initiating cells. Cancer Res. 2014 Aug 1;74(15):4183–4195. doi:10.1158/0008-5472.CAN-14-0404.
- McKeown SJ, Stamp L, Hao MM, et al. HSCR disease: a developmental disorder of the enteric nervous system. Wiley Interdiscip Rev Dev Biol. 2013 Jan-Feb;2(1):113–29. doi: 10.1002/wdev.57. Epub 2012 Apr 24. doi:10.1002/wdev.57.
- Momoh JT. HSCR's disease: problems of diagnosis and treatment. Ann Trop Paediatr. 1982;2:31–35. doi:10.1080/02724936.1982.11748220. PMID:6186188
- Ellis BC, Molloy PL, Graham LD. CRNDE: a long non-coding RNA involved in Cancer, neurobiology, and development. Front Genet. 2012;3:270. doi:10.3389/fgene.2012.00270. PMID:23226159
- Lin N, Chang KY, Li Z, et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell. 2014;53:1005–1019. doi:10.1016/j.molcel.2014.01.021. PMID:24530304
- Yang M, Zhai X, Xia B, et al. Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumour Biol. 2015 Sep;36(10):7615–7622. doi: 10.1007/s13277-015-3465-4. Epub 2015 Apr 29. doi:10.1007/s13277-015-3465-4.
- Yang Y, Shao Y, Zhu M, et al. Using gastric juice lncRNA-ABHD11-AS1 as a novel type of biomarker in the screening of gastric cancer. Tumour Biol. 2016 Jan;37(1):1183–1188. doi: 10.1007/s13277-015-3903-3. Epub 2015 Aug 18. doi:10.1007/s13277-015-3903-3.
- Chen T, Xie W, Xie L, et al. Expression of long noncoding RNA lncRNA-n336928 is correlated with tumor stage and grade and overall survival in bladder cancer. Biochem Biophys Res Commun. 2015 Dec 25;468(4):666–670. doi: 10.1016/j.bbrc.20150. Epub 2015 Nov 6. doi:10.1016/j.bbrc.2015.11.013.
- Liu HS, Xiao HS. MicroRNAs as potential biomarkers for gastric cancer. World J Gastroenterol. 2014;20:12007–12017. doi:10.3748/wjg.v20.i34.12007. PMID:25232237
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi:10.1016/j.cell.2013.02.012. PMID:23498938
- Zhao J, Ohsumi TK, Kung JT, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40: 939–953. doi:10.1016/j.molcel.2010.12.011. PMID:21172659
- Nagano T, Mitchell JA, Sanz LA, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi:10.1126/science.1163802. PMID:18988810
- Liu L, Yang J, Zhu X, et al. Long noncoding RNA H19 competitively binds miR-17–5p to regulate YES1 expression in thyroid cancer. FEBS J. 2016 Jun;283(12):2326–2339. doi: 10.1111/febs.13741. Epub 2016 May 12. doi:10.1111/febs.13741.
- Sun J, Chu H, Ji J, et al. Long non-coding RNA HOTAIR modulates HLA-G expression by absorbing miR-148a in human cervical cancer. Int J Oncol. 2016 Sep;49(3):943–952. doi: 10.3892/ijo.2016.3589. Epub 2016 Jun 30.
- Li CY, Liang GY, Yao WZ, et al. Integrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric cancer. Int J Oncol. 2016 May;48(5):1965–1976. doi: 10.3892/ijo.2016.3407. Epub 2016 Feb 24. doi:10.3892/ijo.2016.3407.
