ABSTRACT
Circular (circ)RNA is a special type of endogenous RNA consisting of a covalently closed loop structure without 5‘ to 3‘ polarity and a polyadenylated tail. Accumulating evidence suggests that circRNAs play important roles in the development and progression of human cancers. However, the role of circRNAs in the progression of hepatocellular carcinoma (HCC) is largely unknown. This was addressed in the present study using high-throughput sequencing to identify aberrantly expressed circRNAs in HCC patient tissue and cell lines. We found that circ-baculoviral IAP repeat-containing (BIRC)6 was upregulated in HCC tissue samples and cells; this was associated with the overall survival of HCC patients. circ-BIRC6 knockdown reduced HCC cell proliferation, migration, and invasion and enhanced their apoptosis. Additionally, circ-BIRC6 overexpression negatively regulated the expression of microRNA miR-3918, which was identified as an inhibitor of B cell lymphoma (Bcl)2. The tumor-suppressive effect of circ-BIRC6 deletion was abrogated by inhibiting miR-3918. These results indicate that circ-BIRC6 functions as a competing endogenous RNA that regulates Bcl2 expression by sponging miR-3918, and may serve as a prognostic biomarker and therapeutic target for the treatment of HCC.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies [Citation1] and the second-leading cause of cancer-related death worldwide [Citation2]. Despite recent progress in the development of therapeutic strategies, the prognosis of most HCC patients remains poor because they are typically diagnosed at advanced stages of the disease [Citation3]. Clarifying the mechanisms underlying the malignant progression of HCC and identifying suitable biomarkers for early detection and for monitoring therapeutic efficacy can improve patient outcome.
Circular (circ)RNAs are a large class of endogenous non-coding (nc)RNAs that covalently link 5‘ and 3‘ ends of RNA to generate a circular structure [Citation4], and play important roles in many biological processes [Citation5]. Recent developments in high-throughput sequencing technology and bioinformatics approaches have permitted the identification and characterization of many circRNAs in human cells [Citation6], with most being up- or downregulated in specific tissues [Citation7]. circRNAs play an important role in the progression of various malignancies [Citation8] including pancreatic [Citation9], bladder [Citation10], and cervical [Citation11] cancers. circRNAs contribute to cancer development by acting as micro (mi)RNA sponges to regulate gene and protein expression [Citation12], and may serve as biomarkers for diagnosing and predicting the prognosis of cancer patients [Citation13]. As such, elucidating the mechanisms underlying circRNA function can provide a basis for the development of novel cancer treatments.
To this end, in this study we compared the expression profiles of circRNAs in HCC and normal tissues by high-throughput sequencing, and show that the circRNA circ-baculoviral IAP repeat-containing (BIRC)6 is upregulated in HCC tissues, which is associated with HCC patient prognosis. According to circBase [Citation14], circ-BIRC6 (ID hsa_circ_0003288) is located on chromosome 2 (32703702–32718734) and is generated by back-splicing of the BIRC6 transcript (NM_016252). We found that circ-BIRC6 knockdown inhibited HCC cell proliferation, migration, and invasion and promoted their apoptosis; moreover, circ-BIRC6 acted as a sponge of the miRNA miR-3918, which targets the 3‘ untranslated region (UTR) of B cell lymphoma (Bcl)2 transcript. These results indicate that circ-BIRC6 enhances Bcl2 expression by sponging miR-3918 and is a potential biomarker and therapeutic target for HCC treatment.
Materials and methods
Patient samples
HCC and matched adjacent non-tumor tissues (n = 55) were obtained from Qianfoshan Hospital of Shandong Province with patient consent and Ethics Committee approval. HCC was diagnosed by pathological examination. None of patients received chemotherapy, radiotherapy, or any other medical intervention before surgery. The tissue samples were flash frozen and stored in liquid nitrogen until use.
Cell culture and transfection
HepG2, Bel-7402, and Huh-7 human HCC cell lines were obtained from the American Type Culture Collection (Rockville, MD, USA). SMMC-7721 human hepatocarcinoma cells and the L02 normal human hepatic cell line were purchased from the Typical Culture Reserve Center of China (Shanghai, China). 293T cells were obtained from Chinese Academy of Science Typical Culture Collection (Shanghai, China). The cells were cultured in Dulbecco’s Modified Eagle’s medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin in a humidified incubator at 37°C and 5% CO2.
Small interfering (si)RNA against circ-BIRC6 (5‘-CUGAAAGGUUCUUGCACGCTT-3‘) and a negative control siRNA along with circ-BIRC6 overexpression sequence (supplementary Table1), miR-3918 inhibitor (5‘-AGUCUCCAUCUGCGGCCCUGU-3‘) and mimic (5‘-ACAGGGCCGCAGAUGGAGACU-3‘) were purchased from GenePharma (Shanghai China). The siRNAs and miRNAs were transfected into HCC cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol.
Cell proliferation assay
Cell proliferation was evaluated with Cell Counting Kit (CCK)-8 (Dojindo Laboratories, Kumamoto, Japan). Transfected cells were seeded in a 96-well plate at a density of 5 × 103 cells/well and cultured for 24, 48, 72, and 96 h. A 10-μl volume of CCK-8 reagent was added to each well. The absorbance at 450 nm was measured on a Spectra Max 190 instrument (Molecular Devices, Sunnyvale, CA, USA). Experiments were performed with triplicate samples.
Transwell migration and invasion assays
A transwell cell migration system (8-μm pore size; BD Biosciences, Franklin Lakes, NJ, USA) and 24-well transwell plates with Matrigel (BD Biosciences) were used to evaluate cell migration and invasion, respectively. Transfected cells were resuspended in serum-free medium to a density of 2.0 × 105cells/ml. A 200-μl aliquot of cells was added to the upper chamber of the transwell insert, and 600 μl complete medium containing 10% FBS were added to the lower well, followed by incubation at 37°C and 5% CO2 for 24 h. The cells were fixed with methanol and stained with 0.1% crystal violet. The cells that passed through the filter were photographed and counted under a microscope.
Flow cytometry analysis
After transfection for 48 h, HepG2 and Bel7402 cells (2 × 105 cells per well) were collected and stained with Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) (Dojindo Laboratories) according to the manufacturer’s instructions. Apoptosis was assessed by flow cytometry (FACSAria II; BD Biosciences).
Fluorescence in situ hybridization (FISH)
FISH was performed to detect the presence of circ-BIRC6 using a Cy3-labed probe (5ʹ-CY3-ACTTTCAACTGAAAGGTTCTTGCACGCAT-3ʹ) and miR-3918 using a FAM-labed probe (5ʹ-FAM-AGTCTCCATCTGCGGCCCTGT-FAM-3ʹ) respectively. FISH probes were designed by Servicebio (Wuhan, China). Cells were cultured on coverslips until they reached 60%–70% confluence, then washed three times with phosphate-buffered saline (PBS) for 5 min and fixed with 4% paraformaldehyde for 15 min at room temperature. After washing with PBS, the cells were incubated overnight at 37°C with the FISH probes in hybridization buffer in a humid chamber according to the manufacturer’s protocol. The following day, the cells were washed and stained with 4‘,6-diamidino-2-phenylindole for 10 min in the dark. Images were obtained using a fluorescence microscope (Hitachi, Tokyo, Japan).
Reverse transcription and quantitative real-time (qrt)-pcr
RNA was extracted from tissues or cells using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Total RNA was reverse transcribed using the FastQuant RT kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s protocol, and qRT-PCR analysis of circ-BIRC6, Bcl2, and miR-3918 expression was performed using SYBR Select Master Mix (Tiangen Biotech) on an ABI ViiA7 system (Applied Biosystems, Foster City, CA, USA), with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 used as internal controls. The following forward and reverse primers were used for qRT-PCR:
circ-BIRC6, 5‘-TGAAAGGTTCTTGCACGCAT-3‘ and 5‘-GCTGGGGTTCGTTCACAATC-3‘; BIRC6 mRNA, 5ʹ-TCAACCTCCGCCTCACCAGTC-3ʹ and 5ʹ- ACCGCCTTCCATCCACCTCTTC-3ʹ;Bcl2 mRNA, 5‘-CGACTTCGCCGAGATGTCCAG-3‘ and 5‘-CGGTTCAGGTACTCAGTCATCCAC-3‘; GAPDH, 5‘-CAGAACATCATCCCTGCCTCTAC-3‘ and 5‘-ATGAAGTCAGAGGAGACCACCTG-3‘; miR-3918, 5‘-ACAGGGCCGCAGATGGAG-3‘; and U6, 5‘-ACGCAAATTCGTGAAGCGTTC-3‘.
Luciferase reporter assay
The sequences of circ-BIRC6 and Bcl2 3‘ UTR containing wild-type (WT) or mutant (Mut) miR-3918 binding sites were cloned into the luciferase vector (Promega). The generated vectors were transfected into 293T cells along with miR-NC and miR-3918 mimic using Lipofectamine 2000. The Dual-luciferase Reporter Assay (E1910; Promega, Madison, WI, USA) was used to measure luciferase activity in cell lysates 48 h after transfection.
Western blot analysis
HCC cells were lysed in cold radioimmunoprecipitation lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) containing 1 nM phenylmethylsufonyl fluoride. Protein concentration was quantified with a BCA Protein Assay kit (Beyotime Institute of Biotechnology). Equal amounts of protein were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA) that was incubated overnight at 4°C in a solution of non-fat milk and antibody (1:1000 dilution; Abcam, Cambridge, MA, USA), followed by incubation with horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody (Beyotime Institute of Biotechnology) for 2 h at room temperature. Protein bands were visualized with enhanced chemiluminescence detection reagent (Millipore), and signal intensity was quantified using Image Lab software (Bio-Rad, Hercules, CA, USA).
Rna-binding protein immunoprecipitation (RIP) assay
The RIP assay was performed using the EZMagna RIP kit (Millipore) according to the manufacture’s protocol. Cells were lysed in RIP lysis buffer, followed by incubation with RIP buffer containing magnetic beads conjugated with anti-human Argonaute (Ago)-2 antibody (Millipore) or normal mouse IgG (Millipore) as a negative control. Immunoprecipitated proteins were digested with proteinase K and the associated RNA was purified and analyzed by qRT-PCR using appropriate primers.
Tumor xenograft model
Animal experiments were approved by the Animal Care and Use Committee of Shandong University. Short hairpin RNAs (shRNAs) targeting circBIRC6 (5ʹ-TTTCAACTGAAAGGTTCTTGCAC-3ʹ) , negative control (NC) (5ʹ-TTCTCCGAACGTGTCACGTTTC-3ʹ) , overexpressed circ-BIRC6 (oe-circ-BIRC6) (supplementary Table1) and corresponding negative control (Vector) were cloned into the lentiviral vectors and lentiviral supernatant was collected to infected Bel7402 cells. Stably transfected Bel7402 cells were subcutaneously injected into the flank of 4-week-old BALB/c nude mice. Tumor size was measured every 3 days using calipers and tumor volume was calculated using the formula length × width2/2. After 4 weeks, mice were sacrificed and tumor weight was measured.
Table 1. Relationship between circ-BIRC6 expression and clinicopathological factors
Statistical analysis
Data were analyzed with SPSS v.17.0 software (IBM, Armonk, NY, USA) and are shown as mean ± SD. The Student’s t test was used to compare the means of two groups, and one-way analysis of variance was used for multiple-group comparisons. The χ2 test or Fisher’s exact test was used to analyze the relationship between circ-BIRC6 and miR-3918 expression levels and the clinicopathological features of HCC patients. Correlations were evaluated using Pearson’s correlation coefficient. Survival was evaluated by Kaplan-Meier analysis and compared with the log-rank test. P < 0.05 was considered statistically significant.
Results
Circ-BIRC6 upregulation in HCC is associated with HCC patient prognosis
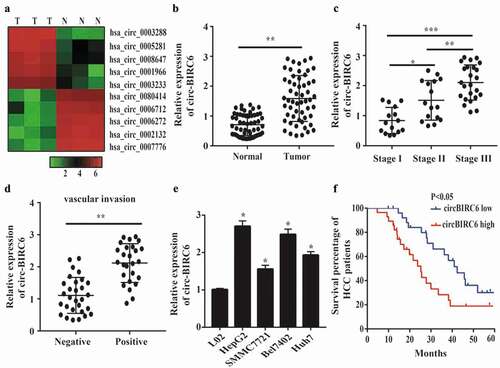
To investigate the relationship between circRNAs and HCC progression, we performed high-throughput sequencing and found that many circRNAs were up- or downregulated in HCC as compared to adjacent normal tissue (). One of these, circ-BIRC6, was overexpressed in HCC tissue; this was confirmed by qRT-PCR analysis of HCC and normal tissues (). To further examine the correlation between circ-BIRC6 expression and HCC severity, we assessed circ-BIRC6 expression in HCC at different stages of the disease. circ-BIRC6 level was associated with tumor-node-metastasis (TNM) stage in HCC (), and was higher in samples with as compared to those without vascular invasion (). A qRT-PCR analysis confirmed that circ-BIRC6 expression was higher in HCC cell lines – i.e., HepG2, Bel-7402, SMMC-7721, and Huh7 cells – than in normal L02 cells (). Given its upregulation in HCC tissue, we investigated whether circ-BIRC6 expression is associated with HCC patient prognosis. The Kaplan-Meier analysis using median circ-BIRC6 level as the cut-off point showed that HCC patients with high circ-BIRC6 expression had shorter overall survival (OS) than those with low circ-BIRC6 expression (). These data indicate that circ-BIRC6 is aberrantly expressed in HCC tissue and may serve as a biomarker for poor prognosis.
Figure 1. circ-B1RC6 is upregulated in HCC. (a) Heat map showing differently expressed circRNAs in three pairs of HCC and normal tissues based on high-throughput sequencing analysis. N, normal tissue; T, tumor tissue. (b) Relative expression of circ-BI RC6 in 55 pairs of HCC and normal tissue, as determined by qRT-PCR. (c) Relative expression of circ-BIRC6 in HCC tissues of different TNM stages. (d) Relative expression of circ-BI.RC6 in HCC tissue with and without vascular invasion, as determined by qRT-PCR. (e) Relative expression of circ-BIRC6 in HCC cell lines (HepG2, SMMC7721, Bel7402, and Huh7 cells) compared to normal (L02) cells, as determined by qRT-PCR. (f) Kaplan-Meier survival curves showing the prognost i c sign ificance of circ-BIR C6 expression for HCC patients (with median value used as the cut-off point). Data are presented as mean :1: SD. *P < 0.05, ***P < 0.0 I, ***P < 0.00 I.
Relationship between circ-BIRC6 expression and clinicopathological features of HCC patients
Given that high circ-BIRC6 expression was correlated with poor prognosis in HCC patients, we analyzed the relationship between circ-BIRC6 level and clinicopathological features of HCC patients including age, gender, serum alpha-fetoprotein level, tumor size, vascular invasion, and TNM stage. circ-BIRC6 expression was associated with tumor stage (P = 0.0143) and vascular invasion (P = 0.0151), but was unrelated to other clinical characteristics ().
Uni- and multivariate Cox regression analyses of clinical variables showed that circ-BIRC6 expression (P = 0.005), vascular invasion (0.006), tumor size (0.045), Edmondson-Steiner histological grade (P = 0.024), and TNM stage (P = 0.001) were associated with the OS of HCC patients (). We performed a multivariate analysis to assess whether circ-BIRC6 expression is an independent prognostic factor and found that circ-BIRC6 level along with vascular invasion and TNM stage was associated with OS. These data imply that circ-BIRC6 contributes to HCC progression.
Table 2. Univariate and multivariate cox regression analyses circ-BIRC6 for OS of patients in the cohort
Circ-BIRC6 knockdown inhibits cell proliferation, migration, and invasion and promotes apoptosis in HCC cells
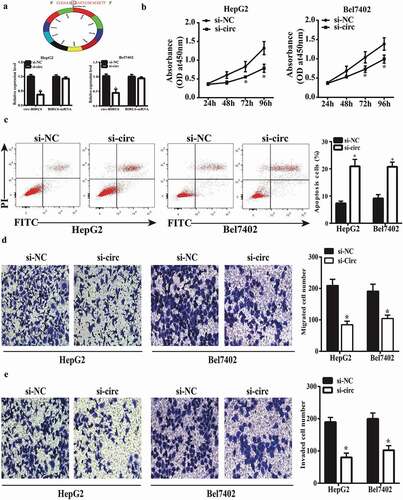
We investigated the function of circ-BIRC6 in HCC using HepG2 and Bel7402 cells transfected with siRNA against circ-BIRC6. A qRT-PCR analysis showed that circ-BIRC6 expression was downregulated in transfected HepG2 and Bel7402 cells, while BIRC6 mRNA had no significant changes, which indicated si-circ-BIRC6 specifically targeted the circRNA transcript but not the linear RNA one (). We also evaluated cell proliferation with the CCK-8 assay and found that circ-BIRC6 knockdown suppressed cell proliferation (). Additionally, circ-BIRC6 silencing enhanced HepG2 and Bel7402 cell apoptosis, as determined by flow cytometry () and inhibited migration and invasion in the transwell assays (). These results indicate that circ-BIRC6 plays an oncogenic role in HCC by positively regulating cancer cell proliferation, migration, and invasion and negatively regulating apoptosis.
Figure 2. circ-BIRC6 knockdown represses HCC cell proliferation, migration, and invasion and promotes apoptosis. (a) Relative expression of circ-BIRC6 and BlRC6 mRNA in HepG2 and Bel7402 cells transfected w ith an si-circ against BIRC6 (si-circ-BTRC6) or negative control siRNA (si-NC), as determined by qRT-PCR. (b) circ-BIRC6 deletion inhibits the proliferation of HepG2 and Bel7402 cells, as shown with the CCK-8 assay. (c) circ-BIRC6 silencing increases apoptosis in HepG2 and Bel7402 cells, as detected by flow cytometry. (d, e) ci.rc-BIRC6 knockdown represses HepG2 and Bel7402 cell migration and invasion, as determined with the transwell assay. Data are presented as mean ± SD. *P < 0.05, **P < 0.0 I, ***P < 0.001.
Circ-BIRC6 overexpression promotes proliferation, migration, and invasion while inhibiting apoptosis in HCC cells
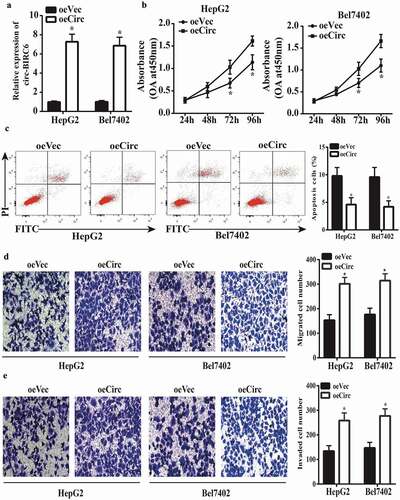
Based on the above results, we speculated that circ-BIRC6 overexpression in HepG2 and Bel7402 cells would enhance their malignant character. Upregulation of circ-BIRC6 in cells transfected with a circ-BIRC6 overexpression vector was confirmed by qRT-PCR (). circ-BIRC6 overexpression increased cell proliferation () as well as migration and invasion () while inhibiting apoptosis (), as determined with the CCK-8 assay, transwell assays, and by flow cytometry analysis, respectively. These results confirm that circ-BIRC6 has an oncogenic function in HCC.
Figure 3. circ-BIRC6 overexpress ion promotes HCC cell pro! iferation, migration, and invasion and inhibits apoptosis. (a) Relative expression of circ-BIRC6 in HepG2 and Bel7402 cells transfected with overexpression and empty vector. oeCirc, cirGBIRC6 overexpression vector; oeVec, control overexpression vector. (b) ciro-BIRC6 overexpression promotes the proliferation in HepG2 and Be17402 cells, as determined with the CCK-8 assay. (c) Cell apoptosis was inhibited in the circ-BlRC6 overexpression group relative to control cells iJl the flow cytometry analysis. (d, e) Migration and invasion of HepG2 and Bel7402 cells lransfected witb oeCirc or oeVec were evaluated with the transwell assay. Data are presented as mean± SD. *P < 0.05, **P < 0.0 I, ***P < 0.00 1.
Circ-BIRC6 directly targets miR-3918
circRNAs can act as an miRNA sponge in cancer [Citation15]. To investigate the mechanism by which circ-BIRC6 promotes HCC progression, we screened potential targets of circ-BIRC6 by in silico analysis and identified miR-3918 as a candidate molecule. FISH experiments showed that circ-BIRC6 and miR-3918 were primarily localized in the cytoplasm of HepG2 cells (), suggesting that the two interact. A putative miR-3918 binding site was identified in the circ-BIRC6 sequence by bioinformatics analysis (); miR-3918 mimic suppressed luciferase activity in 293T cells transfected with the WT circ-BIRC6 sequence, while mutation of the predicted circ-BIRC6 binding site abrogated this effect ().
Figure 4. circ-BJRC6 directly targets miR-3918. (a) Colocalization of circ-BlRC6 (red) and miR-3918 (green) in HepG2 cells was observed by FISH. Cell nuclei were counterstained with 4’,6-diamidino-2-pbenyl .indol e (blue). (b) Potential miR-3918 binding sites in circ-BJRC6. (c) Co-traostection with miR −3918 mim ic reduces the luciferase activity of circ-BLRC6 WT but not circ-BIRC6 Mut in 293T cells, as determined with the dual-luciferase reporter assay. (d) circ-BIRC6 and miR-3918 were enriched in Ago2 as compared to control [gG] immunoprecipitates. (e) circ-BIRC6 IOlockdown promotes miR-3918 expression in HepG2 and Bel7402 cells, as determined by qRT-PCR. (f) mi.R-3918 was downregulated in HCC as compared to adjacent norma l tissue. (g) Con·elation between circ-BIRC6 and miR −3918 levels in HCC tissue (n = 55). Data are presented as mean ± SO. *P < 0.05, **P < 0.01, ***P < 0.001.
![Figure 4. circ-BJRC6 directly targets miR-3918. (a) Colocalization of circ-BlRC6 (red) and miR-3918 (green) in HepG2 cells was observed by FISH. Cell nuclei were counterstained with 4’,6-diamidino-2-pbenyl .indol e (blue). (b) Potential miR-3918 binding sites in circ-BJRC6. (c) Co-traostection with miR −3918 mim ic reduces the luciferase activity of circ-BLRC6 WT but not circ-BIRC6 Mut in 293T cells, as determined with the dual-luciferase reporter assay. (d) circ-BIRC6 and miR-3918 were enriched in Ago2 as compared to control [gG] immunoprecipitates. (e) circ-BIRC6 IOlockdown promotes miR-3918 expression in HepG2 and Bel7402 cells, as determined by qRT-PCR. (f) mi.R-3918 was downregulated in HCC as compared to adjacent norma l tissue. (g) Con·elation between circ-BIRC6 and miR −3918 levels in HCC tissue (n = 55). Data are presented as mean ± SO. *P < 0.05, **P < 0.01, ***P < 0.001.](/cms/asset/677c76dd-fb96-4d1f-8f07-8a800dc84785/kccy_a_1601477_f0004_oc.jpg)
miRNAs suppress gene expression by forming the RNA-induced silencing complex (RISC) that includes Argonaute (Ago)2. We investigated whether circ-BIRC6 and miR-3918 are present in the same RISC complex with the RIP assay and found that both molecules were enriched in the complex immunoprecipitated using the anti-Ago2 antibody as compared to control IgG (). Furthermore, circ-BIRC6 knockdown enhanced miR-3918 expression in HepG2 and Bel7402 cells, as determined by qRT-PCR (). These results imply that circ-BIRC6 and miR-3918 directly interact.
We then analyzed miR-3918 expression in HCC and normal tissues and found that it was reduced in the former relative to the latter (). Moreover, miR-3918 level was inversely correlated with that of circ-BIRC6 in HCC tissue specimens (). We also analyzed the correlation between miR-3918 expression and clinicopathological features of HCC and found that it was associated with tumor stage (P = 0.0306), although it was unrelated to other clinical features (Supplementary Table 2).
miR-3918 plays an anti-tumorigenic role in HCC
To confirm the role of miR-3918 in HCC, HepG2 and Bel7402 cells were transfected with miR-3918 inhibitor or the negative control. The efficiency of miR-3918 silencing was confirmed by qRT-PCR (Supplementary Figure 1A). miR-3918 inhibition enhanced cell proliferation (Supplementary Figure 1B) as well as migration and invasion (Supplementary Figure 1D, E) while inhibiting apoptosis (Supplementary Figure 1C), which is similar to the effects of circ-BIRC6 overexpression.
To confirm the role of miR-3918 in HCC, HepG2 and Bel7402 cells were transfected with miR-3918 mimic (Supplementary Figure 2A). The results of the CCK-8 assay, flow cytometry analysis, and transwell assay showed that miR-3918 overexpression suppressed cell proliferation (Supplementary Figure 2B) and cell migration and invasion (Supplementary Figure 2D, E) while promoting apoptosis (Supplementary Figure 2C). These results indicate that miR-3918 and circ-BIRC6 have opposing functions in HCC.
circ-BIRC6 promotes Bcl2 expression by sponging miR-3918
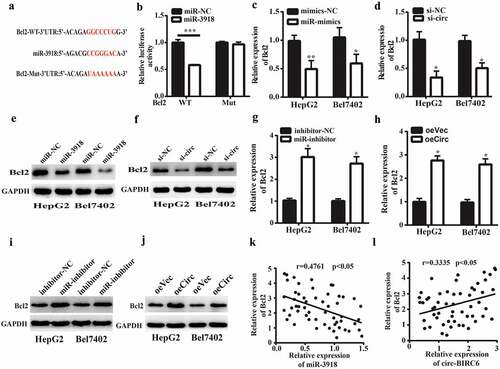
miRNAs regulate gene expression by targeting the 3‘ UTR of mRNAs. We identified Bcl2 as a potential target of miR-3918 by bioinformatics analysis based on the presence of a putative miR-3918 binding site in the 3‘ UTR of the Bcl2 transcript (). miR-3918 overexpression reduced the luciferase activity of the WT Bcl2 3‘ UTR in 293T cells but had no effect when the predicted binding site was mutated (). Furthermore, miR-3918 overexpression and circ-BIRC6 silencing reduced Bcl2 mRNA () and protein () levels in HepG2 and Bel-7402 cells, as determined by qRT-PCR and western blotting, respectively. Conversely, miR-3918 inhibition combined with circ-BIRC6 overexpression had the opposite effects (). Furthermore, Bcl2 mRNA expression was negatively correlated with that of miR-3918 and positively correlated with that of circ-BIRC6 in HCC tissues (). These data suggest that circ-BIRC6 promotes Bcl2 expression by sponging miR-3918.
Figure 5. Circ-BIRC6 promotes Bcl2 expreSSIOn by spongmg miR-3918. (a) miR-3918 binding sites m the 3’ UTR of Bcl2 transcript. (b) Co-transfection with roiR-391 8 mimic decreases the luciferase activity of WT 3’ UTR ofBcl2 transcript in 293T cells, as determined with the dual-luciferase reporter assay. (c-f) miR-3918 overexpression and circ-BIRC6 knockdown inhibit Bcl2 mRNA (c, d) and protein (e, f) levels in HepG2 and Bel −7402 cells, as determined by qRT-PCR and western blotting, respectively.(g-j) miR-3918 inhibitor and circ-BTRC6 overexpression produced the opposite etfect on Bcl2 mRNA (g, h) and protein (i, j) levels. (k, l) Negative correlation between miR-3918 and Bcl2 levels (k) and positive con·elation between Bcl2 and circ-BrRC6 levels (l) in HCC tissues (n = 55). Data are present ed as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
miR-3918 inhibition reverses the effect of circ-BIRC6 silencing
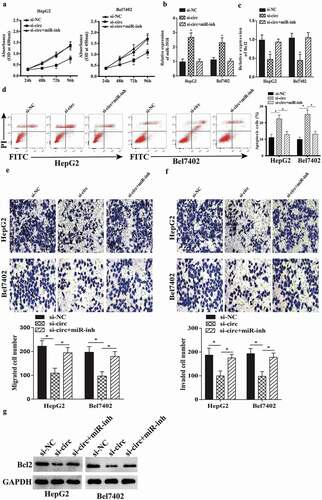
To verify whether circ-BIRC6 promotes HCC progression via regulation of miR-3918 and Bcl2, miR-3918 expression was suppressed in HCC cells transfected with an siRNA against circ-BIRC6. miR-3918 inhibition increased cell proliferation in circ-BIRC6-depleted HepG2 and Bel7402 cells (). miR-3918 inhibition decreased the expression of miR-3918 (); this was accompanied by an upregulation on mRNA and protein level of Bcl2 (). Moreover, miR-3918 inhibition abolished the effects of circ-BIRC6 knockdown on cell migration, invasion, and apoptosis (). These data demonstrate that circ-BIRC6 promotes the malignant progression of HCC by modulating the miR-3918/Bcl2 axis.
Figure 6. miR-3918 inhibition reverses the effect of circ-BlRC6 depletion. (a) Cell proliferation was evaluated with the CCK-8 assay. (b, c) miR-3918 and Bcl2 expression in HepG2 and Bel7402 cells, as determined by qRT-PCR. (d) Detection of apoptosis by flow cytometry. (e, f) Mig1·ation and invasion of HepG2 and Bel7402 cells, as determined with the transwell assay. (g) The protein expression of Bcl2 in HepG2 and Bel7402 cells, as determined by western blotting. Data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
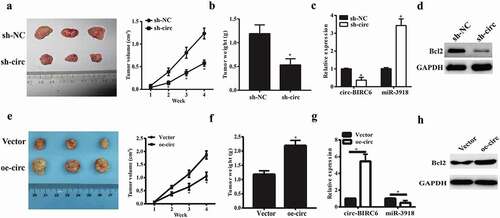
circ-BIRC6 promotes tumor growth in vivo
To confirm the oncogenic role of circ-BIRC6 in vivo, we established a mouse xenograft model of HCC by subcutaneous inoculation of Bel7402 cells stably transfected with shRNA against circ-BIRC6, negative control shRNA, circ-BIRC6 overexpressing vector and empty vector. circ-BIRC6 knockdown inhibited tumor growth in mice (), as evidenced by the lower tumor weights at the endpoint of the experiment (). Besides, circ-BIRC6 expression was downregulated in sh-Circ-BIRC6-derived tumor tissues while miR-3918 was upregulated (). Furthermore, circ-BIRC6 deletion decreased the protein expression of Bcl2 (). However, the stable overexpression of circ-BIRC6 promoted tumor growth in mice (). Moreover, circ-BIRC6 expression was increased in oe-circ-BIRC6-derived tumor tissues while miR-3918 was decreased (). The protein expression of Bcl2 was also increased ().These results provide evidence that circ-BIRC6 promotes HCC progression.
Figure 7. Effect ofcirc-BIRC6 knockdown and circ-BIRC6 overexpressionon tumor growth in vivo. Xenografts formed by subcutaneous injection of Bel7402 cells stably transfected with sh-NC (negative control shRNA), sh-circ(sh-Circ-BfRC6), Vector(empty vector), oe-circ(overexpressed circ-BTRC6 vectory into the flanks of nude mice.(a.e) Tumor growth curve based on measured tumor volumes. (b.f) Tumor weight at the endpoint of the experiment. (c.g) Relatwe expression of circ-BIRC6 and miR-3918 in tumors derived l1·om Bel7402 cellstransfected with sh -eire, sh-NC, vector and oe-circ respectively. (d.h) Protein level of Bcl2 was examined in the resected tumors by western blot. Data are presented as mean± SD. *P < 0.05, **P < 0.0 1, ***P < 0.001.
Discussion
circRNAs have been implicated in a variety of biological processes [Citation16] and human cancers [Citation17]; they have been shown to play an important role in cancer initiation and progression and may function as molecular markers for cancer diagnosis and treatment [Citation18]. For instance, the circRNA protein arginine methyltransferase 5 promotes metastasis of urothelial carcinoma of the bladder by sponging miR-30c to induce epithelial-to-mesenchymal transition [Citation19], while RNA hsa_circ_0000977 silencing suppressed pancreatic ductal adenocarcinoma progression by stimulating miR-874-3p and inhibiting polo-like kinase 1 expression [Citation20]. Downregulation of hsa_circ_0011946 suppressed the migration and invasion of MCF-7 breast cancer cells by targeting replication factor C subunit 3 [Citation21]. In HCC, circMTO1 acts as a sponge of the miRNA miR-9 to suppress HCC progression [Citation22]. Our study identified a novel circRNA – circ-BIRC6 – that plays an important role in HCC progression.
HCC is a common and aggressive cancer that is linked to a large number of cancer-related deaths worldwide [Citation2]. Despite therapeutic advances, the prognosis of patients remains poor. In our study, we found that circ-BIRC6 is upregulated in HCC tissue and cell lines. Importantly, this was associated with the prognosis of HCC patients, with a multivariate Cox analysis revealing that circ-BIRC6 was an independent factor predicting poor outcome. circ-BIRC6 silencing inhibited the proliferation, migration, and invasion of HCC cells while promoting their apoptosis; circ-BIRC6 overexpression had the opposite effects. circRNAs function as miRNA sponges that modulate mRNA stability and regulate gene transcription or protein translation [Citation12]. Previous studies showed that miRNAs plays a critical role in the development and progression of gastric [Citation23], lung [Citation24], colorectal [Citation25], and cervical [Citation26] cancers. We investigated whether circ-BIRC6 acts as a sponge of miR-3918, which was identified as a potential target of circ-BIRC6. We found that circ-BIRC6 and miR-3918 are enriched in the cytoplasm of HCC cells by FISH analysis, suggesting a functional interaction. The luciferase activity reporter and RIP assays confirmed the direct binding between circ-BIRC6 and miR-3918. circ-BIRC6 silencing also increased miR-3918 level, as determined by qRT-PCR.
Although its precise function is unknown, miR-3918 was downregulated in HCC tissue and there was an inverse correlation between circ-BIRC6 and miR-3918 expression: when miR-3918 was overexpressed, HCC cell proliferation, migration, and invasion were suppressed and apoptosis was enhanced, whereas miR-3918 inhibition had the opposite effects, suggesting that miR-3918 has a tumor-suppressive role. We identified Bcl2 as a potential target of miR-3918, and confirmed their interaction with the luciferase reporter assay. circ-BIRC6 knockdown and miR-3918 overexpression reduced Bcl2 mRNA and protein levels, and miR-3918 inhibition reversed the decreases in HCC cell proliferation, migration, and invasion and increase in apoptosis induced by circ-BIRC6 silencing. Thus, circ-BIRC6 promotes HCC progression via a regulatory axis that includes miR-3918 and Bcl2.
In conclusion, our study demonstrates for the first time that circ-BIRC6 is upregulated in HCC tissues and is associated with poor prognosis of HCC patients, and promotes HCC progression by targeting the miR-3918/Bcl2 axis. Based on these findings, circ-BIRC6 may be a useful prognostic biomarker in HCC and a promising therapeutic target for disease treatment.
Ethical approval
This study was approved by the Ethics Committee on Research Involving Human Subjects of Faculty of Medicine, Qianfoshan Hospital, Shandong University.
Informed consent
Informed consent was obtained from all participants in the study.
Supplemental Material
Download Zip (9.2 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplementary data for this article can be accessed here.
Additional information
Funding
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386.
- Vardy J, Rourke S, Tannock IF. Evaluation of cognitive function associated with chemotherapy: a review of published studies and recommendations for future research. J Clin Oncol. 2007;25(17):2455–2463.
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338.
- Lasda E, Parker R. Circular RNAs: diversity of form and function. Rna. 2014;20(12):1829–1842.
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984.
- Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6(5):563–579.
- Chen Y, Li C, Tan C, et al. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53(6):359–365.
- Yang F, Liu DY, Guo JT, et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23(47):8345–8354.
- Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17(1):19.
- Ma HB, Yao YN, Yu JJ, et al. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am J Transl Res. 2018;10(2):592–604.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
- Chen B, Huang S. Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41–50.
- Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. Rna. 2014;20(11):1666–1670.
- Cheng J, Zhuo H, Xu M, et al. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med. 2018;16(1):216.
- Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–111.
- He J, Xie Q, Xu H, et al. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144.
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
- Chen X, Chen R, Wei WS, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24(24):6319–6330.
- Huang WJ, Wang Y, Liu S, et al. Silencing circular RNA hsa_circ_0000977 suppresses pancreatic ductal adenocarcinoma progression by stimulating miR-874-3p and inhibiting PLK1 expression. Cancer Lett. 2018;422:70–80.
- Zhou J, Zhang WW, Peng F, et al. Downregulation of hsa_circ_0011946 suppresses the migration and invasion of the breast cancer cell line MCF-7 by targeting RFC3. Cancer Manag Res. 2018;10:535–544.
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164.
- Yang L, Liang H, Wang Y, et al. MiRNA-203 suppresses tumor cell proliferation, migration and invasion by targeting Slug in gastric cancer. Protein Cell. 2016;7(5):383–387.
- Ma N, Zhang W, Qiao C, et al. The tumor suppressive role of miRNA-509-5p by targeting FOXM1 in non-small cell lung cancer. Cell Physiol Biochem. 2016;38(4):1435–1446.
- Baltruskeviciene E, Schveigert D, Stankevicius V, et al. Down-regulation of miRNA-148a and miRNA-625-3p in colorectal cancer is associated with tumor budding. BMC Cancer. 2017;17(1):607.
- Xia YF, Pei GH, Wang N, et al. miR-3156-3p is downregulated in HPV-positive cervical cancer and performs as a tumor-suppressive miRNA. Virol J. 2017;14(1):20.
