ABSTRACT
Nowadays gene manipulation techniques (“DNA therapy”) undergo progressive development and become widely used in industry and medicine. Since new advances in mRNA technologies are capable for obtaining particles with increased stability and translational efficiency, RNA become an attractive alternative for advancement of DNA therapy. For the past years studies have been conducted to explore different modification in mRNA cap structure and its effect on RNA properties. Recently we have shown that modification of the cap structure at the N2 position of 7-methylguanosine leads to an enhancement in translation inhibition. Currently, we have decided to exploit translational properties of mRNA capped with the ARCA (anti-reversed cap) analogs modified within N2 position of purine moiety s. We designed and synthesized three new dinucleotide cap analogs and investigated them in the rabbit reticulocyte lysate (RRL) and the human embryonic kidney derived HEK293 cell line, in vitro translational model systems. The obtained data indicate that, in both translational assays, the cap analogs synthesized by us when incorporated into mRNA improved its translational properties compared to the ARCA capped transcripts. Furthermore, the introduced modifications enhanced stability of the capped transcripts in HEK293 cells, which become higher compared to that of the transcripts capped with regular cap or with ARCA. Additionally one of the synthesized cap analogs revealed strong translation inhibition potency in RRL system, with IC50 value 1.7 µM.
Introduction
RNA-based biopharmaceuticals, which include therapeutics and vaccines, are a relatively new way of treatment for a number of chronic and rare diseases. Currently, over 700 DNA and RNA therapeutics are in the drug-development phase; most of them are still in the early stages of development, and nearly 35% are in the field of oncology [Citation1]. Unfortunately, DNA-based therapeutics have some serious disadvantages such as toxicity in high doses and possible integration into the host genome [Citation2]. In contrast to DNA, RNA-based drugs are a better choice since new advanced technologies help to reduce their high immunogenicity and enhance the translational stability [Citation3–Citation5]. Over the past years, studies have been conducted on the therapeutic use of mRNAs protected by a cap structure that is naturally present at the 5ʹ end of eukaryotic mRNAs [Citation5–Citation7]. Recognized by highly specialized cap-binding proteins, the cap is involved in a number of processes, including maturation, nuclear export, initiation of translation and turnover of mRNA [Citation8,Citation9]. In the case of protein synthesis, interaction of eIF4E with the mRNA cap structure is a crucial and rate-limiting step [Citation10].
A defining feature of many cancers is deregulated translational control. Overexpression of eIF4E has a profound effect on cell growth and phenotype, causing accelerated cell division and malignant transformation [Citation11,Citation12]. A long-term research has shown that chemically-modified cap analogs have promising anti-cancer properties. To date, numerous modified cap analogs have been tested as inhibitors of cap-dependent translation in vitro by competing with the native-cellular capped-mRNA for the binding site of eIF4E. Among them some exhibit very interesting biological properties [Citation13–Citation15]. An attractive group of analogs are these that are modified at the exocyclic amine group within the methylated guanine moiety. It was previously shown that a single substitution at the N2 position of the first (methylated) guanine moiety leads to increased translation inhibition in vitro [Citation16]. Starting from analogs with a simple N2-methyl, various derivatives (such as those with aliphatic, cyclic, aromatic substituents) were evaluated in order to correlate their properties with the efficacy of translation inhibition [Citation17]. The most interesting were the aromatic substituents such as benzyl, p-methoxybenzyl or triazole. As N2-modified cap analogs exhibit superior inhibitory properties, we postulate that this type of compounds may be useful for preparing mRNA transcripts with high translational activity. Thus far, the capped RNAs synthesized by in vitro transcription have found wide application for studying mRNA function and metabolism and for in vitro protein synthesis, as well as expression of exogenous mRNAs in living cells for biotechnological and medicinal purposes [Citation5].
Synthesis of RNA is achieved by transcribing DNA template with either bacterial or bacteriophage RNA polymerase in the presence of all four NTPs and a cap dinucleotide such as m7GpppG [Citation6]. These polymerases initiate transcription via nucleophilic attack of the 3′-OH of the guanosine (Guo) in m7GpppG on the α-phosphate of the next nucleoside triphosphate specified by the DNA template. Unfortunately, this attack can also occur by the 3′-OH of the m7Guo, producing one-third to one-half of transcripts capped in a reverse orientation, i.e., Gpppm7GpNpN… instead of m7GpppGpNpN [Citation18]. Such reverse-capped transcripts decrease the overall translational activity of mRNA. This problem was overcome by introduction of anti-reverse cap analogs (ARCAs) bearing either 3′-O-methyl, 3′-H, or 2′-O-methyl modification in the m7Guo, ensuring 100% correct orientation [Citation19–Citation21]. ARCA-capped mRNAs were shown to have higher translational efficiency in vitro as well as in cultured cells.
Thus, two sets of cap analogs have been developed so far, one with high affinity to eukaryotic translation initiation factor 4E (containing N2 substituents) and second that allows proper incorporation of dinucleotide cap analog during in vitro transcription (ARCAs).
The main aim of the present work was to synthesize dinucleotide cap analogs with either 3ʹ- or 2ʹ-O-methyl and N2 modifications (benzyl or 4-methoxybenzyl). Subsequently, the synthesized analogs were used to obtain capped mRNA transcripts in order to examine dependence of translational efficiency on three parameters: (i) ability of the cap analog to inhibit cell free translation, (ii) degree to which RNAs synthesized in vitro are capped in the presence of various cap analogs, and (iii) stability of the modified transcripts.
Results and discussion
Chemistry
Within the framework of our studies, synthesis and biological evaluation of three new dinucleotide cap analogs with a metoxy group within ribose moiety (ARCA modification) and aromatic substituent at the N2 position of 7-methylguanine was planned (, 9–11). Synthesis of ARCA and N2-modified cap analogs separately have already been described. According to the literature, preparation of 2ʹ or 3ʹ ARCA analogs [Citation19] begins with protection of O6 position of guanosine by treatment of the nucleoside with diazoethane in DMF and subsequent alkylation using diazomethane in the presence of SnCl2 to give a mixture of 2ʹ(3ʹ)-O-methyl-O6-ethylguanosine. The obtained mixture is fractionated and subjected to strong basic environment in order to remove protecting groups. The obtained 2ʹ-O- and 3ʹ-O-methyl nucleosides are further 5ʹ-phosphorylated and then methylated at the N7 position. Finally, 2ʹ-O,7- and 3ʹ-O,7-dimethylated guanosine monophosphates are coupled with guanosine 5ʹ-diphosphate. In the case of N2-modified guanosine derivatives [Citation16], synthesis requires blocking of ribose hydroxyl groups by acetylation and of guanosine carbonyl moiety with p-nitrophenylethyl (NPE) group via Mitsunobu reaction (labile in basic conditions). In the next step, fully protected guanosine is transformed into a 2-fluoro intermediate, substituted with a proper amine via nucleophilic aromatic substitution (SNAr) and deprotected. N2-modified nucleosides obtained in this manner are further converted into dinucleotide cap analogs.
Figure 1. Structure and synthesis of new cap analogs. (i) MeCN, DMAP, Et3N, Ac2O, MeOH; (ii) NPE-OH, PPh3, DIAD, toluene, (iii) HF/pyridine, tBuONO, pyridine; (iv) RNH2, DMSO; (CH3)2NH/EtOH; (v) POCl3, trimethyl phosphate; (vi) MeI, DMSO; (vii) imGDP, ZnCl2, DMF
In order to synthesize the planned compounds, it was necessary to combine above mentioned methodologies. We believe that we chose the most efficient path starting with ribose alkylation (ARCA). The obtained 2ʹ-O- and 3ʹ-O-methyl-O6-ethylguanosine were used directly for the synthesis of N2 modified cap analogs avoiding Mitsunobu reaction. Such a solution eliminates the removal of one and insertion of the second protective group at the guanosine O6 position, which allows the synthesis to be shortened. Further synthesis steps included acetylation, diazotization, fluorination, and substitution with benzylamine or 4-methoxybenzylamine. All these reactions were carried out without difficulties and led to fully protected compounds containing ARCA and N2 modifications. Unfortunately, it turned out that the last step, removal of the ethyl group under alkaline conditions leading to N2 modified, 2ʹ-O- or 3ʹ-O-methyl nucleosides was the most problematic and isolation of the products was unsuccessful.
Due to the failure of the shortened strategy, a full procedure was applied ()). Thus, the synthesis of ARCA analogs was terminated by the removal of O6 ethyl group and 2ʹ-O- or 3ʹ-O-methylguanosine (1 a-b) was subjected to acetylation and subsequent protection of O6 carbonyl group via the Mitsunobu reaction with p-nitrophenylethyl alcohol (NPE), diisopropyl azodicarboxylate (DIAD) and triphenylphosphine (TPP) in anhydrous toluene. It is worth mentioning that the latter step was the most problematic and limited the overall yield of the synthesis. Subsequently, purified compounds were transformed into 2-fluoroinosine intermediates (2 a-b) and then substituted with benzylamine (3, 5) or 4-methoxybenzylamine (4). After completion of the substitution reaction, dimethylamine in ethanol was added in order to remove protecting groups. Deprotected compounds were 5ʹ-phosphorylated with phosphorus oxide trichloride in trimethyl phosphate and further methylated at the N7 position with methyl iodide in DMSO (6–8). The obtained analogs were purified using ion-exchange chromatography on DEAE-Sephadex A-25 (HCO3− form). As a result, ARCA nucleotides methylated at the N7 and substituted at the N2 position – bn2m27,3ʹOGMP (6), (pOCH3bn)2m27,3ʹOGMP (7) and bn2m27,2ʹOGMP (8), were used in the final step of the synthesis of dinucleotide cap analogs – coupling reaction with 5ʹ-diphosphate guanosine imidazolide. Products: bn2m27,3ʹOGpppG (9), (pOCH3bn)2m27,3ʹOGpppG (10) and bn2m27,2ʹOGpppG (11) were isolated by ion-exchange chromatography on DEAE – Sephadex A-25 (HCO3− form) and additionally purified by HPLC on a C18 column to give products as the ammonium salts. The structure and homogeneity of final products were confirmed by HPLC, mass spectrometry, 1H NMR and 31P NMR (see Supplemental Material).
Biology
Translation inhibition
Previously synthesized N2-modified monophosphate cap analogs containing aromatic substituents [Citation16,Citation17] efficiently inhibit the translation process in vitro. Hence, the newly synthesized dinucleotides bearing benzyl or p-metoxybenzyl substituent within the N2 position and ARCA modification were tested in in vitro translation system for translation inhibition. The applied method requires simultaneous addition of a mixture of ARCA-capped mRNA and various concentrations of a cap analog to a pre-incubated micrococcal nuclease-treated rabbit reticulocyte lysate. After 60 min of the translation reaction, luciferase synthesis was measured by luminometry. Comparison of IC50 values for all investigated cap analogs () and the m7GpppG control showed that bn2m27,2ʹOGpppG (11) compound exhibited the highest inhibitory properties with an IC50 of 1.7 ± 0.16 µM. Interestingly, the change of methyl group position from 2ʹ-O to 3ʹ-O of ribose resulted in almost three fold increase of IC50 value, with an IC50 of 4.46 ± 0.53 µM (compound 9 vs. 11). Further introduction of the electron-donor substituent (the metoxy group) in the benzene ring, resulted in almost the same score of inhibitory potential: IC50 4.12 ± 0.35 µM (compound 10 vs. 9). These data were unexpected because previously published inhibition constants (Ki) for m27,2ʹ-OGpppG and m27,3ʹ-OGpppG showed identical values regardless of whether the methyl group was added at 2ʹ-O or 3ʹ-O position of ribose [Citation21]. Based on the above data, we assume that lower inhibitory potential of bn2m27,3ʹ-OGpppG (9) is not connected with the methyl group placed in 3ʹ-O rather than on 2ʹ-O position but probably is related to the simultaneous presence of two substituents within the cap structure. We postulate that N2-benzyl and 3ʹ-O-methyl groups are closer to each other than N2-benzyl and 2ʹ-O-methyl groups and that simultaneous presence of these substituents in such an alignment is no longer beneficial.
Table 1. Inhibition of capped luciferase mRNA translation in RRL lysate by N2-modified cap analogs. Data represent the means ± SD of at least three independent repeats. Statistical significance between m7GpppG and N2-modified cap analogs was determined using ANOVA and Tukey’s multiple comparison tests using GraphPad Prism 6
Translational properties
The cap structure is important for proper recruitment of mRNA to the translational apparatus. Introduction of modification in the cap structure has significant impact on overall translational potential of the engineered mRNA. Therefore, we tested mRNAs capped with the newly synthesized cap analogs for translation efficiency in Rabbit Reticulocyte Lysate (RRL). Transcripts encoding firefly luciferase were prepared in in vitro transcription reaction using SP6 polymerase in the presence of a DNA template, four NTPs, and a dinucleotide (9–11). Differently capped RNAs were translated in RRL over 60 min and effectiveness of the synthesis was measured by the luciferase luminometry. The results were compared with the control mRNA capped with m7GpppG while cap-independent translation was estimated with the ApppG capped mRNA and subtracted from the measured values. The overall translational efficiencies and cap-dependent translational efficiencies are shown in . The relative translation efficiency for control m27,2ʹOGpppG and m27,3ʹOGpppG-capped RNA were 1.59-fold higher than that for the m7GpppG-capped transcript, which is consistent with previously obtained values for ARCA capped mRNAs [Citation21] and was explained by correct incorporation of cap analogs resulting in 100% translationally active mRNA. An introduction all new compounds to mRNA provided effective translation of the capped transcripts. The highest score of relative translation was obtained for bn2m27,2ʹ-OGpppG capped RNA, 1.72 ± 0.28 as compared to m7GpppG-capped transcript, which is slightly higher than that for ARCA-capped mRNAs, whereas both bn2m27,3ʹ-OGpppG and (p-OCH3bn)2m27,3ʹ-OGpppG capped transcripts showed slightly lower translation efficiency. The obtained differential results could be partially explained by efficiency of incorporation of newly synthesized cap analogs into the RNA. We observed that both compounds, bn2m27,3ʹ-OGpppG (9) and (p-OCH3bn)2m27,3ʹ-OGpppG (10), were incorporated with about 50% efficiency (based on ), capping efficiency determined at time 0), while bn2m27,2ʹ-OGpppG (11) was very well accepted by RNA polymerase. We estimated that in case of bn2m27,2ʹ-OGpppG. capping efficiency was around 100%, though a band below the capped position but above the uncapped triphosphate mRNA was observed. This intermediate as well as the upper migrating bands are connected to the T7 polymerase inherent tendency to product some pool of the transcripts without the last nucleotide or transcripts containing one or more additional non-template nucleotides at the 3ʹ end [Citation22].
Table 2. Translational properties of differently capped mRNA in RRL and HEK293 cells. Data represent the means ± SD of at least three independent repeats
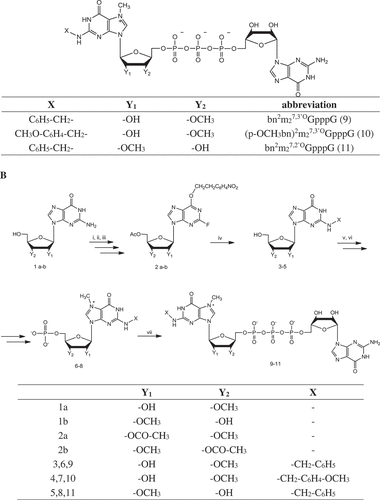
Figure 2. Translation efficiency of analog-capped mRNAs in HEK293 cells. (a) Luciferase activity measured in HEK293 cells transfected with mRNAs capped with following analogs: bn2m27,3ʹOGpppG, bn2m27,2ʹOGpppG, (p-OCH3bn)2m27,3ʹOGpppG, m27,3ʹ-OGpppG, m7GpppG. Luciferase activity was normalized to the protein concentration of samples and then expressed relatively to controls. The graph shows the mean of 4 independent experiments with ± SD. (b) Immunofluorescence staining of luciferase protein. Expression level of luciferase was detected after 24 h of mRNA transfection of HEK293 in use of anti-luciferase antibodies followed by anti-mouse FITC antibody (green signal). Cell nuclei were visualized with DAPI (blue color)
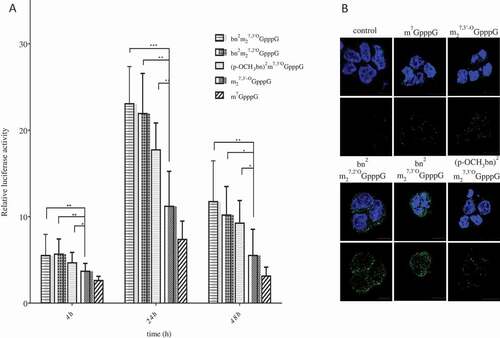
Figure 3. Stability of analog-capped mRNAs. (a) SpDcp1/2 hydrolysis of capped 34nt-RNAs. The transcripts were synthesized with T7 polymerase in use of an EcoRI digested pSP-luc+ template. Samples were resolved on 16% denaturing polyacrylamide/urea gel. Stained with SYBR gold. (b) Effect of the 5ʹ cap analogs on half–life of mRNAs in HEK293 after lipofection. RNA was isolated from cells 4, 24 and 72 h after transfection. Luciferase transcripts levels were quantified by qRT-PCR. ACTB gene was used as a reference gene. Data represent the means ± SD of three independent repeats
In the next step, we tested engineered mRNAs for translation in more complex cellular environment. We transfected equal amounts of luciferase-encoding mRNA capped with analogs 9–11, to the HEK293 cells and measured luciferase activity at 4, 24 and 48 h post transfection ()). The relative total luciferase expression was defined as the integral of the curve normalized to the value obtained for m7GpppG-capped RNA ( and Supplemental Material). mRNAs capped with bn2m27,2ʹ-OGpppG and bn2m27,3ʹ-OGpppG showed total relative protein expression 2.87 ± 0.5 and 3.3 ± 0.86, respectively. RNA capped with (p-OCH3bn)2m27,3ʹ-OGpppG provided a slightly lower total protein expression with score 2.42 ± 0.3 which was still higher than for ARCA or m7GpppG capped transcripts. The expressed luciferase was also visualized with use of specific anti-firefly luciferase antibody and confocal microscopy ()). We observed much more intensive green signal in cells transfected with bn2m27,3ʹ-OGpppG and bn2m27,2ʹ-OGpppG capped mRNA and a slightly stronger in cells containing (p-OCH3bn)2m27,3ʹ-OGpppG RNA, as compared to cells with ARCA and m7GpppG capped RNA. These data perfectly reflect results of luciferase activity measurement in HEK293 cells.
It is well known that translational efficiency is tightly dependent on cap affinity to eukaryotic translation initiation factor 4E. Cap recognition depends on the presence of m7G on the 5ʹ end of mRNA, which is stacked between two tryptophan residues W56 and W102 [Citation23,Citation24]. Guanine methylation at N7 introduces a positive charge into the ring near N7 [Citation25]. This charge distribution can be provided by any substituent at N7 and leads to a change in the character of the interaction between the guanine moiety and the W56 and W102 indol rings, from π-π into more effective cation-π stacking [Citation26]. It was shown that this cation-π interaction is a precondition for formation of three Watson-Crick-like hydrogen bonds involving N2 amine group and N1 of guanine together with carboxylate atoms of E103 [Citation26]. The region of W102 and E103 undergoes the largest secondary structure changes upon cap structure binding, rotating into the cap binding site [Citation27,Citation28]. In the case of currently examined compounds, N2 aromatic group is very close to the region directly engaged in cap binding and it may influence conformation of the protein. We assume that presence of aryl groups in this region of eIF4E could cause W102 and E103 conformational changes favoring tighter binding of a modified cap to eIF4E protein. Similar effect was previously observed for mRNA caps modified at the N7 position with benzyl substituent [Citation29]. This finding was explained by extrusion of additional water molecules from the eIF4E-cap interface causing a higher entropy change on binding.
Stability
Since overall protein synthesis depends, among others, on half-life of the protein encoding transcript, we decided to scrutinize susceptibility of the engineered mRNAs to Dcp2 decapping process. In an in vitro test, we incubated short (34 nt) RNAs with incorporated cap analogs together with Schizosaccharomyces pombe Dcp1/Dcp2 complex. The treated samples were resolved on 16% high resolution denaturing polyacrylamide gel. After staining with SYBR Gold dye, capped and non-capped RNA bands were visualized and quantified densitometrically. shows susceptibility of differently capped 34-nt RNAs to SpDcp1/Dcp2 complex. These values were calculated ()) as percentage loss of the capped bands normalized by the total quantity of material in capped and non-capped bands. The obtained results showed that presence of modification within the N2 position of guanine decreases stability of engineered mRNAs, as compared to ARCA cap. After 5 min reaction 35–65% of mRNAs capped with studied cap analogs were decapped comparing to about 14% of ARCA capped transcript. However, after 30 min of hydrolysis, differences between tested and control mRNAs were less significant, i.e. 46–72% decapped transcripts in case of N2-modified ARCA capped RNA compared to 48% of ARCA capped RNA. Since newly synthesized cap analogs do not protect mRNA from decapping enzymes hydrolysis, we conclude that higher translational efficiency was a results of cap influence on translational apparatus.
Table 3. Susceptibility of differently capped mRNAs to decapping by SpDcp1/Dcp2 complex
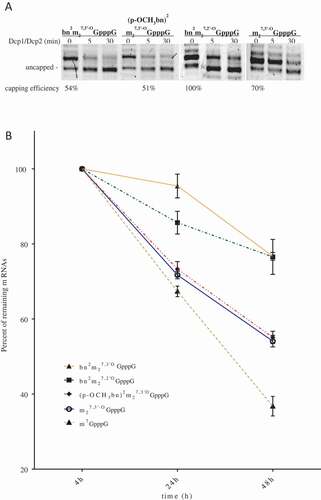
We also tested mRNAs stability in HEK293 cells ()). Transcripts encoding firefly luciferase capped with various cap analogs were synthesized in vitro, polyadenylated and purified. mRNAs were transfected to cells in complex with TransIT transfecting agent over 4h. At time points 4, 24 and 48h after transfection total RNA was extracted from the cells and quantity of luciferase mRNA was estimated by RT-qPCR. Values obtained 4h post transfection were referred as 100% of transfected mRNAs. Detailed data are presented in . The transcripts capped with bn2m27,2ʹOGpppG (11) and bn2m27,3ʹOGpppG (9) provided similar results and were more stable than other mRNAs. 48h after induction about 76% of these RNAs remained in cells. At the same time, transcript capped with (p-OCH3bn)2m27,3ʹ-OGpppG (10) was present in about 55%, which is almost the same as for ARCA capped transcript. The data rising from this experiment are inconsistent with the results obtained in in vitro studies with Dcp1/Dcp2 complex. Since percentage of remaining mRNAs reflected translational efficiency of engineered mRNA, it is more likely that stability of the transcripts is influenced by the translational activity of each mRNA and particularly by complex formed with eIF4E. It is known that eIF4E protein plays a role in multiple biochemical processes. Among others, eIF4E was found in ribonucleoparticles granules known as processing bodies (P-bodies). It was proposed that fraction of eIF4E residing in P-bodies protect mRNAs from decapping-dependent degradation through binding cap structure and preventing association with decapping enzymes [Citation30]. Given the above, we conclude that increased stability of modified mRNAs was probably the result of increased affinity to eIF4E and its protective character for decapping.
Table 4. Stability of differently capped mRNAs in HEK293 cells
Biophysics
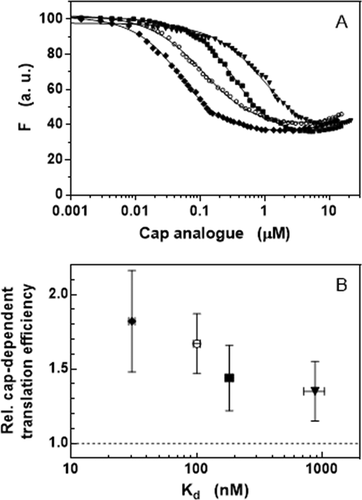
To support the hypothesis that the enhanced translational efficiency could result from the increased affinity of different N2-modified cap analogs to eIF4E, we performed direct binding assays [Citation31] based on the intrinsic protein fluorescence quenching ()). The results are gathered in and show that the nano- to micromolar affinity of the newly synthesized cap analogs strongly depends on the particular functional group of the compounds. Moreover, the comparison with reveals that the relative cap-dependent translation efficiency is correlated with the equilibrium dissociation constant in a semi-logarithmic manner ()). The value of Kd for the unmodified m7GpppG cap analogue at the same conditions was 334 ± 35 nM [Citation26]. The obtained Kd data do not fully explain increased stability of modified mRNAs in HEK293 cells so it seems that other factors may influence it. As it has been observed earlier, mRNAs capped with two diasteroeoisomers of β-S-ARCA resistant to cleavage by Dcp2 behave differently in different cells types [Citation32]. This may reflect the fact that the level and spectrum of decapping enzymes may depend on the type and developmental stage of the cell. Further studies are necessary to explain and understand this complex issue.
Table 5. Binding affinities of N2-modified cap analogs to eIF4E derived from fluorescence titrations in HEPES/KOH 50 mM pH 7.2, KCl 200 mM, DTT 1 mM, EDTA 0.5 mM, at 20°C
Figure 4. Increased affinity to eIF4E of N2-modified cap analogs. (a) Binding curves obtained by intrinsic protein fluorescence quenching upon the complex formation with (◆) bn2m27,2ʹOGpppG, (○) m27,2ʹ-OGpppG, (■) bn2m27,3ʹOGpppG, and (▼) (p-OCH3bn)2m27,3ʹOGpppG. (b) Correlation between the affinity to eIF4E and the relative cap-dependent translation efficiency
In conclusion, we have obtained three dinucleotide cap analogs with interesting biological properties, improved in comparison to ARCA cap which, as shown by us before [Citation19] is considered as an excellent substrate to yield capped transcripts. Analog containing N2 benzyl and C2ʹmethoxy substituents was the most promising as it showed increased translational efficiency in vitro in RRL and HEK293 cells. What is more, the latter analog indirectly influenced stability of capped transcripts compared to transcripts capped with m7GpppG or ARCA as it also turned out to be one of the strongest inhibitor of cap-dependent translation, with IC50 1.7 uM. The present investigation may serve as a useful way towards developing new ARCA type analogs with superior properties.
Materials and methods
Chemical synthesis is described in Supplementary Material
Inhibition of translation
Inhibition of cap-dependent translation in RRL (Flexi Rabbit Reticulocyte Lysate, Promega) by newly synthetized cap analogs was performed in 12,5 µL volume in conditions determined for cap-dependent translation as described previously [Citation33]. The samples were preincubated for 60 min at 30°C. After this time mix of m27,3΄-OGpppG-capped luciferase and investigated cap analog (inhibitor) was added and incubated for next 60 min at 30°C. Luciferase activity was measured using Glomax luminometer (Promega). The IC50 values were determined by nonlinear regression analyses of experimental data using GraphPad Prism 6. IC50 values are averages ± SD of at least three independent repeats. Statistical significance between m7GpppG and N2-modified cap analogs was determined using ANOVA and Tukey’s multiple comparison tests.
Synthesis of luciferase encoded mRNAs
Capped mRNAs, for translation studies in RRL system or HEK293 cells, were in vitro synthesized on dsDNA template with use of SP6 RNA polymerase or T7 RNA polymerase (Thermo Fisher Scientific) like described previously in [Citation34]. Briefly in vitro transcription reaction contained transcription buffer, 600 ng of DNA template, 1U/µL Ribolock Ribonuclease Inhibitor, 0.5 mM ATP/CTP/UTP, 0.1 mM GTP and 0.5 mM dinucleotide cap analog (molar ration cap: GTP was 5:1). The IVT reaction mixture was incubated at 37°C for 1 h in a thermomixer. To remove the template DNA, 2 μl DNase (Thermo Scientific) was added to the IVT reaction mixture and incubated for 20 min at 37°C. Then, the reaction mixture was purified using NucleoSpin® RNA (Macherey-Nagel) according to manufacturer’s instructions.
Translation efficiency in RRL
Luciferase mRNAs capped with newly synthetized cap analogs were tested for translational efficiency in Flexi RRL (Promega) as described previously [Citation35]. Briefly, 10 µL of translation reactions containing 40% of flexi RRL lysate (2 mM endogenous Mg2+ concentration), 0.01 mM of a mixture of amino acids, 0.7 mM magnesium acetate and 190 mM potassium acetate were prepared. Samples were preincubated during 60 min at 30°C. After this time four different concentrations of each 5ʹ-capped transcript were added and incubated for next 60 min in the same temperature. The activity of produced luciferase was measured in a Glomax luminometr and depicted as a function of the capped luciferase mRNA concentration. After linear fitting to the obtained data points (Excel software), the translation efficiency of tested transcripts was normalized to the m7GpppG-capped luciferase mRNA.
Synthesis of short capped mRNAs
Short capped transcripts 34 nt in length were performed in use of pSP-luc+ vector linearized at EcoRI site as described previously [Citation36]. In vitro transcription was run for 45 min at 37°C with T7 polymerase, 0.1 mM nucleoside triphosphate, 0.5 mM various cap dinucleotides in buffer appropriate for RNA polymerase. Plazmid DNA was removed from samples by RQ1 RNase-Free DNase (Promega) treatment. Short RNAs were purified with RNA Clean & Concentrator TM-5 kit (Zymo Research) and estimated for quality and quantity on the DeNovix Spectrophotometer and on 16% of Urea-PAGE.
Decapping assay
Fraction of 20 ng of capped transcripts were subjected to 50 nM S. pombe Dcp1/2 complex hydrolysis in 10 mM Hepes pH 7.5, 100 mM potassium acetate, 2 mM magnesium acetate, 2 mM dithiothreitol at 30°C for indicated time periods. Decapping reactions were stopped by cooling on ice and adding Loading Buffer followed by denaturation for 3 min at 55°C. 16% RNA sequencing gels were run at 600 V for 3–4 hours. The gels were stained with SYBR Gold Nucleic Acid Gel Stain (Life technologies) and visualized with ChemiDoc™ MP Imaging System (Bio-Rad). Densitometric analysis was performed with Image Lab Software (Bio-Rad). The data represent the means ± SD from two experiments.
Cell culture
HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Biowest) supplemented with 10% fetal bovine serum (Biowest) and penicillin-streptomycin (Biowest) at 37°C in a humidified atmosphere of 5% CO2 .
mRNA transfection
Transfections were carried out with TransiT (Mirus Bio LLC). The cells were seeded on 96- and 24-well-plates (15 000 cells or 150 000 cells/well) 24 h prior to the experiment. The culture medium was changed to fresh (without antibiotics) before transfection. 0.1 or 0.5 µg of capped-mRNAs were diluted in 10 or 50 µl OptiMem. Next, 0.18 or 1 µl TransiT and Boost reagents were added and mixtures were incubated 5 minutes at RT. In the next step, components were added to cells and incubated 4, 24 and 48 h.
Luciferase reporter assays
Cells from 96-well plate were lysed 4, 24 and 48 h after transfection with 20 µl of Cell Culture Lysis Reagent (Promega) and used to luciferase activity measurement and estimation of protein concentration. The protein extract sample (17 µl) was mixed with luciferase assay reagent (Promega, USA) (40 µl), and measured on Synergy H1MFDG Microplate Reader (BioTek, USA). HEK293 cells treated with transfection reagent without mRNA and non-transfected HEK293 served as controls. The protein lysates concentration were determined with Roti-Quant Protein quantitation assay according to Bradford (Roth, USA) using BSA as standard.
RNA extraction and cDNA synthesis
Total RNA was extracted from transfected and controls HEK293 cells using Universal Purification Kit (EURx), followed by on-columne DNAse (Thermo Scientific) digestion. Complementary DNA (cDNA) was transcribed from mRNA using High-Capacity cDNA Reverse Transcription Kit (Life Technologies), according to the manufacturer’s protocols.
Real-time quantitative PCR
Quantity of luciferase mRNA was estimated by RT-qPCR using the cDNAs as a template in reactions containing the double-stranded DNA-specific dye SYBR Green I and Maxima Fluorescein RT-qPCR Master Mix (Thermo Scientific) and specific oligonucleotide primers: luciferase (LUC) – forward: 5ʹ-CAGTATGGGCATTTCGCAGC-3ʹ and reverse: 5ʹ-AGCGACACCTTTAGGCAGAC-3; ß-actin (ACTB) – forward: 5ʹ-GCCGAGGACTTTGATTGC-3ʹ and reverse:5ʹ-CTGTGTGGACTTGGGAGAG-3ʹ. PCR reactions were done in triplicates with the following conditions: 95°C/30 s, 40 cycles of 95°C/5 s, 58°C/15s and 72°C/10s on LightCycler® 480 Instrument II (Roche). The Ct values obtained for genes were normalized by the corresponding Ct values of β-ACTB.
Immunofluorescence
Transfected cells grown for 24 h on glass coverslips were washed with PBS and fixed with 4% paraformaldehyde in RT for 15 min. Next, the cells were permeabilized for 10 min with 0,25% Triton X-100 in PBS and blocked with 2% BSA in PBST (0,05% Tween 20) for 1h. The slides were incubated overnight at 4°C with specificity primary antibody anti-luciferase (cat no. MA1-16880, Thermo Scientific) resuspended in PBST with 2% BSA. After several washes with PBST, the cells were incubated with Goat anti-Mouse IgG (H+L) Secondary Antibody, DyLight 488 (cat no. 35502, Thermo Scienticic) diluted 1:500 in 2% BSA/PBST for 1 h in dark. Nuclei staining were performed with 4,6-diamidino-2-phenylindole – DAPI (Sigma-Aldrich) for 2 min. The slides were visualized with Axio Imager Z2 LSM 700, Zeiss, confocal microscope using a 63 x oil-immersion lens and processed with ZEN software, respectively (Zeiss). Data are reported as means ± SD. Statistical analysis was performed in use the nonparametric Mann-Whitney U test (GraphPad, Prism). A P value of < 0,05 was considered statistically significant.
eIF4E binding studies
Full length human eIF4E was expressed and purified as described previously [Citation37]. The protein concentration was determined from the absorption spectrum recorded on Cary 50 UV-VIS spectrophotometer (Agilent), based on the molar extinction coefficient at 280 nm as 53 000 cm^(−1)M^(−1).
Fluorescence titration experiments were performed essentially as described previously [Citation26,Citation31]. The measurements were run on Fluorolog 3.11 (Horiba) spectrofluorometer for eIF4E at 0.2 μM in HEPES/KOH 50 mM pH 7.2, KCl 200 mM, DTT 1 mM, EDTA 0.5 mM, in a thermostated semi-micro cuvette (Hellma) at 20°C, with magnetic stirrer, with excitation wavelength of 280 nm and emission wavelength of 337 nm. A suitable correction for the inner-filter effect was applied. The experimental data were analyzed by non-linear regression using GraphPad Prism 6, as described previously [Citation31].
Supplemental Material
Download PDF (513.3 KB)Acknowledgments
We thank Prof John Gross for SpDcp1/2 encoding plasmids and Dr Joanna Zuberek and Agnieszka Stelmachowska for SpDcp1/2 protein preparation. This work was supported by grant from the National Science Centre, Poland (DEC-2013/11/B/ST5/02226 to MJ-A, DEC-2016/21/D/ST5/01654 to KP, UMO/2013/08/A/NZ1/00866 to ED) and from National Centre of Research and Development, Poland (STRATEGMED1/235773/19/NCBR/2016 to ED).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Gousseinov E, Kozlov M, Scanlan C, et al. RNA-based therapeutics and vaccines. GEN Exclusive. 2015 Sep 15.
- Raper SE, Chirmule N, Lee FS, et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol Genet Metab. 2003;80(1–2):148–158.
- Diken M, Kranz LM, Kreiter S, et al. mRNA: a versatile molecule for cancer vaccines. Curr Issues Mol Biol. 2017;22:113–128.
- Weissman D. mRNA transcript therapy. Expert Rev Vaccines. 2015;14(2):265–281.
- Vallazza B, Petri S, Poleganov MA, et al. Recombinant messenger RNA technology and its application in cancer immunotherapy, transcript replacement therapies, pluripotent stem cell induction, and beyond. Wiley Interdiscip Rev RNA. 2015;6(5):471–499.
- Rhoads RE. Synthetic mRNA: production, introduction into cells, and physiological consequences. Methods Mol Biol. 2016;1428:3–27.
- Grudzien-Nogalska E, Kowalska J, Su W, et al. Synthetic mRNAs with superior translation and stability properties. Methods Mol Biol. 2013;969:55–72.
- Furuichi Y. Discovery of m(7)G-cap in eukaryotic mRNAs. Proc Jpn Acad Ser B Phys Biol Sci. 2015;91(8):394–409.
- Topisirovic I, Svitkin YV, Sonenberg N, et al. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA. 2011;2:277–298.
- Merrick WC. eIF4F: a retrospective. J Biol Chem. 2015;290(40):24091–24099.
- Karaki S, Andrieu C, Ziouziou H, et al. The Eukaryotic Translation Initiation Factor 4E (eIF4E) as a Therapeutic Target for Cancer. Adv Protein Chem Struct Biol. 2015;101:1–26.
- Carroll M, Borden KL. The oncogene eIF4E: using biochemical insights to target cancer. J Interferon Cytokine Res. 2013;33(5):227–238.
- Jia Y, Chiu TL, Amin EA, et al. Design, synthesis and evaluation of analogs of Initiation Factor 4E (eIF4E) cap-binding antagonist Bn7-GMP. Eur J Med Chem. 2010;45:1304–1313.
- Cai A, Jankowska-Anyszka M, Centers A, et al. Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry. 1999;38:8538–8547.
- Chen X, Kopecky DJ, Mihalic J, et al. Structure-guided design, synthesis, and evaluation of guaninę-derived inhibitors of the eIF4E mRNA-cap interaction. J Med Chem. 2012;55:3837–3851.
- Piecyk K, Davis RE, Jankowska-Anyszka M. Synthesis of N2-modified 7-methylguanosine 5ʹ-monophosphates as nematode translation inhibitors. Bioorg Med Chem. 2012;20:4781–4789.
- Piecyk K, Lukaszewicz M, Darzynkiewicz E, et al. Triazole-containing monophosphate mRNA cap analogs as effective translation inhibitors. RNA. 2014;20:1539–1547.
- Pasquinelli AE, Dahlberg JE, Lund E. Reverse caps in 5ʹ RNAs made in vitro by phage RNA polymerases. RNA. 1995;1:957–967.
- Stepinski J, Waddell C, Stolarski R, et al. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3ʹ-O-methyl)GpppG and 7-methyl (3ʹ-deoxy)GpppG. RNA. 2001;7(10):1486–1495.
- Grudzien-Nogalska E, Stepinski J, Jemielity J, et al. Synthesis of Anti-Reverse Cap Analogs (ARCAs) and their applications in mRNA translation and stability. Methods Enz. 2007;431:203–227.
- Jemielity J, Fowler T, Zuberek J, et al. Novel “anti-reverse” cap analogs with superior translational properties. RNA. 2003;9(9):1108–1122.
- Milligan JF, Groebe DR, Witherell GW, et al. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15(21):8783–8798.
- Marcotrigiano J, Gingras AC, Sonenberg N, et al. Cocrystal structure of the messenger RNA 5ʹ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89(6):951–961.
- Shen X, Tomoo K, Uchiyama S, et al. Structural and thermodynamic behavior of eukaryotic initiation factor 4E in supramolecular formation with 4E-binding protein 1 and mRNA cap analogue, studied by spectroscopic methods. Chem Pharm Bull. 2001;49:1299–1303.
- Ruszczynska K, Kamienska-Trela K, Wojcik J, et al. Charge distribution in 7-methylguanine regarding cation-pi interaction with protein factor eIF4E. Biophys J. 2003;85(3):1450–1456.
- Niedzwiecka A, Marcotrigiano J, Stepinski J, et al. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5ʹ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J Mol Biol. 2002;319(3):615–635.
- Volpon L, Osborne MJ, Borden KL. NMR assignment of human eukaryotic translation initiation factor 4E (eIF4E) in its cap-free form. J Biomol NMR. 2006;36(Suppl 1):65.
- Rutkowska-Wlodarczyk I, Stepinski J, Dadlez M, et al. Structural changes of eIF4E upon binding to the mRNA 5ʹ monomethylguanosine and trimethylguanosine Cap. Biochemistry. 2008;47(9):2710–2720.
- Niedzwiecka A, Darzynkiewicz E, Stolarski R.Thermodynamics of mRNA 5' ca bimding by eukaryotic translation initiation factor eIF4E. Biochemistry. 2004;43(42):13305-137.
- Borden KL. The eukaryotic translation initiation factor eIF4E wears a “cap” for many occasions. Translation (Austin). 2016;4(2):e1220899.
- Niedzwiecka A, Stepinski J, Antosiewicz JM, et al. Biophysical approach to studies of cap-eIF4E interaction by synthetic cap analogs. Methods Enzymol. 2007;430:209–245.
- Kuhn AN, Diken M, Kreiter S, et al. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 2010;17(8):961–971.
- Kowalska J, Lukaszewicz M, Zuberek J, et al. Phosphorothioate analogs of m7GTP are enzymatically stable inhibitors of cap-dependent translation. Bioorg Med Chem Lett. 2009;19(7):1921–1925.
- Rydzik AM, Lukaszewicz M, Zuberek J, et al. Synthetic dinucleotide mRNA cap analogs with tetraphosphate 5ʹ,5ʹ bridge containing methylenebis(phosphonate) modification. Org Biomol Chem. 2009;7(22):4763–4776.
- Strenkowska M, Kowalska J, Lukaszewicz M, et al. Towards mRNA with superior translational activity: synthesis and properties of ARCA tetraphosphates with single phosphorothioate modifications. New J Chem. 2010;34(5):993–1007.
- Strenkowska M, Grzela R, Majewski M, et al. Cap analogs modified with 1,2-dithiodiphosphate moiety protect mRNA from decapping and enhance its translational potential. Nucleic Acids Res. 2016;44(20):9578–9590.
- Modrak-Wojcik A, Gorka M, Niedzwiecka K, et al. Eukaryotic translation initiation is controlled by cooperativity effects within ternary complexes of 4E-BP1, eIF4E, and the mRNA 5ʹ cap. FEBS Lett. 2013;587(24):3928–3934.
