Cell-type specific expression of genes is directly determined by the epigenetic state of a cell. Epigenetic information in mammalian cell is stored in two ways: as chemical alterations of DNA on cytosines, and as covalent modifications of histone proteins that are arranged into nucleosomes to package the genome. Both types of modifications are integral parts of the epigenetic code [Citation1,Citation2]. Enhancers are critical regulatory elements that integrate genomic and epigenomic information to orchestrate cell fate transitions and direct cell specific gene regulation programming, both in cell models and in vivo. Additionally, cell-type-specific enhancers are marked by cell-type specific transcription factors [Citation3,Citation4]. The activation of enhancers is key to understanding the mechanisms by which numerous cell types emerge from stem cells harboring identical genomes [Citation5].
In embryonic stem (ES) cells the regulatory regions of ES specific genes are decorated by classical combination of histone marks (e.g. H3K4me2, H3K4me1 and H3K27Ac) and by combination of several ES-specific factors. It is however still unknown how the cell-type restricted regulatory regions are selectively enabled in the process of differentiation, since no specific epigenetic mark have been discovered to label them in the undifferentiated state.
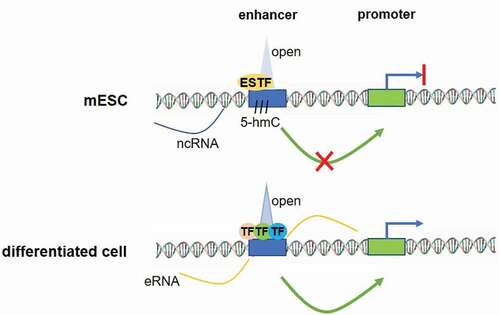
In the work published in Nature in April 2018, we have [Citation6] shed some light on the potential mechanism of this cell-type restricted enhancer activation. We have shown that, instead of a full combination of ES specific transcription factors (TFs), cell-type-restricted enhancers are occupied by a limited number of ES TFs and they do not exhibit traditional epigenetic marks associated with activated enhancers (). In addition, these binding sites are decorated by cohesin molecules and by 5-hydroxymethyl cytosines and exhibit low level of eRNA expression, which suggest their limited functionality in ES cells. Indeed, these premarked regions did not interact with their cognate promoters in ES cells, in contrast to the final cell-type where the robust interaction is indispensable for gene activation. Interestingly, we have shown that this pre-marking is not only required for future activation of the regulatory element in cell-type-specific manner, but also for the subsequent robustness of cell-type-restricted enhancer activity and gene expression. This was demonstrated in macrophage- and neural-restricted enhancers, where we deleted the ES-specific transcription factor binding site from a regulatory element and shown that the activity of the linked gene is dramatically affected.
Figure 1. A model of cell-type-restricted enhancer pre-marked in mESCs. In the ES cells a pre-marked enhancer is bound by a few stem cell TFs (ESTF), is labelled by 5-hmC and shows little extragenic transcription. Surrounding chromatin is in open state, but the enhancer is not active. Conversely, in the terminal cell type, the same enhancer is bound by several cell type-specific transcription factors, eRNAs are expressed at high level, the chromatin structure is open and the enhancer engages in physical interactions with its cognate promoter to assure robust gene activation
Lastly, we investigated a potential mechanism of ‘molecular memory’ which could maintain the pre-marked state of these regulatory elements through cellular differentiation when ES specific factors disappear. Indeed, using openly available genomic data we found that 5-hydroxymethylation of DNA is maintained during differentiation far beyond the disappearance of the ES factors. This suggests that this DNA modification may be used to keep the cell-type-specific enhancers available for cell-type-restricted factors [Citation6].
In sum, delineation of the epigenetic signature of cell-type-restricted enhancers in ESCs has provided a surprising insight into the process of genomic enhancer recognition underlying cell-type-specific transcriptional programs. It appears that cell-type-restricted functional enhancers are pre-marked at an early stem cell stage through binding of a single, or sometimes two, canonical stem cell transcription factors, in varied areas of the enhancers themselves, without activating transcription. At the same time, multiple ES-specific TFs bind ES-specific enhancers within their core sequences to trigger transcriptional activation. The pre-marking pattern of cell-type-restricted enhancers is key to their future availability for subsequently-expressed transcription factors and to gene activation in the differentiated cell.
Our work has also underlined the unexpected complexity in marking of regulatory regions in ES cells. Many unresolved questions remain to be answered. For example, what subset or cell-type-restricted enhancers utilize this type of pre-marking patterns? Is this a general rule or an exception? Are there other marks that have similar function at a different group of regulatory elements? Do other cell-type restricted regulatory regions, such as superenhancers or insulators also require pre-marking for their full functionality? Are the pathogenic sequence variants that correlate with various diseases located in the pre-marked regions rather than located directly in the cell-type restricted enhancer? It also remains unclear how each class of cell-type specific enhancers is activated during the process of development and differentiation.
Taken together, this work is uncovering existence of novel regulatory mechanism – premarking with stem cell factors to assure robust activation in the differentiated tissues. Other studies will have to follow to understand the precise mechanism of premarking and enhancer activation.
Disclosure statement
No potential conflict of interest was reported by the author.
References
- Telese F, Gamliel A, Skowronska-Krawczyk D, et al. “Seq-ing” insights into the epigenetics of neuronal gene regulation. Neuron. 2013;77:606–623.
- Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. 2013;58(7):439–445.
- Skowronska-Krawczyk D, Ballivet M, Dynlacht BD, et al. Highly specific interactions between bHLH transcription factors and chromatin during retina development. Development. 2004;131:4447–4454.
- Skowronska-Krawczyk D, Chiodini F, Ebeling M, et al. Conserved regulatory sequences in Atoh7 mediate non-conserved regulatory responses in retina ontogenesis. Development. 2009;136(22):3767–3777.
- Catarino RR, Stark A. Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes Dev. 2018;32:202–223.
- Kim HS, Tan Y, Ma W, et al. Pluripotency factors functionally premark cell-type-restricted enhancers in ES cells. Nature. 2018;556:510–514.
