ABSTRACT
Glioblastoma (GBM) is a lethal, fast-growing brain cancer, affecting 2–3 per 100,000 adults per year. It arises from multipotent neural stem cells which have reduced their ability to divide asymmetrically and hence divide symmetrically, generating increasing number of cancer stem cells, fostering tumor growth.
We have previously demonstrated that the architectural transcription factor HMGA1 is highly expressed in brain tumor stem cells (BTSCs) and that its silencing increases stem cell quiescence, reduces self-renewal and sphere-forming efficiency in serial passages, suggesting a shift from symmetric to asymmetric division. Since NUMB expression is fundamental for the fulfillment of asymmetric division in stem cells, and is lost or reduced in many tumors, including GBM, we have investigated the ability of HMGA1 to regulate NUMB expression. Here, we show that HMGA1 negatively regulates NUMB expression at transcriptional level, by binding its promoter and counteracting c/EBP-β and at posttranscriptional level, by regulating the expression of MSI1 and of miR-146a. Finally, we report that HMGA1 knockdown-induced NUMB upregulation leads to the downregulation of the NOTCH1 pathway.
Therefore, the data reported here indicate that HMGA1 negatively regulates NUMB expression in BTSCs, further supporting HMGA1 targeting as innovative and effective anti-cancer therapy.
Introduction
The ability of stem cells to maintain lifelong tissue integrity is based on the peculiar capacity to divide asymmetrically, generating one daughter stem cell and one differentiating daughter cell. This process depends on the unequal distribution of key molecular cues at the two poles of the stem cell, before its division. Neural stem cells (NSCs) divide asymmetrically to generate both neural and glial cells. The inability to carry out this odd distribution leads to symmetric division, converting normal stem cells (SCs) into cancer stem cells (CSCs) [reviewed in [Citation1]]. CSCs have lost or reduced their ability to divide asymmetrically and mostly divide symmetrically [reviewed in [Citation2]]. Progenitor cells which have lost the ability to divide asymmetrically have been detected in brain tumors [Citation3]. Conversely, the recovery of asymmetric division is onco-suppressive [reviewed in [Citation1] and [Citation2]]. Therefore, the comprehension of the molecular mechanisms regulating the balance between symmetric and asymmetric division can allow the identification of innovative intervention strategies to treat cancer and brain tumors. Among these, glioblastoma (GBM) is the most aggressive one, characterized by rapid course and unresponsiveness to therapy [Citation4].
HMGA1a and HMGA1b are members of the HMGA family of architectural transcription factors. They bind the minor groove of AT-rich DNA sequences and hence modify chromatin structure and accessibility to transcription factors [reviewed in [Citation5]]. HMGA1 proteins are overexpressed in human malignancies, including GBMs [Citation6], where their presence correlates with cancer recurrence and poor patient outcome [reviewed in [Citation5]]. Several studies have demonstrated their causal role in cell transformation and cancer progression [Citation7–Citation9], through the regulation of crucial oncogenetic pathways, such as the induction of AP-1 [Citation10], the activation of E2F [Citation11], the inhibition of p53 [Citation12], the induction of chromosome instability [Citation13] and EMT [Citation14], impairment of DNA repair [Citation15], and others [reviewed in [Citation5]].
We have previously shown that HMGA1 is enriched in the CD133+ population of cancer stem cells [Citation12] and in brain tumor stem cells (BTSCs) [Citation6]. In these cells, the silencing of HMGA1 expression reduces the CD133+/CD15+ stem cell population [Citation6], suggesting a central role in cancer stem cells. Moreover, we have also demonstrated that HMGA1 silencing increases stem cell quiescence whilst reduces self-renewal and sphere–forming efficiency [Citation6], indicating a shift from symmetric to asymmetric division.
NUMB is a critical player in determining stem cell division mode. Indeed, NUMB is a conserved, multi-task endocytic protein, which promotes asymmetric division and determines cell fate in stem and precursor cells in organisms as distant as worms, flies and mammals [reviewed in [Citation16]]. To accomplish this function, NUMB is asymmetrically partitioned before mitosis and asymmetrically inherited by one daughter cell [reviewed in [Citation16]]. In cortical stem cells and neuroblasts, Numb knockout reduces the number of asymmetric divisions compared to wild type counterparts [Citation17]. Interestingly, in humans, loss of NUMB expression is associated with low differentiation status and poor prognosis in several types of cancer [Citation18–Citation20], including GBM [Citation21]. Besides its role as cell fate determinant, its tumor suppressor activity has been ascribed also to its ability to affect Epithelial-Mesenchymal Transition (EMT), through the regulation of recycling and localization of E-cadherin [Citation22]. Consistently, NUMB knockdown causes a decrease in cell-cell adhesion and an increase in cell motility [Citation23], whereas its overexpression inhibits proliferation, promotes apoptosis and enhances sensitivity to cisplatin of mesothelioma cells [Citation20]. Therefore, the identification of signaling pathways regulating NUMB expression is of central importance in cancer biology and therapy. The mechanisms regulating NUMB expression have not been completely unveiled. In fact, to date, the only identified transcriptional regulator is β-catenin [Citation24]. Conversely, the mRNA binding protein MSI1 and miR-146a are involved in NUMB posttranscriptional regulation [[Citation25] and [Citation26], respectively]. Noteworthy, NUMB exerts its major effect in brain tumorigenesis by targeting NOTCH1 [Citation17,Citation27], which, in turn, promotes brain tumor growth and glioma stem cell proliferation [Citation28].Here, we report that HMGA1 directly modulates NUMB expression at transcriptional level, binding the NUMB promoter, and indirectly, at posttranscriptional level, regulating MSI1 and miR-146a. Accordingly, NUMB upregulation mediates the reduction of NOTCH1 activity in HMGA1-knockdown BTSCs. Taken together, our findings provide evidence of a new mechanism regulating NUMB expression and suggest that HMGA1 targeting may represent a new therapeutic strategy.
Materials and methods
Cell lines and culture conditions
BTSC#83C1, BTSC#83shA1, BTSC#30pC1 and BTSC#30shA1, as well as their culture conditions, have been previously described [Citation6].
HEK293 cells were maintained in DMEM medium containing 10% fetal calf serum (Invitrogen).
HeLa cells were grown in RPMI medium plus 10% fetal bovine serum.
Plasmids
The hairpin RNA interference plasmid for human HMGA1 (pLKO.1-HMGA1, TRCN0000018949) and the scramble control pLKO.1-Puro plasmid (SHC002) were obtained from Sigma-Aldrich and have been already described [Citation12]. The 2 BTSC lines were also transfected with the HMGA1 antisense pRC/CMVanti-A1 construct, carrying the HMGA1 cDNA in antisense orientation [Citation7].
The pCDNA3.1-HMGA1 and the c/EBP-β expression vectors were previously described [[Citation29] and [Citation30], respectively].
The NUMB promoter luciferase reporter construct, containing 875 bp of the human NUMB promoter, and the corresponding empty vector (LightSwitch Promoter Reporter GoClone), were purchased from SwitchGear Genomics.
The shRNA constructs for NUMB, as well as the corresponding scramble vector, were purchased from Sigma Aldrich.
The human MSI1-HA expressing construct, carrying G418 resistance, and the corresponding empty vector (EGFP-M07) were purchased from Genecopoeioa.
Transfections
Transfections of BTSC#83 and BTSC#30p have been already described [Citation6]. For transfections of HMGA1-KD BTSCs with the shRNA for NUMB constructs, BTSC#83antiA1 cells (carrying resistance to G418) were mechanically disaggregated and 1 × 106 cells were electroporated with 30 μg of shRNA construct or the corresponding scramble vector, using the Neon® Transfection System (Invitrogen) (1400 V, 20 msec; 1 pulse). After 48 h, cells were selected with G418 (1.2 μg/μl) and puromycin (1 μg/μl). For transfection of HMGA1-KD BTSCs with the MSI1-expressing construct or the corresponding empty vector, BTSC#83shA1 cells (carrying resistance to puromycin) were transfected as described above and selected with G418 and puromycin.
Stable transfections in HeLa cells were performed using Lipofectamine 2000 (Invitrogen), following manufacture’s procedures. After 24 h from transfection, cells were selected with puromycin (1 μg/ml) and single clones were picked and grown separately.
miR-146a transfections
miR-146a miRNA mimic was purchased from Ambion. For transfection, 6 × 105 cells were transfected with 60 nM miR-146a or negative control (NEG1; Ambion), by using SIPORT transfection reagent (Ambion), following manufacture’s instructions. Cells were collected after 48 h and protein was extracted.
Luciferase assays
For luciferase assays, 2 × 105 HEK293 cells were transiently transfected with 100 ng of luciferase reporter construct or the corresponding empty vector, 800 ng of HMGA1-expressing construct or the corresponding empty vector, and 700 ng of c/EBP-β or the corresponding empty vector. In total, 10 ng of β-gal expressing construct were co-transfected as internal control.
Luciferase activity was measured using the “LightSwitch Luciferase Assay Reagent” (Switchgear Genomics) and normalized for β-gal activity.
Protein extraction and western blot
Single cell suspensions were obtained by mechanical disaggregation of the spheres and total proteins were extracted 48 h later, as already described [Citation7]. After separation by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis, proteins were blotted on nitrocellulose membranes (GE Healthcare Europe Gmb) and hybridized with the following antibodies: anti-NUMB (kindly donated by Prof. Pece), anti-HMGA1 [already described in [Citation6]], anti-NOTCH1 (Abcam), anti-α-actin, anti-c/EBP-β, anti-vinculin, anti-MSI1, anti-HA, anti-HES1, anti-SOX2, anti-HEY1 and anti-GAPDH (SantaCruz Biotechnologies). Densitometric analyses of Western blot were performed using the ImageJ Software.
RNA extraction and qRT-PCR analyses
Single cell suspensions were obtained by mechanically disaggregating the spheres and RNA was extracted 48 h later, as described [Citation6]. qRT-PCR analyses for mRNAs and miRNAs were performed as described [[Citation12] and [Citation6], respectively]. Primers for NUMB (recognizing all the 4 NUMB isoforms) and for all the other genes are listed in Supplemental Table 1.
The 2–ΔΔCt formula was used to calculate the differential gene expression.
Chromatin immunoprecipitation
Chromatin Immunoprecipitation (ChIP) on BTSCs and HeLa cells was performed as described [Citation12]. Primers for each region of the NUMB promoter are listed in Supplemental Table 2. Primers for MSI1 and miR-146a promoters are listed in Supplemental Table 3. ChIP data are reported as mean values ± SD.
Statistical analyses
Data were analyzed by one of the following tests (as indicated in the respective figure legends): Student’s t-test, ANOVA, Kruskal–Wallis test followed by Conover post hoc test, using the statistical software GraphPad Prism v.6.0. ChIP data were tested for normal distribution using D’Agostino & Pearson test for all variables.
Results
HMGA1 silencing induces NUMB upregulation in BTSCs
We previously generated two HMGA1-knockdown (HMGA1-KD) BTSC lines by using short-hairpin RNA constructs and demonstrated that HMGA1 knockdown inhibits the Sphere Forming Efficiency (SFE) increase in serial passages [Citation6], favors BTSCs differentiation [Citation6] and increases the asymmetric localization of NUMB [Citation12]. These data led us to speculate that HMGA1-silencing may elicit asymmetric division in CSCs.
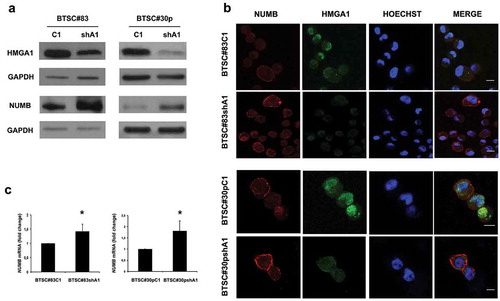
Given the role of NUMB in the establishment of the division mode, as well as in human glioblastoma stem cell (GSC) tumor sphere formation and in regulating the growth and differentiation of primary human GSCs in vitro and in vivo [Citation21], we evaluated NUMB expression in the two HMGA1-KD BTSCs (BTSC#83 and BTSC#30p). As shown in , HMGA1 knockdown increases NUMB expression at protein level (Panel A) in both the BTSC cell lines with respect to the scramble-transfected cells. This reverse correlation between HMGA1 and NUMB expression was confirmed by immunofluorescence () and qRT-PCR (). A similar NUMB upregulation was observed also when HMGA1 was stably silenced by using an antisense construct (not shown) (BTSC#83antiA1 and BTSC#30pantiA1), compared to the empty vector-transfected cells (BTSC#83CMV and BTSC#30pCMV), indicating that this effect is specifically due to HMGA1 downregulation.
Figure 1. HMGA1 knockdown increases NUMB expression. (A) Western blot analyses for HMGA1 (upper panel) and NUMB (lower panel) in scramble (C1) and HMGA1-knockdown (shA1) BTSC#83 (left) and BTSC#30p (right). GAPDH was used as loading control. (B) Immunofluorescence analyses for HMGA1 and NUMB in control and HMGA1-KD BTSC#83 and BTSC#30p cells. Scale bar = 10 μm. C) qRT-PCR for NUMB expression in control (C1) and HMGA1-silenced (shA1) BTSC#83 (left panel) and #30p cells (right panel). Fold changes are shown relative to scramble-transfected cells (C1), with respect to the G6PD expression in each sample. Data are the mean value of 4 independent experiments performed in duplicate or triplicate (*p ≤ 0.05, Student’s t-test).
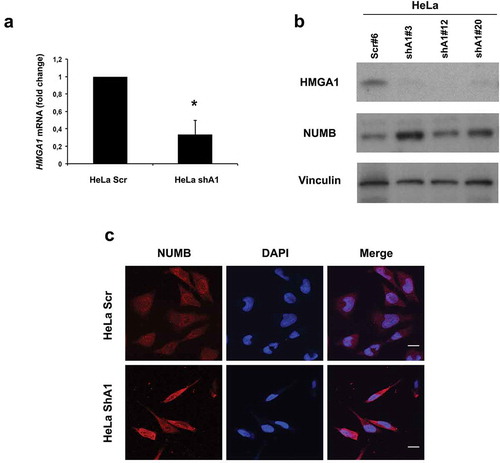
Interestingly, three different clones of HeLa cells, stably interfered for HMGA1 (), expressed higher levels of NUMB with respect to the scramble-transfected cells, as demonstrated by Western blot and immunofluorescence ( and C, respectively), indicating that HMGA1 reduction correlates with increased NUMB expression, in both stem and non-stem cells.
Figure 2. HMGA1-KD HeLa cells overexpress NUMB. (A) qRT-PCR for HMGA1 performed in scramble- (HeLa scr) and HMGA1 shRNA-transfected HeLa cells (HeLA shA1). Fold changes are shown as mean value ± SD of three different clones, relative to scramble-transfected cells, with respect to actin expression. Representative experiment (*p ≤ 0.05, Student’s t-test). (B) Western blot for HMGA1 and NUMB in one HeLa scramble clone and three different shA1 clones. Vinculin expression was used as loading control. (C) Representative images of immunofluorescence staining for NUMB in HeLa scr and shA1 clones. Nuclei were stained with DAPI. Magnification: 63X.
HMGA1 regulates NUMB at transcriptional level
The increase in NUMB-specific mRNA in both HMGA1-KD BTSC cell lines suggests that HMGA1 controls NUMB expression, at least in part, by transcriptional regulation. To verify the direct regulation of HMGA1 on NUMB promoter, we performed Chromatin Immunoprecipitation (ChIP) assay on chromatin from BTSC#83C1 and BTSC#83shA1 cells, using primers spanning 6 different regions of the NUMB promoter, indicated as regions I-VI (Supplemental Figure S1). Chromatin from control cells revealed HMGA1 occupancy in the NUMB promoter regions I, II and III (), whereas no amplification was observed in the remaining regions (not shown), indicating the specificity of HMGA1 binding to these regions. Lower enrichment was detected in regions I–III with chromatin from BTSC#83shA1 cells () and no enrichment was observed with chromatin immunoprecipitated with IgG. Similarly, ChIP performed on HeLa cells demonstrated the ability of HMGA1 to bind the same NUMB promoter regions (Supplemental Figure S2), indicating that HMGA1 protein binds specific regions of the NUMB promoter in vivo.
Figure 3. HMGA1 regulates NUMB expression at transcriptional level. (A) ChIP assay, revealed by qPCR, detecting the in vivo binding of HMGA1 to the sub-regions I–III in the NUMB promoter in BTSC#83C1 and shA1 chromatin extracts. The relative fold enrichment of the NUMB promoter regions by HMGA1 is indicated as vertical bars. The 2–ΔΔCt formula was used to calculate the relative fold enrichment, normalized to its IgG reference and relative to the calibrator (input). (***p ≤ 0.001, ANOVA test). (B) Luciferase activity of the NUMB promoter (NUMB luc) in HEK293 cells in presence of the HMGA1-antisense (antiA1), the HMGA1-expressing (HMGA1) vectors or the corresponding empty vectors. NUMB luciferase activity is shown as fold induction with respect to the NUMB promoter activity. Data are the mean value of four independent experiments ± SD (* p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001, Kruskal–Wallis test, Conover post hoc test).
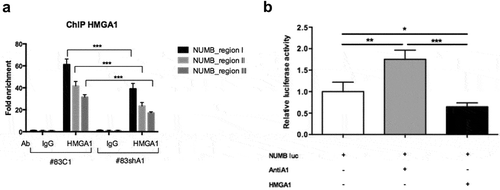
The functional significance of this binding was examined by luciferase assays, performed in HEK293 cells transfected with a NUMB promoter-reporter construct, in presence of a vector expressing the HMGA1 cDNA, either in sense or antisense orientation (see Materials and Methods). As shown in , HMGA1 overexpression reduces luciferase activity, in comparison with the backbone-transfected cells (), whereas the block of HMGA1 expression by an HMGA1 antisense construct (AntiA1) increased the activity of the NUMB promoter ().
Therefore, these results indicate that HMGA1 is able to negatively control NUMB expression at transcriptional level.
In order to identify potential co-regulators of HMGA1 on NUMB promoter, we interrogated the PROMO database, a virtual laboratory for the identification of putative transcription factor binding sites [Citation31]. This search identified 60 high-scoring sites for transcription factors such as c-Jun, E2F, GATA-1, GATA-2, GATA-3, p53, Sp1, p300. Among these factors, c/EBP-β was the most represented binding site (40 binding sites) (Supplemental Figure S1). Moreover, in regions I-III, the database detected 17 c/EBP-β binding sites (Supplemental Figure S1), which were close to each other and in proximity of HMGA1 binding sites in regions I and II. Noteworthy, c/EBP-β is a member of the c/EBP family of transcription factors, regulating several genes involved in proliferation, differentiation and stem cell fate in a variety of tissues [Citation32,Citation33].
To investigate the regulation of NUMB by c/EBP-β and HMGA1, we performed luciferase assays in HEK293 cells by transfecting the NUMB promoter-reporter construct, in the presence of HMGA1 and c/EBP-β expression vectors or their respective empty vectors. As shown in , c/EBP-β increased the luciferase activity of the NUMB promoter, whereas HMGA1 induced its reduction, and transfection with both HMGA1 and c/EBP-β constructs led to an intermediate NUMB promoter activity (). c/EBP-β and HMGA1 overexpression in the transfected cells was confirmed by Western blot (Supplemental Figure S3). This Western Blot also demonstrates that HEK293 cells are almost devoid of both HMGA1 and c/EBP-β; however, c/EBP-β activates, and HMGA1 represses, the NUMB-luc promoter, independently from each other ().
Figure 4. c/EBP-β coregulates the NUMB promoter together with HMGA1. (A) Histogram showing the relative luciferase activity of the NUMB promoter reporter construct in HEK293 cells in the presence of HMGA1 and c/EBP-β-expressing constructs. Luciferase activity was normalized by β-galactosidase activity. Data are the mean value of four independent experiments ± SD (* p ≤ 0.05; Student’s t-test). (B) ChIP assay for c/EBP-β on NUMB promoter regions I–III, using immunoprecipitated chromatin from BTSC#83C1 and BTSC#83shA1 cells. The 2–ΔΔCt formula was used to calculate the relative fold enrichment, normalized to its IgG reference and relative to the calibrator (input). (***p ≤ 0.001, ANOVA test).
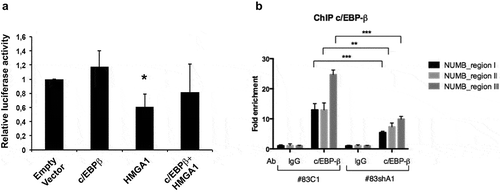
In accordance with the functional data, ChIP analyses demonstrated the occupancy of the regions I-III of NUMB promoter by c/EBP-β in BTSCs (). The reduced binding of c/EBP-β in HMGA1-silenced cells suggests that HMGA1 affects the ability of c/EBP-β to bind the DNA. This result is in agreement with previously reported data demonstrating that HMGA1 favors the binding of C/EBP-β on the insulin receptor (IR) promoter [Citation34]. Re-ChIP analyses, performed by sequentially immunoprecipitating for c/EBP-β and HMGA1, confirmed that the two proteins interact on the NUMB promoter in vivo (Supplemental Figure S4).
All together, these results demonstrate that HMGA1 and c/EBP-β bind cooperatively the NUMB promoter but have opposite effects on its regulation.
HMGA1 regulates NUMB at posttranscriptional level, by acting on MSI1 and miR-146a
The increased NUMB expression in shA1 cells is more evident at protein than at mRNA level, suggesting that HMGA1 might affect NUMB expression also at posttranscriptional level. NUMB mRNA translation is regulated by the mRNA-binding protein Musashi-1 (MSI1) [Citation25] and by the micro-RNA miR-146a [Citation26]. Therefore, we evaluated whether these two regulators could mediate the effect of HMGA1 on NUMB protein expression. As shown in , knockdown of HMGA1 induces a significant reduction of both MSI1 (panel A) and miR-146a (panel B) compared to control cells, seemingly contributing to the increase in NUMB protein levels. Noticeably, the introduction of a MSI1-expressing construct or of a miR-146a mimic in HMGA1-KD BTSCs, reduces NUMB expression levels (), indicating that the upregulation of NUMB expression in HMGA1-KD cells is mediated, at least in part, by these two posttranscriptional regulators. ChIP analyses demonstrate that HMGA1 is able to bind the upstream promoter region of MSI1 and miR-146a in BTSCs and that this binding is reduced upon HMGA1 knockdown ().
Figure 5. HMGA1 silencing affects NUMB expression at posttranscriptional level by regulating MSI1 and miR-146a. (A) qRT-PCR (left) and Western blot (right) evaluating the expression of MSI1 in scramble and HMGA1-KD BTSC#83 and BTSC#30p. Fold change was calculated using the 2–ΔΔCt formula, considering the scramble transfected cells as control. Data are the mean value ± SD of 4 independent experiments performed in duplicate or triplicate. (** p ≤ 0.01; *** p ≤ 0.001, Student’s t-test). (B) Histogram showing the qRT-PCR results for miR-146a expression in scramble and HMGA1-KD BTSC#83 and BTSC#30p. Data are the mean value ± SD of 3 independent experiments performed in duplicate or triplicate. (* p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001, Student’s t-test). (C) Western blots for MSI-HA and NUMB in BTSC#83shA1 cells transfected with a MSI-HA-expressing construct (Msi-HA) or with the corresponding empty vector (EV). Vinculin was used as loading control. Quantification of relative NUMB expression compared to Vinculin expression is indicated. (D) Western blot for NUMB in BTSC#83shA1 cells transfected with an miR-146a mimic or with NEG1 scramble oligonucleotide. Vinculin was used as loading control. Quantification of the relative NUMB expression compared to Vinculin expression is indicated. (E) Histogram of ChIP assays showing the binding of HMGA1 to the MSI1 (left) and miR-146a (right) promoters in BTSC#83C1 and shA1 extracts. The 2–ΔΔCt formula was used to calculate the relative fold enrichment, normalized to its IgG reference and relative to the calibrator (input). (*** p ≤ 0.001, ANOVA test).
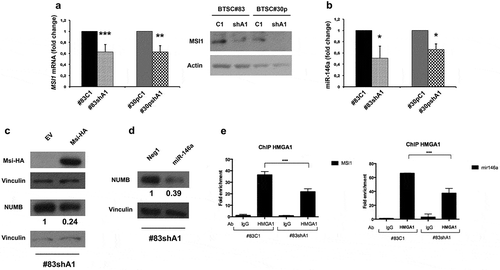
Therefore, these results demonstrate that HMGA1 regulates NUMB at transcriptional and posttranscriptional level.
HMGA1 knockdown induces NOTCH1 downregulation
It has been previously demonstrated that NUMB is able to downregulate NOTCH1 level and function by several mechanisms, such as increase of its ubiquitination, endocytic trafficking and degradation [Citation35,Citation36]. Therefore, to identify the pathway affected by HMGA1 downregulation, we analyzed NOTCH1 expression in the HMGA1-KD BTSCs. Western blot analyses display reduced expression of the 120 kDa intracellular domain (NICD) of NOTCH1 in HMGA1-KD cells of both BTSC lines with respect to control cells (), consistently with the ability of NUMB in participating in NOTCH1 degradation [Citation36].
Figure 6. NOTCH1 is downregulated in HMGA1-knockdown BTSCs. (A) Western blot for NOTCH1 and NOTCH1 targets (HES1, HEY1, SOX2) in extracts from control and HMGA1-KD BTSC#83 and BTSC#30p cells. (B) qRT-PCR analyses for HES1, HEY1 and SOX2 expression in control and HMGA1-KD BTSC#83 and BTSC#30p cells. Data are the mean value, ± SD of 2 independent experiments performed in duplicate or triplicate (*p ≤ 0.05, Student’s t-test). (C) qRT-PCR for NUMB and HES1, HEY1 and SOX2 in BTSC#83antiA1 cells transfected with NUMB shRNA (shNUMB) or with the scramble vector (Scr). Data are the mean value ± SD of 2–3 independent experiments performed in triplicate. (*** p ≤ 0.001, Student’s t-test).
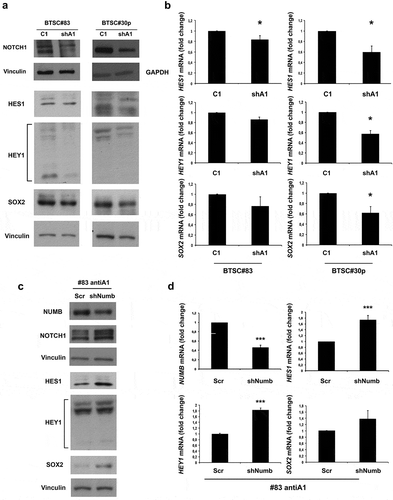
Accordingly, qRT-PCR and Western blot analyses display reduced expression of the NOTCH1 targets HES1, HEY1 and SOX2 in both HMGA1-KD BTSC cell lines ( and B, respectively), in comparison with control cells.
In order to investigate whether this effect is mediated by NUMB, we silenced NUMB expression in HMGA1-KD cells by using a shRNA construct ( and D). As shown in and D, the silencing of NUMB in HMGA1-knockdown BTSCs restores NOTCH1 expression and activity compared to control scramble-transfected cells (Scr), as demonstrated by the recovery of HES1, HEY1 and SOX2 expression.
These results indicate that HMGA1 silencing affects the NUMB/NOTCH1 pathway, and that NUMB mediates the effect of HMGA1 on NOTCH1.
Discussion
GBM arises from stem-like cells which have lost or reduced their ability to divide asymmetrically and are therefore characterized by increased symmetric divisions [Citation2]. We have previously shown that HMGA1 silencing reduces sphere-forming efficiency in cancer stem cells from colon [Citation12] and brain [Citation6] tumors, suggesting a role of HMGA1 in the regulation of CSCs division mode. Accordingly, the reduction of HMGA1 favors differentiation and increases p53 expression while reducing self-renewal, in vivo tumorigenicity and CD133+/CD15+ cancer stem cell population [Citation6,Citation12].
NUMB has a key role in the balance of stem cell symmetric and asymmetric division [reviewed in [Citation16]]. During Drosophila and mammalian cortical neurogenesis, NUMB is necessary for neural stem cell differentiation and its asymmetric segregation inhibits NOTCH activity in one daughter, to induce neuronal differentiation [Citation17], whereas its loss reduces the asymmetric divisions, compromising neurogenesis [Citation17]. In humans, loss of NUMB is associated to aggressive forms of cancer [Citation27], including GBM [Citation21].
Therefore, we evaluated the possible role of HMGA1 in regulating BTSC division mode, through the NUMB-NOTCH1 pathway. NUMB expression was increased in two lines of HMGA1-knockdown BTSCs at mRNA and protein level. Interestingly, the same results were obtained in HeLa cells interfered for HMGA1 expression, suggesting that the control of HMGA1 on NUMB expression is a general event. Accordingly, NUMB is expressed in many different cell types, where it is involved in basic “house-keeping” functions [Citation16] but it is asymmetrically segregated only in some kind of stem cells, skewing its function toward one of the daughter cells.
From a mechanistic point of view, we show that HMGA1 is able to bind to specific regions of NUMB promoter, directly repressing its expression. Interestingly, the negative activity exerted by HMGA1 is competed by c/EBP-β, which binds the same regions of HMGA1, as demonstrated by ChIP analyses. Consistently, previous studies have shown that, together with the other members of the cEBP family proteins, c/EBP-β has important roles in lineage commitment, cell growth, morphogenesis and differentiation [reviewed in [Citation33]]. Therefore, our results suggest that c/EBP-β affects cell fate, at least in part, by regulating NUMB expression. A cooperative binding of c/EBP-β and HMGA1 has been already described in the regulation of insulin receptor (IR) promoter [Citation34]. Our results demonstrate that HMGA1 and c/EBP-β cooperatively bind the NUMB promoter but have opposite effects on its regulation, at odd with the IR promoter, in which the cooperative binding corresponds to a cooperative activation [Citation34].
However, it is not clear how this interaction may influence the genesis of GBM in vivo, since it has been previously shown that decreased expression of c/EBP-β inhibits the growth and migration of GBM cells [Citation37]. Indeed, it is known that the regulation of survival, apoptosis and senescence by c/EBP-β is highly context specific, being this factor able to influence the survival of some transformed cells while inducing growth arrest in others [reviewed in [Citation38]].
Here, we also report that HMGA1 is able to regulate NUMB at posttranscriptional level, by binding the promoters of its posttranscriptional regulators, MSI1 and miR-146a. Indeed, HMGA1 silencing leads to the downregulation of both MSI1 and miR-146a, which bind NUMB mRNA and prevent its translation. Increased expression of MSI1 has been detected in GBMs [reviewed in [Citation39]], as well as in spheroid cultures of tumor cells and in the CD133+ cell population. The targeting of MSI1 by RNA interference-based approaches has shown promising results in preclinical studies [reviewed in [Citation40]].
On the other hand, it has been previously found that miR-146a is induced by SNAIL [Citation26], which is in turn downregulated in HMGA1-KD cells [Citation41], suggesting that HMGA1 can regulate miR-146a expression both directly and indirectly. Its upregulation in neural progenitor cells (NPCs) during neurogenesis [Citation42] and its ability to spur Colon Cancer Stem Cells to undergo symmetric division [Citation26], together with our results, support a relevance in BTSC and, hence, in the development of GBM.
In agreement with the role of NUMB in NOTCH degradation, we also demonstrated that NOTCH1 is reduced in HMGA1-KD cells and that this effect is mediated by NUMB. Consistently, NOTCH1 overexpression has been reported in GBM and its targeting by si-RNA or by γ-secretase inhibitors (GSI) is able to decrease glioma stem cell self-renewal and tumor growth and increase differentiation into mature neural cells [Citation28,Citation43].
The identification of this novel pathway, regulated by HMGA1, may have translational implication not only in GBM therapy, since NUMB has been found downregulated in the most aggressive forms of leukemia [Citation44], non-small cell lung carcinomas [Citation45], breast cancers [Citation27], colon cancers [Citation46], brain tumors [Citation21], in which HMGA1 overexpression has been frequently reported [Citation5,Citation6,Citation47]. Intriguingly, the loss of NUMB expression has been detected in a subgroup of proneural glioblastomas [Citation21], where HMGA1 has been found overexpressed [Citation6].
Noteworthy, NUMB downregulation leads to p53 inactivation and increases tumor aggressiveness [Citation19]; conversely, NUMB overexpression has onco-suppressive function, at least in part because of its ability to stabilize p53 expression [Citation19] and to degrade NOTCH1 [reviewed in [Citation16]].
In conclusion, the results reported here highlight a novel HMGA1-driven regulatory pathway affecting, directly and indirectly, NUMB expression, at both transcriptional and posttranscriptional level. This new pathway links HMGA1 expression to reduced asymmetric divisions and connects this architectural factor to the pathogenesis of brain tumors, originating from asymmetry-defective progenitors [[Citation3]; reviewed in [Citation2]]. Therefore, our data further support the insight that HMGA1 can be an effective target for anti-cancer stem cell therapy.
Supplemental Material
Download Zip (3 MB)Acknowledgments
We thank Prof. Salvatore Pece and Prof. Pier Paolo Di Fiore for providing the anti-NUMB antibody.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol. 2010;11:849–860. Review.
- Lewis KM, Petritsch C. Asymmetric cell division: implications for glioma development and treatment. Transl Neurosci. 2013;4:484–503.
- Sugiarto S, Persson AI, Munoz EG, et al. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell. 2011;20:328–340.
- Bleeker FE, Molenaar RJ, Leenstra S. Recent advances in the molecular understanding of glioblastoma. J Neurooncol. 2012;108:11–27.
- Fedele M, Fusco A. HMGA and cancer. Biochim Biophys Acta. 2010;1799:48–54. Review.
- Colamaio M, Tosti N, Puca F, et al. HMGA1 silencing reduces stemness and temozolomide resistance in glioblastoma stem cells. Expert Opin Ther Targets. 2016;20:1169–1179.
- Berlingieri MT, Manfioletti G, Santoro M, et al. Inhibition of HMGI-C protein synthesis suppresses retrovirally induced neoplastic transformation of rat thyroid cells. Mol Cell Biol. 1995;15:1545–1553.
- Scala S, Portella G, Fedele M, et al. Adenovirus-mediated suppression of HMGI(Y) protein synthesis as potential therapy of human malignant neoplasias. Proc Natl Acad Sci U S A. 2000;97:4256–4261.
- Berlingieri MT, Pierantoni GM, Giancotti V, et al. Thyroid cell transformation requires the expression of the HMGA1 proteins. Oncogene. 2002;21:2971–2980.
- Vallone D, Battista S, Pierantoni GM, et al. Neoplastic transformation of rat thyroid cells requires the junB and fra-1 gene induction which is dependent on the HMGI-C gene product. Embo J. 1997;16:5310–5321.
- Federico A, Forzati F, Esposito F, et al. Hmga1/Hmga2 double knock-out mice display a “superpygmy” phenotype. Biol Open. 2014;3:372–378.
- Puca F, Colamaio M, Federico A, et al. HMGA1 silencing restores normal stem cell characteristics in colon cancer stem cells by increasing p53 levels. Oncotarget. 2014;5:3234–3245.
- Pierantoni GM, Conte A, Rinaldo C, et al. Deregulation of HMGA1 expression induces chromosome instability through regulation of spindle assembly checkpoint genes. Oncotarget. 2015;6:17342–17353.
- Pegoraro S, Ros G, Piazza S, et al. HMGA1 promotes metastatic processes in basal-like breast cancer regulating EMT and stemness. Oncotarget. 2013;4:1293–1308.
- D’Angelo D, Mussnich P, Rosa R, et al. High mobility group A1 protein expression reduces the sensitivity of colon and thyroid cancer cells to antineoplastic drugs. BMC Cancer. 2014;14:851.
- Pece S, Confalonieri S, Romano PR, et al. NUMB-ing down cancer by more than just a NOTCH. Biochim Biophys Acta. 2011;1815:26–43.
- Shen Q, Zhong W, Jan YN, et al. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129:4843–4853.
- Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat Genet. 2005;37:1125–1129.
- Colaluca IN, Tosoni D, Nuciforo P, et al. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80.
- Kang Y, Ding M, Tian G, et al. Overexpression of Numb suppresses tumor cell growth and enhances sensitivity to cisplatin in epithelioid malignant pleural mesothelioma. Oncol Rep. 2013;30:313–319.
- Jiang X, Xing H, Kim TM, et al. Numb regulates glioma stem cell fate and growth by altering epidermal growth factor receptor and Skp1-Cullin-F-box ubiquitin ligase activity. Stem Cells. 2012;30:1313–1326.
- Sato K, Watanabe T, Wang S, et al. NUMB controls E-cadherin endocytosis through p120 catenin with aPKC. Mol Biol Cell. 2011;22:3103–3119.
- Wang Z, Sandiford S, Wu C, et al. Numb regulates cell-cell adhesion and polarity in response to tyrosine kinase signalling. Embo J. 2009;28:2360–2373.
- Liu XH, Wu Y, Yao S, et al. Androgens up-regulate transcription of the Notch inhibitor Numb in C2C12 myoblasts via Wnt/β-catenin signaling to T cell factor elements in the Numb promoter. J Biol Chem. 2013;288:17990–17998.
- Kawahara H, Imai T, Imataka H, et al. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J Cell Biol. 2008;181:639–653.
- Hwang WL, Jiang JK, Yang SH, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol. 2014;16:268–280.
- Pece S, Serresi M, Santolini E, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215–221.
- Wang J, Wang C, Meng Q, et al. siRNA targeting Notch-1 decreases glioma stem cell proliferation and tumor growth. Mol Biol Rep. 2012;39:2497–2503.
- Battista S, Pentimalli F, Baldassarre G, et al. Loss of Hmga1 gene function affects embryonic stem cell lympho-hematopoietic differentiation. Faseb J. 2003;17:1496–1498.
- Battista S, Fedele M, Martinez Hoyos J, et al. High-mobility-group A1 (HMGA1) proteins down-regulate the expression of the recombination activating gene 2 (RAG2). Biochem J. 2005;389(Pt 1):91–97.
- Messeguer X, Escudero R, Farré D, et al. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334.
- LaMarca HL, Visbal AP, Creighton CJ, et al. CCAAT/enhancer binding protein beta regulates stem cell activity and specifies luminal cell fate in the mammary gland. Stem Cells. 2010;28:535–544.
- Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318–324. Review.
- Foti D, Iuliano R, Chiefari E, et al. A nucleoprotein complex containing Sp1, C/EBP beta, and HMGI-Y controls human insulin receptor gene transcription. Mol Cell Biol. 2003;23:2720–2732.
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278:23196–23203.
- McGill MA, Dho SE, Weinmaster G, et al. Numb regulates post-endocytic trafficking and degradation of Notch1. J Biol Chem. 2009;284:26427–26438.
- Aguilar-Morante D, Cortes-Canteli M, Sanz-Sancristobal M, et al. Decreased CCAAT/enhancer binding protein β expression inhibits the growth of glioblastoma cells. Neuroscience. 2011;176:110–119.
- Zahnow CA. CCAAT/enhancer-binding protein beta: its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev Mol Med. 2009;11:e12.
- Glazer RI, Vo DT, Penalva LO. Musashi1: an RBP with versatile functions in normal and cancer stem cells. Front Biosci (Landmark Ed). 2012;17:54–64.
- Kudinov AE, Karanicolas J, Golemis EA, et al. Musashi RNA-binding proteins as cancer drivers and novel therapeutic targets. Clin Cancer Res. 2017;23:2143–2153.
- Shah SN, Cope L, Poh W, et al. HMGA1: a master regulator of tumor progression in triple-negative breast cancer cells. PLoS One. 2013;8:e63419.
- Liu XS, Fan BY, Pan WL, et al. Identification of miRNomes associated with adult neurogenesis after stroke using Argonaute 2-based RNA sequencing. RNA Biol. 2017;14:488–499.
- Hu YY, Zheng MH, Cheng G, et al. Notch signaling contributes to the maintenance of both normal neural stem cells and patient-derived glioma stem cells. BMC Cancer. 2011;11:82.
- Ito T, Kwon HY, Zimdahl B, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768.
- Westhoff B, Colaluca IN, D’Ario G, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A. 2009;106:22293–22298.
- Bu P, Wang L, Chen KY, et al. A miR-34a-numb feedforward loop triggered by inflammation regulates asymmetric stem cell division in intestine and colon cancer. Cell Stem Cell. 2016;18:189–202.
- Donato G, Martinez Hoyos J, Amorosi A, et al. High mobility group A1 expression correlates with the histological grade of human glial tumors. Oncol Rep. 2004;11:1209–1213.
