ABSTRACT
Regulation of the mitochondrial respiratory chain biogenesis is a matter of great interest because of its implications for mitochondrial disease. One of the mitochondrial disease genes recently discovered associated to encephalopathy and mitochondrial complex III (cIII) deficiency is TTC19. Our study of TTC19-deficient human and mouse models, has led us to propose a post-assembly quality control role or ‘husbandry’ function for this factor that is linked to Rieske Fe-S protein (UQCRFS1). UQCRFS1 is the last incorporated cIII subunit, and its presence is essential for enzymatic activity. During UQCRFS1 assembly, the precursor is cleaved and its N-terminal part remains bound to the complex, between the two core subunits (UQCRC1 and UQCRC2). In the absence of TTC19 there is a prominent accumulation of these UQCRFS1-derived N-terminal fragments that proved to be detrimental for cIII function. In this article we will discuss some ideas around the UQCRFS1 processing and assembly and its importance for the regulation of cIII activity and biogenesis.
Complex III overview
The mitochondrial respiratory chain (MRC) is composed of four multimeric enzymes, responsible for the redox reactions that take electrons from NADH and FADH2, ultimately reducing molecular oxygen to water. This electron transfer is coupled to proton pumping from the matrix to the intermembrane space, generating a proton electrochemical gradient, which is the driving force for the condensation of ADP+Pi by the FO-F1 ATP synthase, a fifth complex, which together with complexes I to IV form the oxidative phosphorylation system (OXPHOS).
Complex III (cIII) or ubiquinol:cytochrome c oxidoreductase is the center of the MRC, transferring electrons from coenzyme Q (CoQ or just Q) to cytochrome c.
X-ray diffraction crystal structures of cIII are available from Bos taurus [Citation1], Gallus gallus [Citation2,Citation3] and Saccharomyces cerevisiae [Citation4] as well as a cryo-EM structure that has recently been published from Homo sapiens [Citation5]. All of them are practically identical in shape and subunit composition. In all of these organisms, cIII is a tightly bound symmetrical dimer where one mitochondrial DNA (mtDNA) and nine nuclear genes encode the structural subunits forming each cIII monomer. Of all these subunits, only three possess electron transfer properties: 1) the mtDNA-encoded cytochrome b (MT-CYB in the mammalian nomenclature) that contains the two Q binding sites (Qo or QP and Qi or QN) and two b-type hemes with different redox potentials (bL and bH): 2) cytochrome c1 (CYC1) that contains one c-type heme and 3) the Rieske Fe-S protein (UQCRFS1), whose intermembrane space-facing hydrophilic C-terminus hosts a 2Fe-2S high-potential cluster.
The electron transfer and proton pumping processes within cIII occur according to a ‘Q-cycle’ model [Citation6,Citation7]. UQCRFS1 receives one of the two electrons from oxidation of one ubiquinol molecule, and transfers it to CYC1. During this redox reaction, UQCRFS1 ‘head’ moves, towards the CYC1 subunit of one of the monomers, whereas the N-terminal transmembrane domain is in close contact with MT-CYB of the other monomer [Citation8]. The reason for this configuration may be to provide stability to the dimeric cIII2 structure.
Mammalian complex III assembly process
Most of what is known about the assembly of cIII comes from studies in yeast [Citation9,Citation10]. Based on the structural similarities and the fact that the first and last steps of human cIII assembly are conserved, it is assumed that the in-between process will be the same in yeast and mammals [Citation11].
CIII assembly starts with the synthesis of cytochrome b by dedicated ribosomes, where the yeast translation factors Cbp3 (mammalian UQCC1) and Cbp6 (UQCC2) are bound to the peptide exit tunnel and direct the nascent polypeptide to translocate into the mitochondrial inner membrane [Citation12,Citation13]. A third factor, Cbp4 (UQCC3), is bound to cytochrome b after the bL heme but before the bH heme are incorporated into the apoprotein [Citation14]. Pathological mutations associated with human cIII deficiency have been described in UQCC2 [Citation15] and UQCC3 [Citation16]. These early assembly factors are released when the first nuclear-encoded subunits bind to cytochrome b. Cbp3 and Cbp6 are now liberated and can bind to the mitochondrial ribosomes to act again as translational activators for cytochrome b. The rest of the structural subunits are then incorporated into the nascent complex until only Rip1 (UQCRFS1) and the smallest subunit Qcr10 (UQCR11) are missing, in an already dimeric structure named as pre-cIII2 [Citation10,Citation11]. The incorporation of Rip1/UQCRFS1 can be considered as the crucial cIII2 maturation step because it is then that the enzyme becomes catalytically active. As depicted in , UQCRFS1 is synthesized as a precursor protein in the cytosol and imported to the mitochondrial matrix following the TOM and TIM23 pathway, similarly to other proteins containing cleavable mitochondrial targeting sequences (MTS) [Citation17]. In the matrix, UQCRFS1 binds to MZM1L/LYRM7 [Citation18–20], the LYR-motif containing chaperone responsible for its stabilization and for the recruitment of the Fe-S transfer complex in the matrix [Citation21,Citation22]. Once UQCRFS1 has acquired its 2Fe-2S cluster, BCS1L translocates and incorporates it to pre-cIII2 in the IM [Citation23–25]. This function is therefore essential for cIII2 activity and mutations in BCS1L are the most frequent cause of mitochondrial disease associated to cIII deficiency (reviewed in ref. [Citation11]). Pathological mutations have also been described in LYRM7 [Citation26–29] and in TTC19, encoding a third factor necessary for UQCRFS1 metabolism [Citation30,Citation31], which is not present in yeast and will be discussed in more detail below.
Figure 1. UQCRFS1 pathway from synthesis of the precursor in the cytoplasm, transport into the mitochondrial matrix through the TOM/TIM23 import system, incorporation of the [2Fe-2S] clusters in the matrix and translocation to pre-cIII2 in the inner membrane (IM). According to our model, the N-terminal MTS is cleaved off in situ by the UQCRC1+UQCRC2 MPP activity. See main text for more details.
![Figure 1. UQCRFS1 pathway from synthesis of the precursor in the cytoplasm, transport into the mitochondrial matrix through the TOM/TIM23 import system, incorporation of the [2Fe-2S] clusters in the matrix and translocation to pre-cIII2 in the inner membrane (IM). According to our model, the N-terminal MTS is cleaved off in situ by the UQCRC1+UQCRC2 MPP activity. See main text for more details.](/cms/asset/f43f0858-c864-48d6-b4b4-f313f7d8b3e9/kccy_a_1417707_f0001_c.jpg)
Bovine CIII subunit 9: the UQCRFS1 mitochondrial targeting sequence
One interesting observation that was pointed out as the main difference between the bovine cIII and the yeast and chicken enzymes was that the bovine complex contained eleven subunits instead of the ten found in the latter organisms. This extra subunit with a molecular mass of 8 kDa was called ‘subunit 9’ or SU9 [Citation32,Citation33] and its sequence was determined to be the 78 N-terminal amino acids of the UQCRFS1 precursor, i.e. its MTS [Citation34]. As a norm, proteins directed to the mitochondrial matrix contain an N-terminal amphipathic helix of variable length, which is recognized by the TOM complex and directs the precursor protein to the TIM23 complex to be translocated to the mitochondrial matrix. Here the precursor is then processed by the matrix processing peptidase (MPP), which cleaves off the MTS and gives rise to the mature protein [Citation35]. Normally, the MTS peptides are proteolysed by Cym1/PITRM1 or hPreP, as their accumulation inside mitochondria could have a highly toxic effect [Citation36]. Therefore, the presence of the UQCRFS1 MTS incorporated inside cIII as a structural subunit represents an anomaly and the reason why this happens is unknown.
Yeast Rip1 has a much shorter MTS than mammalian and avian UQCRFS1: 30 amino acids vs. 78 and 76, respectively (). Interestingly, TTC19 orthologs exist in organisms with long UQCRFS1 MTS, while absent in yeast. The processing of yeast Rip1 is also different from bovine UQCRFS1. Rip1 is processed in two steps, first, the 22 N-terminal amino acids are cleaved by the MPP, generating an intermediate form of Rip1 (iRip1), and, in a second step, the mitochondrial intermediate peptidase (MIP) removes the next octapeptide to give rise to the mature form of the Rieske Fe-S protein (mRip1) [Citation37]. This processing was proposed to happen once Rip1 is inserted into cIII2 [Citation38]. In the case of the bovine enzyme, a single-step processing of UQCRFS1 seems to be carried out by MPP after it is incorporated into cIII2 [Citation34], where SU9 remains bound to the complex and localized between the two core subunits (UQCRC1 and UQCRC2), as determined in the crystal structure () [Citation1]. When chicken cIII2 was isolated and crystallized for the first time, no SU9 was identified [Citation2], leading to the idea that there was no such UQCRFS1-derived subunit in the avian enzyme.
Figure 2. Graphic alignment of the Rieske protein precursors from yeast (S. cerevisiae), fruit fly (D. melanogaster), chicken (G. gallus) and mammals (bovine, mouse and human). Numbers indicate relevant residue positions. The two yeast cleavage sites, at positions 22 and 30 are indicated with arrows. The main cleavage site, conserved in all organisms is indicated with the big arrow. Different color MTS indicates the lack of homology of these sequences, while the homology in the mature protein sequences in very high.
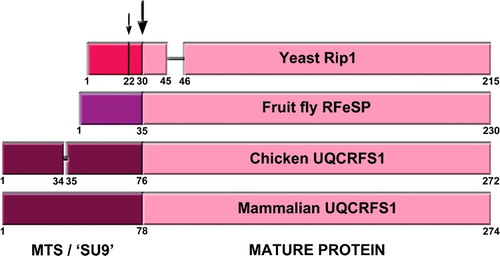
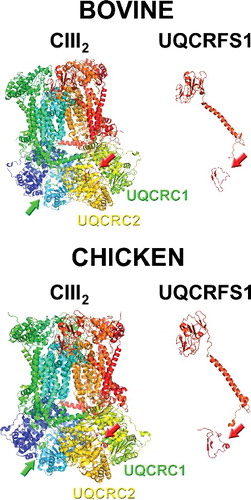
Figure 3. Structures of full complex III dimer (cIII2) and of isolated UQCRFS1 (of only one of the monomers), from bovine (PDB ID: 1BGY) and chicken (PDB ID: 3H1J). The arrows indicate the presence of the UQCRFS1 fragments in the cavity between UQCRC1 and UQCRC2. Images were generated with MacPyMol.
UQCRC1 and UQCRC2 are homologous to the two MPP subunits (β−MPP and α−MPP, respectively), and in plants they still preserve their processing properties [Citation39]. Mammalian cIII2 was considered to have lost its MPP activity. However, when the isolated enzyme was treated with detergents it showed this activity [Citation40]. The same phenomenon was observed by reconstitution of the system with recombinant UQCRC1 and UQCRC2; interestingly, MPP activity was inhibited when stoichiometric amounts of ‘SU9’ were formed by adding N-terminal UQCRFS1 peptides as substrate [Citation41].
Taking all this information together, it is plausible that UQCRFS1 is processed in situ by UQCRC1+UQCRC2 MPP activity when incorporated into pre-cIII2 (). This would explain why the N-terminus of the UQCRFS1 precursor is found between these two subunits in the cIII2 structure and inhibits its MPP activity. This mode of action was also postulated based on the structural features of vertebrate cIII2 [Citation42].
N-terminal UQCRFS1 peptides bound to mammalian and avian CIII2
After the identification of the first pathological mutations in TTC19 [Citation30] our laboratory got interested in determining the molecular function of this factor, related with cIII2 biogenesis. For that reason we have studied TTC19-deficient cells and a knock out (Ttc19−/−) mouse model, whose characterization was recently published [Citation31]. In this report, by importing radioactively labeled UQCRFS1 in isolated mouse liver mitochondria and analyzing its incorporation into cIII2 in a time course, we found that UQCRFS1 is processed right after its import and assembly, and the peptides derived from the N-terminus of the UQCRFS1 precursor remain bound to the complex. However, we did not find evidence of a unique cleavage point, giving rise to only two fragments corresponding to the mature UQCRFS1 and the 78-amino acid long SU9. Contrariwise, we observed the formation of the mature UQCRFS1 and the appearance of three main species of approximately 4, 8 and 12 kDa in size. In the steady state, these small UQCRFS1-derived peptides were readily detectable in the Ttc19−/− tissues while they were practically absent in the Ttc19wt samples. By mass spectrometry we were able to find tryptic peptides spanning from positions 8 to 204 of the UQCRFS1 precursor in the lower part of a SDS-PAGE (corresponding to sizes of 14 kDa and smaller), whereas in the higher part of the gel (30–14 kDa) we could only find peptides within the sequence of the mature protein (from residues 84 to 274) [Citation31].
When checking the twenty-two bovine and sixteen chicken cIII2 crystal structures available in the RCSB PDB protein data bank (https://www.rcsb.org) [Citation43] there are peptides derived from UQCRFS1 MTS in all of them (examples in ). In the original chicken structure, density corresponding to an amino acid chain sitting between UQCRC1 and UQCRC2 was detected and a chain was built in the structure, but the identity of this peptide remained unknown (PBD ID 1BCC, 2BCC, 3BCC) [Citation3]. This issue was resolved in the later chicken structures (see ). Even though all the structures contain these peptides, the sequences in each of them are very variable and in none of them the full UQCRFS1 MTS, from amino acids 1 to 78, is found (). This could reflect heterogeneity in the fragments that are contained in each of the analyzed cIII2 crystals [Citation42].
Table 1. UQCRFS1 MTS sequences found in the published cIII2 structures
In our in organello assays, after import and incorporation into cIII2 of radioactive mouse Uqcrfs1, we observed that the 8 and 12 kDa fragments that are generated immediately after incorporation, disappeared with time and this clearance was much more efficient in wild-type compared to Ttc19−/− mitochondria. The abnormal accumulation of the N-terminal peptides was associated with aberrant electrophoretic mobility of cIII2 in blue-native gel electrophoresis (BNGE), defective enzymatic activity and increased reactive oxygen species (ROS) production. Therefore, we concluded that during the incorporation and in situ UQCRFS1 processing, several peptides containing its MTS are produced and remain bound to cIII2. However, in order to preserve cIII2 structural integrity and function, these peptides (at least those of 8 kDa and 12 kDa) must be removed. TTC19 is involved in the removal of these N-terminal UQCRFS1 peptides, as discussed in the next section.
Role of TTC19 in complex III biogenesis and function
Since the first TTC19 mutations were published in 2011 [Citation30], a significant number of cases have been subsequently reported [Citation44,Citation45]. The biochemical profile of these patients, showing isolated cIII deficiency, underscores the importance of TTC19 for a correct biogenesis of this complex. Analysis of cIII2 assembly state in TTC19-deficient skin fibroblasts and muscle biopsies revealed the presence of UQCRC1 and UQCRC2-containing subassemblies [Citation30,Citation31], which suggested an involvement of TTC19 in the first steps of cIII2 assembly. However, the availability of the Ttc19−/− mice and more detailed kinetic analyses in human cells, revealed that the assembly process per se was not particularly altered in the absence of TTC19, including a normal incorporation of UQCRFS1 [Citation31]. TTC19 is loosely bound to fully assembled cIII2, as this interaction is disrupted when treating the mitochondrial membranes with a relatively mild detergent such as n-Dodecyl-β-D-maltoside (DDM), but not if the samples are exposed to the even milder detergent Digitonin. Moreover, TTC19 only binds to cIII2 if UQCRFS1 is incorporated into the complex [Citation31], suggesting a role in cIII2 biogenesis or maturation after assembly is completed. In fact, because the absence of TTC19 impairs the removal of the small UQCRFS1-derived fragments, we assigned a quality control or “husbandry” role for TTC19, in maintaining the structural and functional integrity of cIII2.
Prospects
The imminent questions derived from our recently published work [Citation31] are: 1) whether UQCRC1+UQCRC2 possess MPP activity in vivo and what is their role in UQCRFS1 processing, 2) what proteases are working in collaboration with TTC19 in the removal of UQCRFS1 N-terminal peptides produced during assembly-processing and 3) how this process intervenes in UQCRFS1 turnover, to warrant cIII2 fitness.
As for the first question, in plants, MPP is integrated into cIII2, thus carrying out the processing of the precursor proteins in addition to act as an MRC complex [Citation39]. However, in S. cerevisiae and in mammals, there is a soluble MPP localized in the matrix. Only two reports cited above [Citation40,Citation41], have demonstrated an MPP activity incorporated into purified bovine cIII2 and performed by UQCRC1+UQCRC2. It would be necessary to demonstrate in vivo whether this activity is necessary for the in situ processing and proper incorporation of UQCRFS1.
Concerning the second question, we performed co-immunoprecipitation studies that revealed specific interactions of TTC19 with cIII2, in the context of the respiratory supercomplexes (i.e. cI+cIII+cIV) [Citation46] and with the components of the so-called SPY complex [Citation47]. This is a mitochondrial quality control “proteolytic hub” that includes both the i-AAA protease YME1L and the rhomboid protease PARL. Logically the SPY complex is the first candidate to be explored in relation to its role in UQCRFS1/cIII2 quality control.
Regarding the third question and taking together the information reported above, this process can be considered as a regulatory mechanism of cIII2 maturation and activity. Even in situations in which UQCRFS1 cannot be incorporated, there is an assembly of pre-cIII2, which contains all the subunits except UQCRFS1 and most probably the smallest UQCR11 [Citation20,Citation25]. Therefore, the pre-cIII2 can be considered as an assembly checkpoint previous to the final activation, which must be somehow regulated. Accordingly, UQCRC1+UQCRC2 could control the amount of UQCRFS1 that must be incorporated into cIII2 by recognizing the UQCRFS1 MTS and processing it upon incorporation. The presence of the N-terminal UQCRFS1 fragments in between UQCRC1 and UQCRC2 would inhibit their MPP activity and the incorporation of any new UQCRFS1. The removal of the UQCRFS1 N-terminal fragments would reactivate cIII2 MPP activity and promote the removal of old, possibly damaged, UQCRFS1 from the pre-cIII2 scaffold, so that new UQCRFS1 can then be processed and incorporated again. This would keep the complex functional. The interchange of catalytically active subunits that can be oxidatively damaged in large enzymatic complexes is a mechanism demonstrated in chloroplast Photosystem II [Citation48–50] and in mammalian mitochondrial cI [Citation51], where there seems to be an interchange of old damaged subunits for newly imported ones. If this model is correct, the turnover of UQCRFS1 should be faster than that of the other cIII2 subunits, which is in fact the case in mouse tissues [Citation52].
To determine whether the rate of UQCRFS1 incorporation and turnover, as well as the N-terminal fragment clearance change depending on the metabolic conditions is a question of great interest to understand the relevance of this process in the adaptation to different energetic demands. This could be tested in differentiated tissues, where mitochondria in general and cIII2 in particular could play a different metabolic role. It is also possible to analyze UQCRFS1 turnover in cultured cells grown in OXPHOS-dependent (galactose as carbon source) and independent (glucose as carbon source) media. If this were to be changed, it would reflect how cIII2 maturation must adapt to an increased MRC activity.
These questions can be experimentally addressed; their elucidation will contribute to clarify the complex regulatory mechanisms of OXPHOS and their implication in health and disease.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Our work was supported by the Core Grant from the MRC; ERC Advanced under Grant FP7-322424 and NRJ-Institut de France Grant (to M.Z.); and by Association Française contre les Myopathies (AFM) under Grant 16086 (to E.F.-V.).
Additional information
Funding
References
- Iwata S, Lee JW, Okada K, et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi:10.1126/science.281.5373.64. PMID:9651245
- Berry EA, Huang LS, Zhang Z, et al. Structure of the avian mitochondrial cytochrome bc1 complex. J Bioenerg Biomem. 1999;31:177–190. doi:10.1023/A:1005459426843. PMID:10591524
- Zhang Z, Huang L, Shulmeister VM, et al. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi:10.1038/33612. PMID:9565029
- Hunte C, Koepke J, Lange C, et al. Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000;8:669–684. doi:10.1016/S0969-2126(00)00152-0. PMID:10873857
- Guo R, Zong S, Wu M, et al. Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2. Cell. 2017;170:1247–1257, e12.
- Cramer WA, Hasan SS, Yamashita E. The Q cycle of cytochrome bc complexes: a structure perspective. Biochim Biophys Acta. 2011;1807:788–802. doi:10.1016/j.bbabio.2011.02.006. PMID:21352799
- Crofts AR, Holland JT, Victoria D, et al. The Q-cycle reviewed: How well does a monomeric mechanism of the bc(1) complex account for the function of a dimeric complex? Biochim Biophys Acta. 2008;1777:1001–1019.
- Iwata M, Bjorkman J, Iwata S. Conformational change of the Rieske [2Fe-2S] protein in cytochrome bc1 complex. J Bioenerg Biomem. 1999;31:169–175. doi:10.1023/A:1005407410005. PMID:10591523
- Zara V, Conte L, Trumpower BL. Biogenesis of the yeast cytochrome bc1 complex. Biochim Biophys Acta. 2009;1793:89–96.
- Smith PM, Fox JL, Winge DR. Biogenesis of the cytochrome bc(1) complex and role of assembly factors. Biochim Biophys Acta. 2012;1817:276–286. doi:10.1016/j.bbabio.2011.11.009. PMID:22138626
- Fernandez-Vizarra E, Zeviani M. Nuclear gene mutations as the cause of mitochondrial complex III deficiency. Front Genet. 2015;6:134. doi:10.3389/fgene.2015.00134. PMID:25914718
- Gruschke S, Kehrein K, Rompler K, et al. Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J Cell Biol. 2011;193:1101–1114. doi:10.1083/jcb.201103132. PMID:21670217
- Gruschke S, Rompler K, Hildenbeutel M, et al. The Cbp3-Cbp6 complex coordinates cytochrome b synthesis with bc(1) complex assembly in yeast mitochondria. J Cell Biol. 2012;199:137–150. doi:10.1083/jcb.201206040. PMID:23007649
- Hildenbeutel M, Hegg EL, Stephan K, et al. Assembly factors monitor sequential hemylation of cytochrome b to regulate mitochondrial translation. J Cell Biol. 2014;205:511–524. doi:10.1083/jcb.201401009. PMID:24841564
- Tucker EJ, Wanschers BF, Szklarczyk R, et al. Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome b Protein Expression. PLos Genet. 2013;9:e1004034. doi:10.1371/journal.pgen.1004034. PMID:24385928
- Wanschers BF, Szklarczyk R, van den Brand MA, et al. A mutation in the human CBP4 ortholog UQCC3 impairs complex III assembly, activity and cytochrome b stability. Hum Mol Genet. 2014;23:6356–6365. doi:10.1093/hmg/ddu357. PMID:25008109
- Wasilewski M, Chojnacka K, Chacinska A. Protein trafficking at the crossroads to mitochondria. Biochim Biophys Acta. 2017;1864:125–137. doi:10.1016/j.bbamcr.2016.10.019. PMID:27810356
- Atkinson A, Smith P, Fox JL, et al. The LYR Protein Mzm1 Functions in the Insertion of the Rieske Fe/S Protein in Yeast Mitochondria. Mol Cell Biol. 2011;31:3988–3996. doi:10.1128/MCB.05673–11. PMID:21807901
- Cui TZ, Smith PM, Fox JL, et al. Late-Stage Maturation of the Rieske Fe/S protein: Mzm1 Stabilizes Rip1 but does not facilitate its translocation by the AAA ATPase Bcs1. Mol Cell Biol. 2012;32:4400–4409. doi:10.1128/MCB.00441-12. PMID:22927643
- Sanchez E, Lobo T, Fox JL, et al. LYRM7/MZM1L is a UQCRFS1 chaperone involved in the last steps of mitochondrial Complex III assembly in human cells. Biochim Biophys Acta. 2013;1827:285–293. doi:10.1016/j.bbabio.2012.11.003. PMID:23168492
- Maio N, Kim KS, Singh A, et al. A single adaptable cochaperone-scaffold complex delivers nascent iron-sulfur clusters to mammalian respiratory chain complexes I-III. Cell Metab. 2017;25:945–953, e6. doi:10.1016/j.cmet.2017.03.010. PMID:28380382
- Maio N, Singh A, Uhrigshardt H, et al. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab. 2014;19:445–457. doi:10.1016/j.cmet.2014.01.015. PMID:24606901
- Cruciat CM, Hell K, Folsch H, et al. Bcs1p, an AAA-family member, is a chaperone for the assembly of the cytochrome bc(1) complex. Embo J. 1999;18:5226–5233. doi:10.1093/emboj/18.19.5226. PMID:10508156
- Wagener N, Ackermann M, Funes S, et al. A Pathway of protein translocation in mitochondria mediated by the AAA-ATPase Bcs1. Mol Cell. 2011;44:191–202. doi:10.1016/j.molcel.2011.07.036. PMID:22017868
- Fernandez-Vizarra E, Bugiani M, Goffrini P, et al. Impaired complex III assembly associated with BCS1L gene mutations in isolated mitochondrial encephalopathy. Hum Mol Genet. 2007;16:1241–1252. doi:10.1093/hmg/ddm072. PMID:17403714
- Invernizzi F, Tigano M, Dallabona C, et al. A homozygous mutation in LYRM7/MZM1L associated with early onset encephalopathy, lactic acidosis, and severe reduction of mitochondrial complex III activity. Hum Mutat. 2013;34:1619–1622. doi:10.1002/humu.22441. PMID:24014394
- Dallabona C, Abbink TE, Carrozzo R, et al. LYRM7 mutations cause a multifocal cavitating leukoencephalopathy with distinct MRI appearance. Brain. 2016;139:782–794. doi:10.1093/brain/awv392. PMID:26912632
- Kremer LS, L'Hermitte-Stead C, Lesimple P, et al. Severe respiratory complex III defect prevents liver adaptation to prolonged fasting. J Hepatol. 2016;65:377–385. doi:10.1016/j.jhep.2016.04.017. PMID:27151179
- Hempel M, Kremer LS, Tsiakas K, et al. LYRM7 – associated complex III deficiency: a clinical, molecular genetic, MR tomographic, and biochemical study. Mitochondrion. 2017. doi:10.1016/j.mito.2017.07.001. PMID:28694194
- Ghezzi D, Arzuffi P, Zordan M, et al. Mutations in TTC19 cause mitochondrial complex III deficiency and neurological impairment in humans and flies. Nat Genet. 2011;43:259–263. doi:10.1038/ng.761. PMID:21278747
- Bottani E, Cerutti R, Harbour ME, et al. TTC19 plays a husbandry role on UQCRFS1 turnover in the biogenesis of mitochondrial respiratory complex III. Mol Cell. 2017;67:96–105, e4. doi:10.1016/j.molcel.2017.06.001. PMID:28673544
- Borchart U, Machleidt W, Schagger H, et al. Isolation and amino acid sequence of the 8 kDa DCCD-binding protein of beef heart ubiquinol:cytochrome c reductase. FEBS Lett. 1985;191:125–130. doi:10.1016/0014-5793(85)81007-3. PMID:2996928
- Schagger H, Link TA, Engel WD, et al. Isolation of the eleven protein subunits of the bc1 complex grom beef heart. Methods Enzymol. 1986;126:224–237. doi:10.1016/S0076-6879(86)26024-3. PMID:2856130
- Brandt U, Yu L, Yu CA, et al. The mitochondrial targeting presequence of the Rieske iron-sulfur protein is processed in a single step after insertion into the cytochrome bc1 complex in mammals and retained as a subunit in the complex. J Biol Chem. 1993;268:8387–8390.
- Chacinska A, Koehler CM, Milenkovic D, et al. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi:10.1016/j.cell.2009.08.005. PMID:19703392
- Teixeira PF, Glaser E. Processing peptidases in mitochondria and chloroplasts. Biochim Biophys Acta. 2013;1833:360–370. doi:10.1016/j.bbamcr.2012.03.012. PMID:22495024
- Hartl FU, Schmidt B, Wachter E, et al. Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell. 1986;47:939–951. doi:10.1016/0092-8674(86)90809-3. PMID:3022944
- Graham LA, Brandt U, Trumpower BL. Protease maturation of the Rieske iron-sulphur protein after its insertion into the mitochondrial cytochrome bc1 complex of Saccharomyces cerevisiae. Biochem Soc Trans. 1994;22:188–191. doi:10.1042/bst0220188. PMID:8206223
- Glaser E, Dessi P. Integration of the mitochondrial-processing peptidase into the cytochrome bc1 complex in plants. J Bioenergy Biomem. 1999;31:259–274. doi:10.1023/A:1005475930477. PMID:10591532
- Deng K, Zhang L, Kachurin AM, et al. Activation of a matrix processing peptidase from the crystalline cytochrome bc1 complex of bovine heart mitochondria. J Biol Chem. 1998;273:20752–20757.
- Deng K, Shenoy SK, Tso SC, et al. Reconstitution of mitochondrial processing peptidase from the core proteins (subunits I and II) of bovine heart mitochondrial cytochrome bc(1) complex. J Biol Chem. 2001;276:6499–6505. doi:10.1074/jbc.M007128200. PMID:11073949
- Berry EA, De Bari H, Huang LS. Unanswered questions about the structure of cytochrome bc1 complexes. Biochim Biophys Acta. 2013;1827:1258–1277. doi:10.1016/j.bbabio.2013.04.006. PMID:23624176
- Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi:10.1093/nar/28.1.235. PMID:10592235
- Ardissone A, Granata T, Legati A, et al. Mitochondrial complex III deficiency caused by TTC19 defects: report of a novel mutation and review of literature. JIMD Rep. 2015;22:115–120. doi:10.1007/8904_2015_419. PMID:25772319
- Mordaunt DA, Jolley A, Balasubramaniam S, et al. Phenotypic variation of TTC19-deficient mitochondrial complex III deficiency: a case report and literature review. Am J Med Genet A. 2015;167:1330–1336. doi:10.1002/ajmg.a.36968. PMID:25899669
- Schagger H. Respiratory chain supercomplexes of mitochondria and bacteria. Biochim Biophys Acta. 2002;1555:154–159.
- Wai T, Saita S, Nolte H, et al. The membrane scaffold SLP2 anchors a proteolytic hub in mitochondria containing PARL and the i-AAA protease YME1L. EMBO Rep. 2016;17:1844–1856. doi:10.15252/embr.201642698. PMID:27737933
- Aro EM, Suorsa M, Rokka A, et al. Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J Exp Bot. 2005;56:347–356. doi:10.1093/jxb/eri041. PMID:15569703
- De Las Rivas J, Andersson B, Barber J. Two sites of primary degradation of the D1-protein induced by acceptor or donor side photo-inhibition in photosystem II core complexes. FEBS Lett. 1992;301:246–252. doi:10.1016/0014-5793(92)80250-K. PMID:1577160
- Komayama K, Khatoon M, Takenaka D, et al. Quality control of photosystem II: cleavage and aggregation of heat-damaged D1 protein in spinach thylakoids. Biochim Biophys Acta. 2007;1767:838–846. doi:10.1016/j.bbabio.2007.05.001. PMID:17543883
- Lazarou M, McKenzie M, Ohtake A, et al. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol Cell Biol. 2007;27:4228–4237.
- Karunadharma PP, Basisty N, Chiao YA, et al. Respiratory chain protein turnover rates in mice are highly heterogeneous but strikingly conserved across tissues, ages, and treatments. FASEB J. 2015;29:3582–3592. doi:10.1096/fj.15-272666. PMID:25977255
- Gao X, Wen X, Esser L, et al. Structural basis for the quinone reduction in the bc1 complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc1 with bound substrate and inhibitors at the Qi site. Biochemistry. 2003;42:9067–9080. doi:10.1021/bi0341814. PMID:12885240
- Gao X, Wen X, Yu C, et al. The crystal structure of mitochondrial cytochrome bc1 in complex with famoxadone: the role of aromatic-aromatic interaction in inhibition. Biochemistry. 2002;41:11692–11702. doi:10.1021/bi026252p. PMID:12269811
- Esser L, Gong X, Yang S, et al. Surface-modulated motion switch: capture and release of iron-sulfur protein in the cytochrome bc1 complex. PNAS. 2006;103:13045–13050. doi:10.1073/pnas.0601149103. PMID:16924113
- Esser L, Quinn B, Li YF, et al. Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex. J Mol Biol. 2004;341:281–302. doi:10.1016/j.jmb.2004.05.065. PMID:15312779
- Huang LS, Cobessi D, Tung EY, et al. Binding of the respiratory chain inhibitor antimycin to the mitochondrial bc1 complex: a new crystal structure reveals an altered intramolecular hydrogen-bonding pattern. J Mol Biol. 2005;351:573–597. doi:10.1016/j.jmb.2005.05.053. PMID:16024040
- Xia D, Yu CA, Kim H, et al. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277:60–66. doi:10.1126/science.277.5322.60. PMID:9204897
- Capper MJ, O'Neill PM, Fisher N, et al. Antimalarial 4(1H)-pyridones bind to the Qi site of cytochrome bc1. PNAS. 2015;112:755–760. doi:10.1073/pnas.1416611112. PMID:25564664
- Esser L, Zhou F, Zhou Y, et al. Hydrogen bonding to the substrate is not required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III. J Biol Chem. 2016;291:25019–25031. doi:10.1074/jbc.M116.744391. PMID:27758861
- McPhillie M, Zhou Y, El Bissati K, et al. New paradigms for understanding and step changes in treating active and chronic, persistent apicomplexan infections. Sci Rep. 2016;6:29179. doi:10.1038/srep29179. PMID:27412848
- Berry EA, Huang LS, Lee DW, et al. Ascochlorin is a novel, specific inhibitor of the mitochondrial cytochrome bc1 complex. Biochim Biophys Acta. 2010;1797:360–370. doi:10.1016/j.bbabio.2009.12.003. PMID:20025846
- Crowley PJ, Berry EA, Cromartie T, et al. The role of molecular modeling in the design of analogues of the fungicidal natural products crocacins A and D. Bioorg Med Chem. 2008;16:10345–10355. doi:10.1016/j.bmc.2008.10.030. PMID:18996700
- Hao G-F, Yang, S.-G., Huang, W., et al. Rational design of highly potent and slow-binding cytochrome bc1 inhibitor as fungicide by computational substitution optimization. To be published 2015.
- Hao GF, Wang F, Li H, et al. Computational discovery of picomolar Q(o) site inhibitors of cytochrome bc1 complex. J Am Chem Soc. 2012;134:11168–11176. doi:10.1021/ja3001908. PMID:22690928
- Huang L, Berry, E.A. Famoxadone and related inhibitors bind like methoxy acrylate inhibitors in the Qo site of the BC1 compl and fix the rieske iron-sulfur protein in a positio close to but distinct from that seen with stigmatellin and other "distal" Qo inhibitors. To be published 2010.
