ABSTRACT
Hepatocellular carcinoma (HCC) is one of the most common causes of cancer-related death worldwide. In China, the situation is even worse as cancer incidence and mortality continue to increase rapidly. Although tremendous progress has been made toward HCC treatments, the benefits for liver cancer patients are still limited. Therefore, it is necessary to identify and develop novel therapeutic methods. Neuronally expressed developmentally downregulated 4 (NEDD4), an E3 ubiquitin ligase, plays a critical role in the development and progression of various types of human cancers. In our study, NEDD4 acts as an oncoprotein in both QGY7703 and SMMC7721 liver cancer cell lines. We found that depletion of NEDD4 by siRNA transfection led to inhibition of cell growth, invasion and migration, and promotion of apoptosis. In contrast, overexpression of NEDD4 via plasmid transfection resulted in facilitated cell proliferation, invasion and migration, and decreased apoptosis. Importantly, we observed that tumor suppressor LATS1, also a core component of Hippo pathway, was negatively regulated by NEDD4 in liver cancer cells. Our findings suggested that NEDD4 may be involved in the HCC progression via regulating LATS1 associated signaling pathway. Therefore, targeting NEDD4-LATS1 signaling could be a potential therapeutic option for HCC treatment.
KEYWORDS:
Introductions
Hepatocellular carcinoma (HCC) is one of the most common causes of cancer death worldwide [Citation1–3]. For decades, cancer incidence and mortality continue to increase rapidly in China, and liver cancer has become the most commonly diagnosed cancer and the leading cause of cancer death in men before the age of 60 years [Citation4]. Current treatments for HCC include surgical resection, radiotherapy and chemotherapy. However, surgical treatment is only available in the initial stage of HCC. In most cases, liver cancer patients are already in an advanced disease stage upon diagnosis, which limited the efficacy of radiotherapy and chemotherapy [Citation5,Citation6]. Although rapid development and wide application of imaging technology, pathology and molecular genetics have benefited HCC patients, death rates are still increasing rapidly for liver cancers [Citation7–11]. Therefore, progressive fights against cancers need consistent clinical and basic research to improve treatment outcome and benefit survival.
Accumulating data has demonstrated that ubiquitination plays an important role in HCC development [Citation12]. Ubiquitin proteasome system (UPS) regulates various cellular post-translational modification processes such as cell growth, migration, invasion, and cell cycle progression through ubiquitination and degradation of different molecular targets [Citation13–15]. Ubiquitination process contains three coordinated strides: the ubiquitin-activating enzyme (E1) activates ubiquitin molecule, the ubiquitin-conjugating enzyme (E2) subsequently transfers the activated ubiquitin, and ubiquitin ligase (E3) finally recruits the ubiquitin moieties to the substrate protein [Citation16]. Among these UPS members, the E3 ubiquitin ligase possesses a high variability of their components and interacts directly with ubiquitin substrates and determines the substrate specificity [Citation17]. Neuronally expressed developmentally downregulated 4 (NEDD4, often known as NEDD4-1), a homologue to E6-AP C-terminus (HECT) family of E3 ubiquitin ligase, plays a critical role in the development and progression of many human cancers [Citation18–21]. Amodio et al. demonstrated that NEDD4 facilitated non-small-cell lung carcinomas (NSCLC) cell lines proliferation through inactivation of PTEN [Citation22]. Recently, curcumin was identified to exert its tumor suppressive function via inhibition of NEDD4 oncoprotein in glioma and pancreatic cancer cells [Citation23,Citation24]. In hepatic cancer cells, NEDD4 was reported to associate with low density lipoprotein receptor class A domain containing 4 (LDLRAD4) and exhibited an oncogenic role in tumorigenesis [Citation25].
The Hippo pathway, consists of a core kinase cascade, transcription coactivators, and DNA-binding partners, has been a subject extensively studied in mammals in the last several years [Citation26]. Large tumor suppressor kinase 1 (LATS1)/ LATS2, a putative serine/threonine kinase, is the major kinase components of the Hippo-LATS/Warts pathway that suppress tumor growth by regulating cell proliferation, cell growth, and cell death [Citation27]. LATS1/2 is always downregulated in many human cancer types and control tumor development [Citation28–30], through various molecular mechanisms and signaling pathways [Citation31,Citation32].
In this study, we aimed to determine the function of NEDD4 in liver cancer cells. We measured the effect of NEDD4 on the proliferation, apoptosis, invasion and migration of two liver cancer cell lines after deletion or overexpression of NEDD4. We further identified the molecular signal pathway that could be involved in the regulating of liver cancer initiation or progression.
Results
Knocking down of NEDD4 inhibited liver cancer cells proliferation
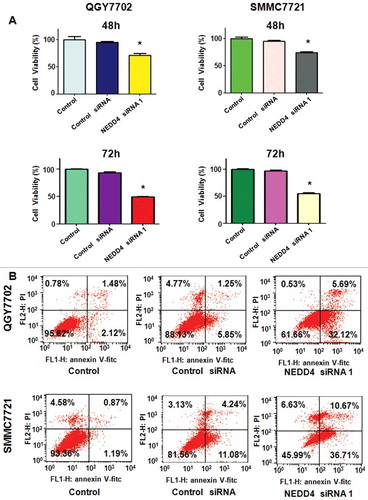
Three pairs of siRNAs targeting human NEDD4 sequences were transfected in liver cancer cell lines, respectively. As shown in , NEDD4 was well down-regulated at both mRNA and protein levels. We used NEDD4 siRNA 1 for the following studies. MTT assay was performed in QGY7703 and SMMC7721 cells after NEDD4 siRNA transfection to determine whether the modulation of NEDD4 expression could affect cell growth. Our results showed that knocking NEDD4 expression inhibited liver cancer cell proliferation in a time-dependent manner ().
Figure 1. Knockdown of NEDD4 in liver cancer cells. A. mRNA level of NEDD4 was detected by q-PCR in liver cancer cell lines after transfected with NEDD4 siRNAs. B. Protein level of NEDD4 was detected by Western blotting analysis in liver cancer cells after transfected with NEDD4 siRNAs. * P<0.05 vs control.
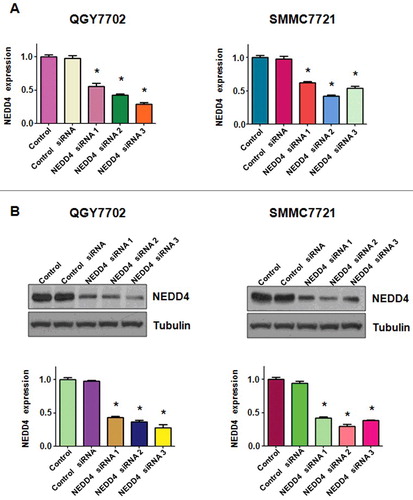
Figure 2. Knockdown of NEDD4 inhibited liver cancer cells growth and promoted apoptosis. A. MTT assay was performed in liver cancer cells after 48 h and 72 h of NEDD4 depletion. B. Cell apoptosis in NEDD4 siRNA transfected liver cancer cells was accessed by Flow cytometry. Lower penal: Quantitative results are illustrated. * P<0.05 vs control.
Knocking down of NEDD4 induced liver cancer cells apoptosis
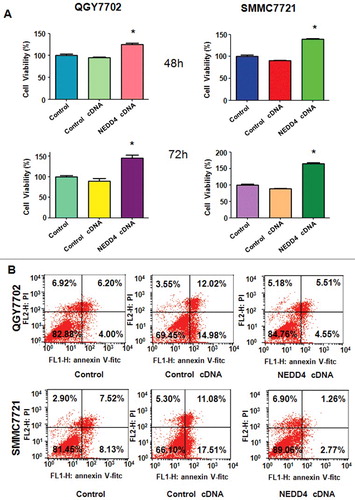
After transfected with NEDD4 siRNA for 48h, PI-FITC-annexin assay was performed to measure the apoptotic cell death in both liver cancer cell lines. We found that knocking down of NEDD4 significantly induced cell apoptosis, compared with control cells (). These findings indicated that knocking down of NEDD4 promoted apoptosis in liver cancer cells, which could partly responsible for NEDD4 depletion-induced cell proliferation suppression.
Knocking down of NEDD4 inhibited liver cancer cells invasion
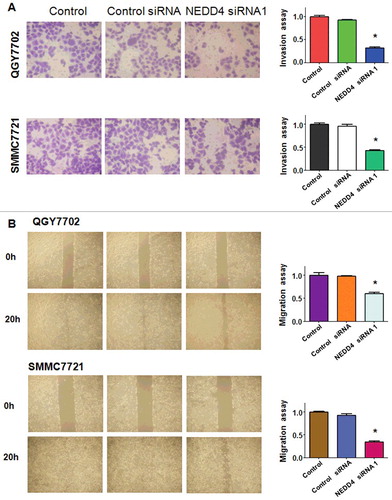
To further demonstrate whether NEDD4 depletion could inhibit cell invasive ability, transwell invasion assay was performed in a 24-well transwell chamber. Those cells that thread the pores of matrigel-coated membrane were markedly decreased in both QGY7703 and SMMC7721 liver cancer cell lines (). The result suggested that knocking down of NEDD4 had anti-invasive potential in liver cancer cells.
Figure 3. Knockdown of NEDD4 inhibited cell migration and invasion in liver cancer cells. A. Left panel: Invasion assay was conducted to measure the invasive capacity of liver cancer cells after NEDD4 depletion. Right panel: Quantitative results are illustrated for panel A. B. Left panel: Wound healing assays were used to detect the motility in liver cancer cells after NEDD4 depletion. Right panel: Quantitative results are illustrated for panel B. * P<0.05 vs control.
Knocking down of NEDD4 inhibited liver cancer cells migration
In order to determine whether NEDD4 depletion could inhibit cell migration, wound healing assay was applied in both QGY7703 and SMMC7721 cells after transfected with NEDD4 siRNA. We found that NEDD4 depletion significantly inhibited cell migration in both liver cancer cell lines ().
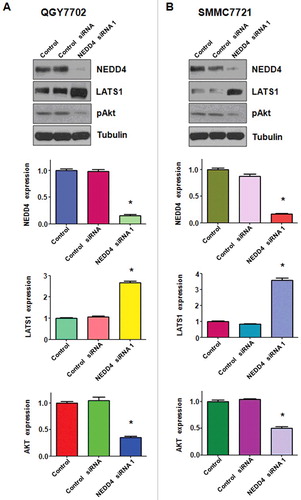
Knocking down of NEDD4 promoted LATS1 expression in liver cancer cells
Accumulated data has well characterized LATS1 as a tumor suppressor in various cancers including liver cancer by regulating cell proliferation, cell growth, and cell death. In this study, Western blotting was performed to demonstrate whether NEDD4 depletion could regulate the expression of LATS1 in liver cancer cells. As shown in , our results showed a significantly increased expression of LATS1 in NEDD4 siRNA transfected liver cancer cells. Moreover, the protein level of pAkt, a typical downstream target of LATS1, was down-regulated after NEDD4 depletion (). Taken together, these results validate that NEDD4 plays as an oncoprotein in liver cancer cells, and NEDD4 depletion induced cell growth inhibition, apoptosis and migration suppression at least partially by promoting LATS1 expression.
Figure 4. Knockdown of NEDD4 inhibited LATS1 expression. A. Upper panel: Western blotting analysis was used to detect the expression of LATS1 and its target pAkt in QGY7703 liver cancer cell line after NEDD4 depletion. Lower panel: Quantitative results are illustrated for panel A. B. Upper panel: Western blotting analysis was performed to measure the expression of LATS1 and pAkt in SMMC7721 cells after NEDD4 depletion. Lower Panel: Quantitative results are illustrated for panel B. * P<0.05 vs control.
Over-expression of NEDD4 promoted liver cancer cells proliferation and inhibited apoptosis
We further determined whether NEDD4 displayed as an oncoprotein in liver cancer cells. NEDD4 cDNA was transfected into QGY7703 and SMMC7721cells, empty vector was also transfected as control cDNA. Our results showed that over-expression of NEDD4 significantly promoted both liver cancer cells proliferation (). Moreover, we measured whether over-expression of NEDD4 could decrease apoptosis. As shown in , NEDD4 over-expression observably reduced the percentage of apoptotic cells in both liver cancer cells, compared with empty vector transfection. The results suggested that NEDD4 exhibited its oncoprotein characteristics in liver cancer cells.
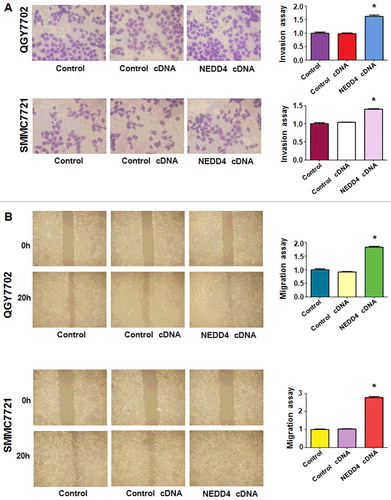
Over-expression of NEDD4 promoted liver cancer cells motility
In order to detect whether NEDD4 over-expression could affect the invasive capacity of liver cancer cells, transwell assay was performed after NEDD4 cDNA thranfection. The results showed a significantly increased number of invaded cells in NEDD4 overexpressed liver cancer cells (), which demonstrated that the invasive capacity of liver cancer cells could be greatly promoted by NEDD4. Then, the migration potential of liver cancer cells was further measured by wound healing assay after NEDD4 overexpression. We found that NEDD4 significantly promoted cell migration in both liver cancer cell lines ().
Figure 6. Overexpression of NEDD4 enhanced cell invasion and migration in liver cancer cells. A. Left panel: Invasion assay was conducted by Transwell chambers assay to measure the invasive capacity of liver cancer cells after NEDD4 overexpression. Right panel: Quantitative results are illustrated for panel A. B. Left panel: Wound healing assays were used to detect the motility. Right panel: Quantitative results are illustrated for panel B. * P<0.05 vs control.
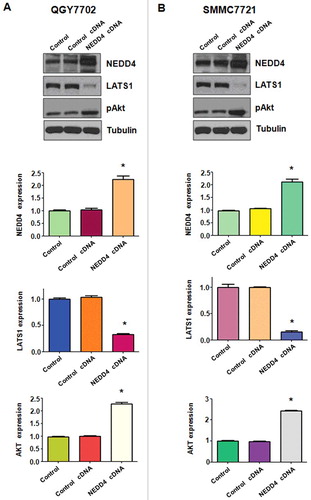
Over-expression of NEDD4 suppressed LATS1 expression in liver cancer cells
We further determined the changes of downstream targets of NEDD4 following its cDNA transfection. We found that over-expression of NEDD4 suppressed its downstream target LATS1 in QGY7703 liver cancer cells (). Further, pAkt was up-regulated after NEDD4 over-expression. Similar findings were detected in SMMC7721 cells (). These results suggest that NEDD4 exerts its oncoprotein function partially by inactivation of LATS1 signaling in liver cancer cells.
Figure 7. Overexpression of NEDD4 increased LATS1 expression. A. Upper panel: Western blotting analysis was used to detect the expression of LATS1 and pAkt in QGY7703 liver cancer cell line after NEDD4 cDNA transfection. Lower panel: Quantitative results are illustrated for panel A. B. Upper panel: The expression of LATS1 and pAkt in SMMC7721 cells were detected by Western blotting after NEDD4 overexpression. Lower Panel: Quantitative results are illustrated for panel B. * P<0.05 vs control.
Discussion
As one of the leading causes of cancer-related deaths worldwide, HCC has attracted much attention. Although great efforts in liver cancer treatments have been made, the benefits for HCC patients are still limited [Citation1,Citation4]. Therefore, it is necessary to identify and develop novel therapeutic methods. These years, a large number of molecules have been identified to regulate the initiation and/or progress of HCC. In this regard, molecular therapies are promising treatment options.
A mount of evidence has indicated that ubiquitination plays an important role in HCC development. It was reported that Rho-associated coiled-coil containing protein kinase 2 (Rock2) and Cdc25A were aberrantly upregulated and revealed a significantly positive correlation in HCC tissues, and Rock2 regulated Cdc25A ubiquitination and promoted its degradation via directly binding [Citation33]. Rock2 was also identified to promote the invasion and metastasis of hepatocellular carcinoma by modifying MMP2 ubiquitination and degradation [Citation34]. Ubiquitin C-terminal Hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence after curative resection, promotes cell migration and invasion via interacting and deubiquitinating PRP19 [Citation35]. As an HECT-domain E3 ligase, NEDD4 plays an important role in neuronal development [Citation36] and is an essential protein for animal development and survival [Citation37]. NEDD4 was also identified to regulate a large number of molecules that modulate signaling pathways to control cell growth and cancer development [Citation19,Citation38]. It was reported that NEDD4 is frequently overexpressed in many types of human cancers and always negatively regulate tumor suppressor PTEN [Citation18,Citation24,Citation39–41]. One study showed that depletion of NEDD4 inhibited cell proliferation in bladder cancer cells [Citation42]. Our study here demonstrated that NEDD4 overexpression could favorite HCC cells proliferation, migration and invasion, and inhibited apoptosis. Recently, Huang et al. found that overexpression of NEDD4 was responsible for adverse clinical outcomes in patients with HCC and depletion of NEDD4 resulted in inhibition Huh7 cell proliferation, migration and invasion possibly by the up-regulation PTEN signaling [Citation43].
The Hippo pathway plays paramount roles to determine and maintain proper organ size and stem cell homeostasis via modulating cell proliferation and apoptosis. Serine-threonine kinase LATS1 is a tumor suppressor and a central player to regulate the activity of Hippo pathway. It contains two PPxY motifs that are able to bind to WW domain-containing proteins with high affinity [Citation44,Citation45]. LATS1 participates in controlling the cell cycle and apoptosis and negatively regulates tumor development through several different mechanisms [Citation32,Citation46]. Hao and colleagues demonstrated that LATS1 can bind to and phosphorylate transcription regulator and oncogene YAP in vitro and in vivo [Citation27]. Downregulation of LATS1/2 has been frequently observed in different kinds of human cancers, including sarcomas, ovarian sarcomas, breast cancers, lung adenocarcinoma, astrocytomas, prostate cancer, glioma and acute lymphoblastic leukemia [Citation29,Citation30,Citation47–51].
As LATS1 is one of the main core components of such important Hippo pathway, there is a badly need to understand how LATS1 activity is modulated. It was reported that NEDD4 directly interacts with LATS1, leading to ubiquitination and decreased levels of LATS1 and, increased transcriptional activity of YAP [Citation52]. Bae et al. demonstrated that NEDD4 ubiquitylates and destabilizes LATS and WW45 kinase, which results in the suppression of Hippo signaling pathway and the promotion of intestinal stem cell renewal in Drosophila [Citation53]. Their study shed light on a new mechanism by which NEDD4 controls coordination of cell proliferation and apoptosis. However, whether LATS1 is regulated by NEDD4 in HCC has not been discussed yet. In fact, in our present study, we identified LATS1 was also negatively regulated by NEDD4 in both liver cancer cells. Taken together, these findings suggested that NEDD4 may be involved in the HCC progression via regulating LATS1 associated signaling pathway. Therefore, targeting NEDD4-LATS1 signaling could be a potential therapeutic option for HCC treatment.
Materials and methods
Cell culture and reagents
Human liver cancer cell lines QGY7703 and SMMC7721 were purchased from the Chinese Academy of Sciences (Shanghai, China) and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37˚C with 5% CO2. DMEM and FBS were purchased from Gibco (MGC803; Grand Island, NY). Penicillin/streptomycin was purchased from HyClone™ (Logan, Utah, USA). MTT (3-4,5-dimethyl-2- thiazolyl-2, 5-diphenyl-2-H-tetrazolium bromide, CAS number 57360-69-7), obtained from Sigma-Aldrich (St.Louis, MO, USA), was diluted in DMSO and stocked at −20°C. TRIzol, Lipofectamine 2000 and plus reagents were purchased from Invitrogen (Carlsbad, CA). We acquired RevertAid First Strand cDNA Synthesis Kit and SYBR® Select Master mix from Thermo Fisher Scientific (Waltham, MA, USA). Primary antibodies for NEDD4 (#2740, 1:1000), LATS1 (#3477, 1:1000), pAkt (#4060, 1:1000) were purchased from Cell Signaling Technology (Danvers, MA, USA). Monoclonal anti-Tubulin (T9028, 1:5000) was purchased from Sigma-Aldrich (St.Louis, MO, USA). All secondary antibodies were purchased from Thermo Fisher Scientific.
Transfection
Knockdown the expression of NEDD4 in liver cancer cells was achieved by transfection of small interfering RNAs (siRNAs) of NEDD4 using the Lipofectamine™ 2000 transfection reagent, following the manufacturer's instruments. siRNAs were designed and synthesized by GenePhama (Shanghai, China): Overexpression of NEDD4 was obtained by transfection of pcDNA3.1-NEDD4-1 into liver cancer cells. At least there independent transfections were performed.
Quantitative real-time reverse transcription-PCR (q-PCR) analysis
Total RNAs were extracted with Trizol from liver cancer cells and reversed-transcribed into cDNA by RevertAid First Strand cDNA Synthesis Kit. Q-PCR was performed using Power SYBR Green PCR Master Mix and the results were calculated by 2-ΔΔCt method. The primer sequences according to NEDD4-1 were list as follows: forward primer: 5′-GGA GTT GCC AGA GAA TGG TT-3′; antisense: 5′-TTG CCA TGA TAA ACT GCC AT-3′. And primers for GAPDH: sense: 5′-ACC CAG AAG ACT GTG GAT GG-3′; antisense: 5′-CAG TGA GCT TCC CGT TCA G-3′.
Western blotting analysis
Liver cancer cells were collected and lysed in cell lysis buffer (Cell Signaling Technology, Danvers, MA, USA) supplemented with protease inhibitors. BCA protein assay kit was used to quantify protein concentrations. After boiled at 95˚C for 5 min, equal amount of denatured protein samples were separated by SDS-PAGE electrophoresis and then transferred onto a nitrocellulose filter (NC) membrane. NC membranes were blocked with 5% fat-free milk and then incubated with primary antibody at 4°C overnight. Then, washed the membranes with TBST for three times and incubated with horseradish peroxidase–conjugated second antibody at room temperature for about 1 hour. The expression of target proteins was visualised by electrochemiluminescence (ECL) assay (Pierce, Rockford, IL, USA). Stripped the NC membranes with 0.2M NaOH and reprobed with tubulin primary antibody as the loading control. We used ImageJ software to calculate the ensitometric quantification of the protein bands. After normalization with tubulin, the results were displayed as fold changes relative to the control.
MTT assays
3 × 103 cells/well liver cancer cells were seeded in a 96-well plate and cultured overnight. The cells were transfected with NDEE4 cDNA or siRNAs and incubated for designated time. MTT assays were then performed to determine cell viability, by following the manufacturer's introductions. Briefly, added 10 μl MTT solutions (0.5 mg/ml) to each well and incubated for 4 h at 37°C. The liquid supernatant was then removed from each well. Added 100 μl DMSO to each well and incubated for about 5min at room temperature. Multimode Reader of SpectraMax M5 (Moleucular Devices, Sunnyvale, CA, USA) was used to measure the absorbance of each well at 490 nm. At least three independent experiments were repeated for each experiment.
Cell apoptosis analysis
Liver cancer cells were seeded at a density of 3 × 105 cells/well in a 6-well plate and incubated overnight. After transfections, cells were allowed to continue culture for 48 h. Then, cells were harvested by centrifugation and subsequently resuspended in 500 μl of binding buffer containing 5 μl Propidium iodide (PI) and 5 μl annexin V-FITC. The cells were stained for 15 min under dark conditions and then analyzed using a FACScalibur flow cytometer (BD, USA) to determine effects of NEDD4 on liver cancer cells apoptosis.
Cell invasion assay
Liver cancer cells invasive capacity was measured using a transwell chamber (8 μm pore size, Corning), by following the manufacturer's introductions. Briefly, all filters of the chamber were precoated with 100 μl of Matrigel (BD Biosciences), which was diluted with serum-free medium to the final concentration of 1 mg/ml. Suitable amount of control and transfected cells were suspended in serum-free DMEM and then added into the upper chamber. 500 μL of complete medium (containing 10% FBS) was added into the bottom chamber. About 24 hours after incubation at 37°C in 5% CO2, the non-invasive cells still stayed in the upper chamber were removed with a cotton swab. The invaded cells attached on the outside of the filters were stained with Giemsa. The stained cells in were photographed and counted in at least six randomly-selected images.
Wound healing assay
Liver cancer cells were seeded in a 6-well plate at the concentration of 2 × 106 cells per well and incubated at 37°C. After an overnight incubation, both liver cancer cells grew to 90% confluence. Rectangular lesions on cell monolayers were created with a sterile 100 μl pipette tip. Detached cells were removed by careful washing with PBS. Complete DMEM was added and cells were cultured for the designated time course. Photographic images of the lesion border were photographed using an inverted microscope (Olympus, IX71).
Statistical analysis
All data were analysed using GraphPad Prism 4.0 program (Graph Pad Software, La Jolla, CA) and were presented as mean ± SD of triplicates. Statistical significance values between each group and its control group were evaluated by the 2-tailed Student's t test and differences with a p<0.05 were considered significant.
Disclosure of potential conflicts of interest
There is no conflict of interest.
Acknowledgments
This work was supported by grant from the Key Project of Natural Science Research of Universities of Anhui Province (KJ2015A177).
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi:10.3322/caac.21387. PMID:28055103
- Bruix J, Sherman M, American Association for the Study of Liver D. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–1022. doi:10.1002/hep.24199. PMID:21374666
- Yuksel S, Boylu Akyerli C, Cengiz Yakicier M. Angiogenesis, invasion, and metastasis characteristics of hepatocellular carcinoma. J Gastrointest Cancer. 2017. doi:10.1007/s12029-017-9962-5. PMID: 28785955
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi:10.3322/caac.21338. PMID: 26808342
- Yeung YP, Lo CM, Liu CL, et al. Natural history of untreated nonsurgical hepatocellular carcinoma. Am J Gastroenterol. 2005;100:1995–2004. doi:10.1111/j.1572-0241.2005.00229.x. PMID: 16128944
- Bruix J, Boix L, Sala M, et al. Focus on hepatocellular carcinoma. Cancer Cell. 2004;5:215–219. doi:10.1016/S1535-6108(04)00058-3. PMID: 15050913
- Niu ZS, Niu XJ, Wang WH, et al. Latest developments in precancerous lesions of hepatocellular carcinoma. World J Gastroenterol. 2016;22:3305–3314. doi:10.3748/wjg.v22.i12.3305. PMID: 27022212
- Maida M, Cabibbo G, Brancatelli G, et al. Assessment of treatment response in hepatocellular carcinoma: a review of the literature. Future Oncol. 2013;9:845–854. doi:10.2217/fon.13.33. PMID: 23718305
- Salvaggio G, Furlan A, Agnello F, et al. Hepatocellular carcinoma enhancement on contrast-enhanced CT and MR imaging: response assessment after treatment with sorafenib: preliminary results. Radiol Med. 2014;119:215–221. doi:10.1007/s11547-013-0332-5. PMID: 24297581
- Agnello F, Salvaggio G, Cabibbo G, et al. Imaging appearance of treated hepatocellular carcinoma. World J Hepatol. 2013;5:417–424. doi:10.4254/wjh.v5.i8.417. PMID: 24023980
- Braconi C, Meng F, Swenson E, et al. Candidate therapeutic agents for hepatocellular cancer can be identified from phenotype-associated gene expression signatures. Cancer. 2009;115:3738–3748. doi:10.1002/cncr.24417. PMID: 19514085
- Yamaguchi H, Hsu JL, Hung MC. Regulation of ubiquitination-mediated protein degradation by survival kinases in cancer. Front Oncol. 2012;2:15. doi:10.3389/fonc.2012.00015. PMID: 22649777
- Bassermann F, Eichner R, Pagano M. The ubiquitin proteasome system – implications for cell cycle control and the targeted treatment of cancer. Biochim Biophys Acta. 2014;1843:150–162. doi:10.1016/j.bbamcr.2013.02.028. PMID: 23466868
- Gilberto S, Peter M. Dynamic ubiquitin signaling in cell cycle regulation. J Cell Biol. 2017;216:2259–2271. doi:10.1083/jcb.201703170. PMID: 28684425
- Liu J, Shaik S, Dai X, et al. Targeting the ubiquitin pathway for cancer treatment. Biochim Biophys Acta. 2015;1855:50–60. PMID: 25481052
- Bedford L, Lowe J, Dick LR, et al. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov. 2011;10:29–46. doi:10.1038/nrd3321. PMID: 21151032
- Weissman AM, Shabek N, Ciechanover A. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat Rev Mol Cell Biol. 2011;12:605–620. doi:10.1038/nrm3173. PMID: 21860393
- Chen C, Matesic LE. The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 2007;26:587–604. doi:10.1007/s10555-007-9091-x. PMID: 17726579
- Ye X, Wang L, Shang B, et al. NEDD4: a promising target for cancer therapy. Curr Cancer Drug Targets. 2014;14:549–556. doi:10.2174/1568009614666140725092430. PMID: 25088038
- Weng M, Luo ZL, Wu XL, et al. The E3 ubiquitin ligase NEDD4 is translationally upregulated and facilitates pancreatic cancer. Oncotarget. 2017;8:20288–20296. doi:10.18632/oncotarget.15446. PMID: 28423617
- Zhang X, Li B, Rezaeian AH, et al. H3 ubiquitination by NEDD4 regulates H3 acetylation and tumorigenesis. Nat Commun. 2017;8:14799. doi:10.1038/ncomms14799. PMID: 28300060
- Amodio N, Scrima M, Palaia L, et al. Oncogenic role of the E3 ubiquitin ligase NEDD4-1, a PTEN negative regulator, in non-small-cell lung carcinomas. Am J Pathol. 2010;177:2622–2634. doi:10.2353/ajpath.2010.091075. PMID: 20889565
- Wang X, Deng J, Yuan J, et al. Curcumin exerts its tumor suppressive function via inhibition of NEDD4 oncoprotein in glioma cancer cells. Int J Oncol. 2017;51:467–477. doi:10.3892/ijo.2017.4037. PMID: 28627598
- Su J, Zhou X, Yin X, et al. The effects of curcumin on proliferation, apoptosis, invasion, and NEDD4 expression in pancreatic cancer. Biochem Pharmacol. 2017;140:28–40. doi:10.1016/j.bcp.2017.05.014. PMID: 28535906
- Liu Z, Huo X, Zhao S, et al. Low density lipoprotein receptor class A domain containing 4 (LDLRAD4) promotes tumorigenesis of hepatic cancer cells. Exp Cell Res. 2017;360:189–198. doi:10.1016/j.yexcr.2017.09.005.
- Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi:10.1101/gad.274027.115. PMID: 26728553
- Hao Y, Chun A, Cheung K, et al. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi:10.1074/jbc.M709037200. PMID: 18158288
- Deng J, Zhang W, Liu S, et al. LATS1 suppresses proliferation and invasion of cervical cancer. Mol Med Rep. 2017;15:1654–1660. doi:10.3892/mmr.2017.6180. PMID: 28259899
- Britschgi A, Duss S, Kim S, et al. The Hippo kinases LATS1 and 2 control human breast cell fate via crosstalk with ERalpha. Nature. 2017;541:541–545. doi:10.1038/nature20829. PMID: 28068668
- St John MA, Tao W, Fei X, et al. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet. 1999;21:182–186. doi:10.1038/5965. PMID: 9988269
- Furth N, Bossel Ben-Moshe N, Pozniak Y, et al. Down-regulation of LATS kinases alters p53 to promote cell migration. Genes Dev. 2015;29:2325–2330. doi:10.1101/gad.268185.115. PMID: 26588988
- Visser S, Yang X. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle. 2010;9:3892–3903. doi:10.4161/cc.9.19.13386. PMID: 20935475
- Liu, T, Yu X, Li G, et al. Rock2 regulates Cdc25A through ubiquitin proteasome system in hepatocellular carcinoma cells. Exp Cell Res. 2012;318:1994–2003. doi:10.1016/j.yexcr.2012.04.017. PMID: 22705122
- Huang D, Du X, Yuan R, et al. Rock2 promotes the invasion and metastasis of hepatocellular carcinoma by modifying MMP2 ubiquitination and degradation. Biochem Biophys Res Commun. 2014;453:49–56. doi:10.1016/j.bbrc.2014.09.061. PMID: 25251472
- Fang Y, Fu D, Tang W, et al. Ubiquitin C-terminal Hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence, promotes cell migration and invasion via interacting and deubiquitinating PRP19. Biochim Biophys Acta. 2013;1833:559–572. doi:10.1016/j.bbamcr.2012.11.020. PMID: 23220010
- Kawabe H, Neeb A, Dimova K, et al. Regulation of Rap2A by the ubiquitin ligase Nedd4-1 controls neurite development. Neuron. 2010;65:358–372. doi:10.1016/j.neuron.2010.01.007. PMID: 20159449
- Cao XR, Lill NL, Boase N, et al. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci Signal. 2008;1:ra5. PMID: 18812566
- Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 2004;23:1972–1984. PMID: 15021885
- Liu J, Wan L, Liu P, et al. SCF(beta-TRCP)-mediated degradation of NEDD4 inhibits tumorigenesis through modulating the PTEN/Akt signaling pathway. Oncotarget. 2014;5:1026–1037. PMID: 24657926
- Hong SW, Moon JH, Kim JS, et al. p34 is a novel regulator of the oncogenic behavior of NEDD4-1 and PTEN. Cell Death Differ. 2014;21:146–160. PMID: 24141722
- Shao C, Li Z, Ahmad N, et al. Regulation of PTEN degradation and NEDD4-1 E3 ligase activity by Numb. Cell Cycle. 2017;16:957–967. PMID: 28437168
- Wen W, Li J, Wang L, et al. Inhibition of NEDD4 inhibits cell growth and invasion and induces cell apoptosis in bladder cancer cells. Cell Cycle. 2017;16:1509–1514. PMID: 28745938
- Hang X, Zhu S, Di H, et al. NEDD4 depletion inhibits hepatocellular carcinoma growth via targeting PTEN. Cell Physiol Biochem. 2016;39:768–779. PMID: 27467187
- Sudol M, Harvey KF. Modularity in the Hippo signaling pathway. Trends Biochem Sci. 2010;35:627–633. PMID: 20598891
- Salah Z, Aqeilan RI. WW domain interactions regulate the Hippo tumor suppressor pathway. Cell Death Dis. 2011;2:e172. PMID: 21677687
- Xia H, Qi H, Li Y, et al. LATS1 tumor suppressor regulates G2/M transition and apoptosis. Oncogene. 2002;21:1233–1241. PMID: 11850843
- Hisaoka M, Tanaka A, Hashimoto H. Molecular alterations of h-warts/LATS1 tumor suppressor in human soft tissue sarcoma. Lab Invest. 2002;82:1427–1435. PMID: 12379777
- Fernandez LA, Kenney AM. The Hippo in the room: a new look at a key pathway in cell growth and transformation. Cell Cycle. 2010;9:2292–2299. PMID: 20581446
- Zeng H, Ortiz A, Shen PF, et al. Angiomotin regulates prostate cancer cell proliferation by signaling through the Hippo-YAP pathway. Oncotarget. 2017;8:10145–10160. PMID: 28052036
- Kim MH, Kim YK, Shin DH, et al. Yes associated protein is a poor prognostic factor in well-differentiated lung adenocarcinoma. Int J Clin Exp Pathol. 2015;8:15933–15939. PMID: 26884866
- Ji T, Liu D, Shao W, et al. Decreased expression of LATS1 is correlated with the progression and prognosis of glioma. J Exp Clin Cancer Res. 2012;31:67. PMID: 22909338
- Salah Z, Cohen S, Itzhaki E, et al. NEDD4 E3 ligase inhibits the activity of the Hippo pathway by targeting LATS1 for degradation. Cell Cycle. 2013;12:3817–3823. PMID: 24107629
- Bae SJ, Kim M, Kim SH, et al. NEDD4 controls intestinal stem cell homeostasis by regulating the Hippo signalling pathway. Nat Commun. 2015;6:6314. PMID: 25692647