ABSTRACT
Tumorigenic cancer stem cells (CSCs) exist in various tumors including the cutaneous squamous cell carcinoma (cSCC) as a minor subpopulation and are tightly associated with metastasis and therapeutic resistance. Better understanding of CSCs properties is essential for the novel therapeutic strategy targeted toward these cancers. The cSCC stem cells (cSCCSCs) were enriched from a cSCC cell line A431 by repeated sphere culture, and identified via the expression analysis of stemness marker genes and CD44 proteolysis. MiR-199a-5p was previously reported to be related with the proteolysis modulation of CD44, so the specific regulation mechanisms were verified by overexpression in vitro and in vivo. MiR-199a-5p is under-expressed in cSCCSCs and functions as a tumor suppressive molecule. Overexpression of miR-199a-5p reduced the stemness of cSCCSCs and inhibited cell proliferation. By targeting the deacetylase Sirt1, miR-199a-5p inhibited cellular proteolysis of CD44 and reduced the CD44 intracellular domain (CD44ICD) release and nuclear translocation. Overexpression of CD44ICD reversed the effects of miR-199a-5p overexpression or Sirt1 silencing, and increased the transcriptional expression of stemness genes. Our results revealed that the miR-199a-5p/Sirt1/CD44ICD signaling pathway regulates cSCCSCs progression by affecting its migration ability and tumorigenicity, therefore can be utilized to develop a curative approach for cSCC.
Abbreviations: CSCs: cancer stem cells; cSCC cutaneous squamous cell carcinoma; cSCCSCs: cSCC stem cells; CD44ICD: CD44 intracellular domain; HA: hyaluronic acid; HNSCC: hand and neck squamous cell carcinoma; ESCC: esophageal squamous cell carcinoma;MMPs: matrix metalloproteinases; SFM: sphere formation medium; EGF: epidermal growth factor; bFGF: basic fibroblast growth factor; BSA: bovine serum albumin; CCK-8: cell counting kit-8
Introduction
Cutaneous squamous cell carcinoma (cSCC) is one of the most common malignant tumors worldwide and roughly accounts for 20% of all skin cancer-related deaths [Citation1]. Owing to its high rates of recurrence cSCC represents a deadly threat for many organ transplant recipient patients that are under immunosuppression treatment. Cancer stem cells (CSCs) are a small subpopulation of tumor cells which are identified with higher tumor-initiating capability, and high frequency of CSCs has been demonstrated to associate with therapy resistance and poor prognosis of the cSCC. The stemness properties of CSC, such as long-term self-renewal, differentiation and proliferation abilities, were reported to be governed by a set of stemness genes [Citation2]. During the last ten years CSCs targeting has emerged as a new therapeutic strategy for many solid tumors, although the detailed mechanisms by which the CSC populations drive cSCC tumor growth, regeneration and cause therapeutic resistance, remain unclear.
CD44 is a multifunctional transmembrane glycoprotein that binds to hyaluronic acid (HA) and mediates cell-matrix and cell-cell interactions. CD44 has been used as CSCs enrichment marker across multiple tumor types [Citation3]. Previous studies showed that CD44+ cells increase cancer proliferation in hand and neck squamous cell carcinoma (HNSCC) and give rise to new tumors in vivo [Citation4], while high CD44 expression in esophageal squamous cell carcinoma (ESCC) is correlated with increased colony formation, drug resistance in vitro, as well as enhanced tumor forming ability in vivo, indicating CD44 as a significant biomarker for CSCs in SCC [Citation5]. During tumor progression, CD44 functions as a signal transduction molecule through two-step proteolytic cleavages in the extracellular domain (CD44ECD) and the intramembranous domain (CD44ICD). The proteolytic processing of CD44 is mediated by matrix metalloproteinases (MMPs) [Citation6], presenilin-dependent γ-secretase [Citation7] and acetylation modification [Citation8]. Numerous CD44 isoforms exist due to the alternative splicing of 10 exons at the ectodomain [Citation9–Citation11], while the CD44ICD is equal to all the variants [Citation12]. The proteolytic cleavage product CD44ICD was translocated into the nucleus and regulated directly the tumor cell migration, invasion, angiogenesis, and metastasis by activating the transcriptional machinery [Citation13,Citation14].
MiR-199a precursor is a short non-coding RNA and produces mature products miR-199a-5p and miR-199a-3p. MiR-199a attracted growing interest in cancer research in the last few years, and has been reported with compelling evidences as tumor oncogene [Citation15,Citation16] or tumor suppressor [Citation17,Citation18] in many human cancers. Herein we focus on miR-199a-5p because our previous studies showed that miR-199a-5p is dramatically under-expressed in a cSCC, where it functions as an anti-tumor gene by targeting mitogenic molecules and eventually suppresses cancer cell proliferation and migration [Citation19].
In this study, we report that miR-199-5p targeted directly the expression and function of the deacetylase Sirt1, the knockout of which inhibited the cleavage of CD44 and promoted translocation of CD44ICD into nucleus. In addition, we provide the evidence that the miR-199-5p/Sirt1 signaling pathway moderated the CSC stemness in cSCC by regulating the expression of stemness genes, and therefore plays critical roles in tumor formation and migration.
Materials and methods
Cell culture and human cSCC stem cells enrichment by spheroid formation assays
Human cSCC cell line A431 and normal human skin fibroblast were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). A431 cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS), while HSF cells were cultured in Fibroblast Basal Medium supplemented with 2% FBS. All culture reagents were from Invitrogen (Carlsbad, CA, USA).
To enrich the cSCC stem cells (cSCCSCs), single-cell populations of A431 cells were resuspended in serum-free sphere formation medium (SFM) composed of RPMI-1640 medium, 20 ng/ml of epidermal growth factor (EGF), 10 ng/ml of basic fibroblast growth factor (bFGF) and 2% B27 supplement (w/v), and cultured at 37°C for spheroid formation. When the primary spheroids grow to about 200 μm, spheres were dissociated by TrypLETM Express, gently resuspended and further cultured in the SFM. After the sphere formation procedure was repeated five times, the secondary cellular spheroids were considered as cSCCSCs and used for subsequent experiments.
MiRNA oligonucleotides, adenovirus generation and cell transfection
MiR-199a-5p mimics (5ʹ-CCCAGUGUUCAGACUACCUGUUC-3ʹ) with matched mimics negative control (5ʹ-UCGACUCUCUGCACGAAUCGCUU-3ʹ) or inhibitor (5ʹ-GAACAGGUAGUCUGAACACUGGG-3ʹ) with matched inhibito negative control (5ʹ-GAGCGGUCUGCAUAGAAGUACGA-3ʹ) were designed and synthesized by GenePharma (Shanghai, China), and these oligonucleotides were transiently transfected into cells using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions.
Adenoviruses expressing Short hairpin RNAs of Sirt1 gene (siRNA-1: 5ʹ-GATGATCAAGAGGCAATTAAT-3ʹ; siRNA-2: 5ʹ-ACTTTGCTGTAACCCTGTA-3ʹ; siRNA-3: 5ʹ-CCTCGAACAATTCTTAAAGAT-3ʹ; siRNA-4: 5ʹ-CATGAAGTGCCTCAGATATTA-3ʹ), and cloned CD44ICD (forward primer: 5ʹ- GATCTTAATTAACGCAGTCAACAGTCGAAGAAGG-3ʹ; reverse primer: 5ʹ- GATCGCGGCCGCTTATTACACCCCAATCTTCATGTCCAC-3ʹ) were created using the Adeno-XTM expression system (Clontech, USA) according to the user’s manual. After amplification and purification, the virus titer was determined through plaque assays. Cells were infected with the adenovirus at MOI (multiplicity of infection) of 50 and harvested 48 to 72 hours after infection.
Immunofluorescence assay
After treatment, cells were plated on glass coverslips in a 12-well plate and fixed with 4% formaldehyde for 10 min. Followed by permeabilized using 0.5% Triton X-100 in PBS, the cells were washed three times and unspecific binding was blocked with 1% bovine serum albumin (BSA). The cells were stained with primary antibodies against Sirt1 (1:50, Abcam) or CD44ICD (1:100, Abcam) at 4°C overnight. After washing three times using Tris-buffered saline with Tween 20 (TBST), the cells were further incubated with secondary antibodies conjugated to Alexa Fluor 594 at room temperature for 1 h in the dark. The nuclei were stained with 1 μg/mL 4ʹ,6-diamidino-2-phenylindole (DAPI, 1:100; Sigma-Aldrich, St. Louis, MO, USA) for 5 min at 37°C. The slides were mounted with SlowFade Gold Antifade Reagent (Invitrogen, USA) and the fluorescence images were obtained under a Nikon confocal microscope (Eclipse TE2000U).
Cell proliferation analysis by CCK-8
The cell proliferation was evaluated using cell counting kit-8 (CCK-8) assay. Briefly, the cells were seeded in a 96-well microplate at a density of 5000 cells/well. After incubation for 24, 48, 72 or 96 hours, the culture medium was replaced with 100 µl growth medium and 10 µl CCK-8 solution, followed by further incubation at 37 °C for 3 h. The absorbance at 450 nm was measured by a spectrometer reader (SpectraMax M2, Molecular devices, USA).
Clonal and sphere-formation assays
N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester (DAPT, Selleck, USA) was prepared as a 10 µM stock in DMSO (0.1%, Sigma, USA). Cells were diluted in growth medium by adding miR-199a-5p mimics/negative control mimics (miRNA-NC), 10 µM DAPT or DMSO, and cultured in a 6-well plate at a density of 100 cells per well. The holoclones were enumerated upon crystal violet staining 2 weeks after seeding.
For sphere formation assay, cells were resuspended in sphere formation medium by adding miRNA oligos/adenoviruses or DAPT (10 µM)/DMSO (0.1%) and seeded in an ultra-low attachment 6-well plate at a density of 3000 cells per well. After 1–2 weeks culture, spheres were observed under microscope.
Luciferase reporter assays
The miR-199a-5p recognition motif in Sirt1 transcript 3ʹUTR was identified using TargetScan (targetscan.org). A 638 bp 3ʹUTR sequence of Sirt1 gene (NM_012238.5) that contains the predicted binding sites was amplified from A431 cells by PCR (Forward primer: 5ʹ-CAATCAGCTGTTGGTCAAGACT-3ʹ; Reverse primer: 5ʹ-GATCAATGCAAGCTCTACCACAG-3ʹ) and then cloned into the luciferase reporter vector pMIR-REPORT as previously described [Citation20]. Mutations at the putative binding sites of the Sirt1 3ʹUTR sequence were performed using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). HEK cells were seeded in a 96-well plate and co-transfected with 25 ng Sirt1 luciferase reporter, 15 nM miR-199a-5p mimics or inhibitor oligos and 10 ng Renilla luciferase plasmid pRL-TK using Lipofectamine 2000. The cells were lysed 48 h after transfection and the luciferase activity was quantified using Dual Luciferase Reporter Assay Kit (Promega), according to the manufacturer’s instruction. The activities were normalized to Renilla luciferase activity.
Total RNA extraction and quantitative PCR (qPCR)
The cells were pelleted and resuspended in TRIzol reagent (Invitrogen). Total RNA was extracted using RNeasy Mini Kit (Qiagen, Chatsworth, CA) for mRNA or mirVanaTM miRNA Isolation Kit (Ambion, Austin, TX) for miRNA. Expression of Sirt1, Oct4, Sox2 and Nanog mRNA was detected by SYBR green qPCR assay, using GAPDH as an endogenous expression control. 2 μg total RNA was reverse transcribed into cDNA using MiScript Reverse Transcription kit, and the expression of miR-199a-5p was detected through real time PCR using MiScript SYBR-Green PCR kit, according to the user’s guide. Internal “house keeping” miRNA U6 was used as control. All the primers for qPCR are listed in . Finally, the gene relative expression results were normalized to GAPDH or U6 in the same samples and analyzed with a 2−ΔΔCt methods.
Table 1. Paired primer sequences used in qPCR
Western blot analysis
Cultured cells were lysed on ice in RIPA buffer containing protease inhibitor cocktail. Total protein (30) µg was separated by SDS-PAGE and transferred to nitrocellulose membrane, followed by 1 h incubation with 5% milk powder blocking buffer. The membranes were first immunoblotted with the primary antibodies at 4 °C overnight. β-actin was used as a loading control. All the primary antibodies (Oct4, ab181557; Sox2, ab92494; Nanog, ab109250; CD44, ab157107; Sirt1, ab32441; β-actin, ab8226) were purchased from Abcam (England) and used with a dilution recommended by the instructions, 1:1000 dilution for example. After washing three times in 0.1M PBST the membranes were incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. Then ECL substrates were added to visualize the protein bands under a chemiluminescence system (Bio-Rad, US). Digital images were quantified using ImageJ software (National Institutes of Health, USA).
Xenografts models
Animal experiments were performed according to the institutional ethical guidelines from the Institutional Animal Care and Use Committee of Xiangya Hospital affiliated to Central South University. 6–8 weeks old nude mice BALB/c were subcutaneously injected with A431 cells (1 × 107) to establish cSCC xenograft models. Tumor volume was measured twice per week and allowed to grow to 100 mm3, and then miR-199a-5p mimics or CD44ICD adenoviruses were injected intratumorally every other day. For liver metastasis experiments, A431 cells (1 × 107) were injected via the tail vein of nude mice, and miR-199a-5p mimics or CD44ICD adenoviruses were administrated to the models every other day by intraperitoneal injection. After 30-day treatment, the primary tumors and metastatic liver tissues were isolated from the mice for further analysis. To quantitate the efficiency of liver metastasis, the number of nodules visible to the naked eye on the largest hepatic lobule were counted and statistical analysis.
Histopathological analysis
After fixed with 10% formaldehyde (Sigma, USA) for more than 24h, the isolated tissues samples were embedded with paraffin and sectioned at 5 μm thickness. For hematoxylin&eosin (HE) staining, the sections were stained with hematoxylin solution and Eosin solution in steps. For immunohistochemistry analyis, the sections were incubated with 3% H2O2 in methanol to quench the endogenous peroxidase. After washing, the sections were blocked and firstly probed with indicated primary antibodies (CD44, ab157107; Sirt1, ab32441; 1:500 dilution) at 4°C overnight, then probed with the secondary antibody IgG-HRP for 1 h at room temperature. At last, followed by incubation with DAB substrate for signal development, these sections were washed, dehydrated, sealed by coverslips and imaged by an upright microscopy (Olympus, Japan).
Statistical analysis
All experiments have been repeated independently at least three times. Results are presented as mean ± SD and assessed by Student’s t test or one-way ANOVA analysis using GraphPad Prism 5.0 (GraphPad Software Inc., CA). Differences were considered statistically significant with P < 0.05.
Results
MiR-199a-5p was under-expressed in CD44+ A431 and cSCC stem cells
As a small subpopulation of tumor cells, the cancer stem cells (CSCs) are often isolated from the specific tumor cell populations. According to the stemness characteristics, we used a spheroid formation assay to enrich the cSCC stem cells (cSCCSCs) from the human cutaneous squamous cell line A431. The expression of stemness markers at mRNA and proteins was measured by qPCR and western blot. The results showed that the mRNA and protein levels of Oct4, Sox2 and Nanog were significantly upregulated in the cSCCSCs than that in A431 cells (,)), suggesting that the enriched stem cells cSCCSCs were isolated successfully. CD44 is commonly used as a marker for CSC enrichment [Citation3,Citation5], so in order to identify the cSCC stem cells enriched from A431, flow cytometry analysis of CD44 was performed with a CD44 monoclonal antibody. And results showed that CD44 was positively expressed in both A431 cells and the enriched stem cells cSCCSCs, while the normal HSF cells are CD44 negative; moreover, the side scatter (SSC) value of cSCCSCs was remarkably higher than A431 cells, indicating that more complex cellular granularity and fine cellular structure in cSCCSCs compared to A431 cells ()). CD44 functions as a signal transduction molecule through proteolysis, with CD44ECD released into extracellular matrix and CD44ICD transported into the nucleus. Immunoblotting and immunofluorescence experiments were performed to analyze the cellular distribution of CD44 in these cell lines. Similar to results of flow cytometry analysis, the total protein levels of CD44 in A431 and cSCCSCs cells were remarkably high-positive, but that in HSF cells was very low-expressed ()). What’s more interesting, the levels of CD44ICD protein in cSCCSCs cells were obviously higher than that in A431 cells ()), and the cell immunofluorescence staining of CD44 also showed that, compared to A431 cells, the nuclear protein levels of CD44 were increased in cSCCSCs cells, which mainly distributed on the cell membrane in A431 cells ()), indicating the proteolysis of CD44 and nuclear translocation of CD44ICD may be closely related to the stemness of cSCCSCs cells. As reported previously that miR-199a may be an upstream regulator of CD44 [Citation19], we detected that miR-199a-5p was downregulated in cSCCSCs and A431 cells compared to the normal HSF cells ()), to a certain extent, negatively correlating with the expression levels of CD44, especially the CD44ICD, indicating that miR-199a-5p may be as a upstream regulator for CD44 and its proteolysis process.
Figure 1. MiR-199a-5p was under-expressed in CD44+ cSCCSCs. (a) QPCR analysis and (b) western blot analysis for the relative expression of stemness marker genes Oct4, Sox2 and Nanog in A431 cells and the cSCCSCs isolated from A431 using a spheroid formation assay. (c) Flow cytometry analysis for the positive rates of CD44 expression in A431 cells and cSCCSCs compared with a normal human skin fibroblast HSF. (d) The cellular distribution of CD44 in A431 cells and cSCCSCs was analyzed by immunofluorescence (scale bar = 150μm). (e) Western blot analysis for the protein expression of CD44 and CD44ICD in HSF, A431 cells and cSCCSCs. (f) QPCR analysis for the differential expression of miR-199a-5p in HSF, A431 cells and cSCCSCs. * p < 0.05, ** p < 0.01, *** p < 0.001.
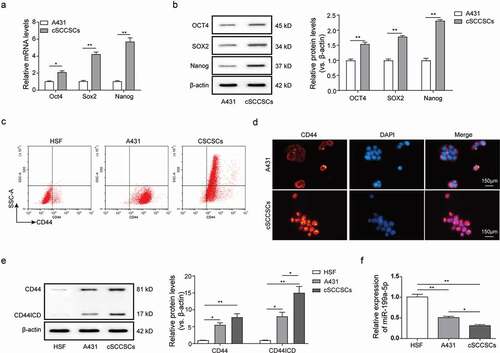
Overexpression of miR-199a-5p significantly suppressed the stemness of cSCCSCs
To investigate the effects and underlying regulatory mechanisms of miR-199a-5p, cSCCSCs were transfected with miR-199a-5p mimics and negative control (NC) oligonucleotides relatively, and qPCR analysis was used to verify the overexpression of miR-199a-5p on high-efficiency in A431 cells ()). Γ-secretase is an essential component for CD44 proteolysis [Citation7,Citation21], and it has been demonstrated previously that treatment of DAPT, a γ-secretase inhibitor, inhibited the self-renewal and stemness properties of CD44+ ovarian cancer stem-like cells [Citation22] and gastric cancer stem cells [Citation23]. In order to confirm whether miR-199a-5p mediates the process of CD44 proteolysis, DAPT was used to treat with A431 cells as a positive control of CD44 proteolysis. Western blot results revealed that the CD44ICD level was suppressed obviously both by miR-199a-5p overexpression and DAPT treatment ()). Although the level of intact CD44 was also decreased after miR-199a-5p overexpression, the rangeability of CD44ICD were significantly greater than the intact CD44 ()), indicating that miR-199a-5p may mainly regulate the CD44 proteolysis process than the primary expression.
Figure 2. Overexpression of miR-199a-5p significantly suppressed the stemness of CSCCSCs. (a) QPCR analysis for overexpression efficiency of miR-199a-5p by mimic transfection in cSCCSCs. (b) Western blot analysis for CD44 and CD44ICD expression in cSCCSCs transfected with miR-199a-5p mimic and negative control (miR-NC), or cSCCSCs treated with 10 μM DAPT (a γ-secretase inhibitor which suppresses CD44 intro-cellular cleavage) and DMSO as a control. (c) CCK-8 assay for the cell growth curve analysis of cSCCSCs as described in B. (d) Colony formation experiments were performed to examine the cell proliferation ability of cSCCSCs as described in B. (e) Sphere formation experiments were performed to analyze the cell self-renewal ability of cSCCSCs as described in B (scale bar = 300μm). (f) QPCR analysis for the expression of stemness genes in cSCCSCs as described in B. * p < 0.05, ** p < 0.01, *** p < 0.001.
Furthermore, the cell proliferation, clonogenic and sphere forming capacities of the cSCCSCs were inhibited by the DAPT treatment or the miR-199a-5p overexpression (–)). To test the effects of miR-199a-5p on cSCCSCs stemness, we then compared the relative expression of Oct4, Sox2 and Nanog ()). Impressively, the expression level of the stemness genes was generally decreased in the cSCCSCs which were treated with DAPT or transfected with miR-199a-5p mimics. Collectively these results suggest that the overexpression of miR-199a-5p significantly inhibited the cell proliferation, self-renewal in cSCCSCs, and decreased their stemness properties via closely mediating the CD44 intracellular cleavage process.
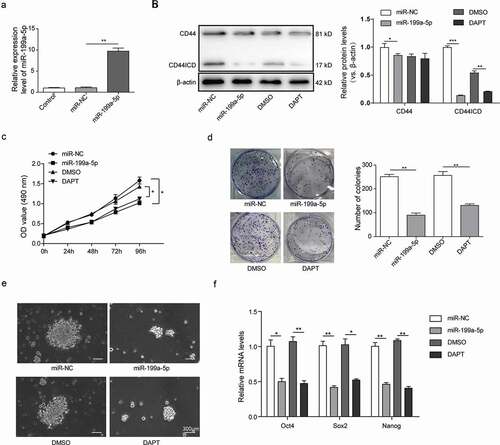
MiR-199a-5p directly targeted the expression of Sirt1
We employed the online target-prediction tools and identified Sirt1 mRNA as a putative binding partner for miR-199a-5p. Sirt1 is a nicotinamide adenine dinucleotide (NAD+) dependent deacetylase [Citation24] and acts as a tumor promoter in acute myeloid leukemia [Citation25], prostate cancer [Citation26], colon cancer [Citation27] and skin cancers [Citation28]. MiR-199a has been reported to inhibit Sirt1 expression in cardiomyocytes [Citation29]. In contrast to miR-199a-5p, the mRNA and protein levels of Sirt1 are much higher in A431 cells and in cSCCSCs than in HSF cells ()). The direct binding of miR-199a-3p to the Sirt1 3ʹ-UTR was further confirmed using a dual-luciferase reporter assay. Notably, the luciferase activities in miR-199a-5p transfected cSCCSCs were significantly reduced, whereas full activities were restored by mutating the miR-199a-5p binding site at Sirt1 3ʹ-UTR ()).
Figure 3. MiR-199a-5p directly targeted the expression of Sirt1. (a) The expression of Sirt1 in HSF, A431 and cSCCSCs were detected by Q-PCR and Western blot respectively. (b) The predicted binding site between miR-199a-5p and Sirt1 3ʹUTR was analyzed using starBase software (Guangzhou, China) and verified by luciferase assay. (c) QPCR analysis for the expression of miR-199a-5p and Sirt1 in A431 cells transfected with miR-199a-5p mimic (miR-199a-5p) or inhibitor (anti-miR-199a-5p) compared with corresponding negative controls (miR-NC and anti-miR-NC). (d) Western blot analysis for Sirt1 expression in A431 cells as described in C. (e) Immunofluorescence analysis for Sirt1 in A431 cells as described in C (scale bar = 150μm). ** p < 0.01, *** p < 0.001.
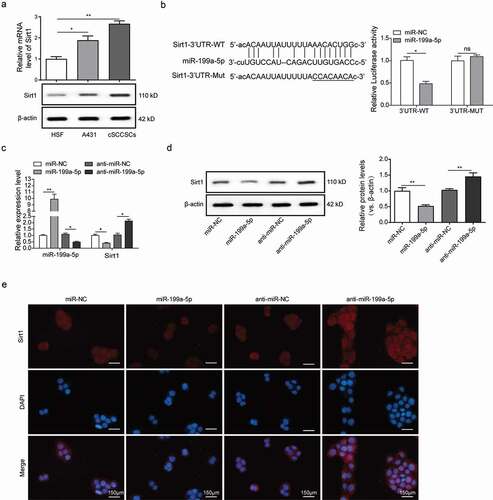
We then transfected A431 cells with miR-199a-5p mimics or inhibitor oligos. Interestingly, the miR-199a-5p mimics inhibited Sirt1 mRNA and protein expression, while the miR-199a-5p inhibitor promoted Sirt1 expression (,)). In concordance, our immunofluorescence assay showed that the fluorescence intensity of Sirt1 in the cells transfected with miR-199a-5p mimics was much lower than the miR-NC group, whereas the cells transfected with miR-199a-5p inhibitor showed higher Sirt1 abundance ()). Taken together, these results indicated that miR-199a-5p overexpression inhibited Sirt1 expression at both mRNA and protein levels, which were conversely promoted by miR-199a-5p inhibitor, exhibiting a significant negative correlation.
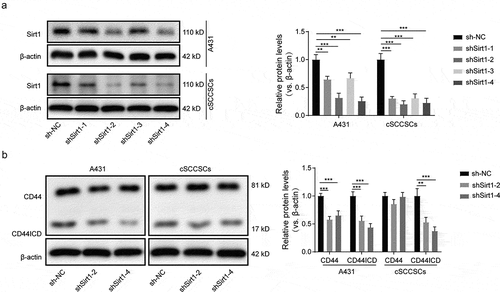
Sirt1 silencing blocked the nuclear translocation of CD44ICD through proteolysis inhibition
We designed three siRNAs against Sirt1 and inserted the corresponding shRNAs to adenoviruses vector. While all three shRNAs reduced Sirt1 expression ()), our following experiments focused on shRNA-2 of Sirt1 (shSirt1-2 in short) which exhibited the highest silencing efficiency. The protein expression levels of CD44 in A431 cells and cSCCSCs treated with shSirt1-2 displayed no significant changes, however, CD44ICD was dramatically downregulated ()), suggesting a positive correlation between Sirt1 and CD44 intracellular proteolysis. Consistent results were observed in our immunofluorescence experiments (Figure S1), indicating that the deficiency of Sirt1 blocked CD44 intracellular cleavage as well as the translocation of the proteolysis product CD44ICD into cell nucleus.
Figure 4. Sirt1 silencing blocked the nuclear translocation of CD44ICD through proteolysis inhibition. (a) Four small interfering RNAs (siRNAs) were designed to silence the expression of Sirt1 and recombinant adenoviruses carrying the synthetic shRNAs were infected to A431 cells and cSCCSCs. The inhibition efficiency was evaluated by western blot analysis. (b) The shSirt1-2 and shSirt1-4 with the more higher inhibition efficiency were selected to infect A431 cells and cSCCSCs. The protein levels of cellular CD44 and CD44ICD were detected by western blot. * p < 0.05, ** p < 0.01.
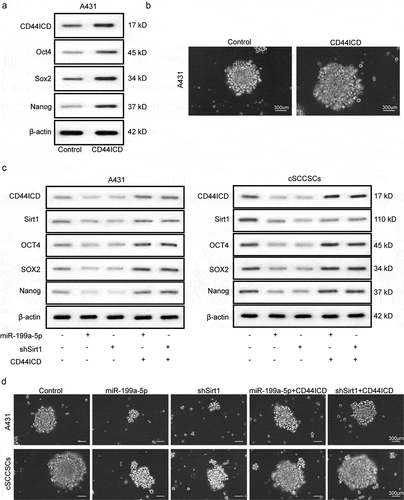
The miR-199a-5p/Sirt1 signaling axis modulated the stemness of cSCCSCs via CD44ICD proteolysis
To examine the role of CD44ICD in cSCCSCs stemness regulation, we transfected the A431 cells with adenovirus that carried CD44ICD coding sequence (CDS). Notably, the protein concentrations of the stemness markers Oct4, Sox2, and Nanog were significantly higher than in control ()). In addition, the CD44ICD overexpressed A431 cells demonstrated higher self-renewal capacity and formed sphere colonies more effectively than in the control cells ()). As demonstrated previously, overexpression of miR-199a-5p or silencing of Sirt1 reduced the molecular levels of CD44ICD as well as the stemness markers. Intriguingly, the stemness inhibiting effect induced by miR-199a-5p and Sirt1 diminished in A431 cells and cSCCSCs which were transfected with CD44ICD adenovirus, resulting in even elevated expression of stemness markers ()). Furthermore, CD44ICD overexpression efficiently restored the sphere formation ability of A431 cells and cSCCSCs, which was significantly inhibited by miR-199a-5p or sh-Sirt1 adenovirus transfection ()).
Figure 5. The miR-199a-5p/Sirt1 signaling axis modulated the stemness of cSCCSCs via CD44ICD proteolysis. The effects of CD44ICD on cSCCSCs stemness were confirmed by overexpressing CD44ICD in A431 cells. (a) Western blot analysis for the expression of stemness markers Oct4, Sox2 and Nanog in A431 cells transfected with CD44ICD adenoviruses or not. (b) Sphere formation of A431 cells as described in A (scale bar = 300 μm). (c) Western blot analysis for the expression of stemness markers Oct4, Sox2 and Nanog in A431 cells and cSCCSCs treated with miR-199a-5p/Sirt1-shRNA combined with or without CD44ICD overexpression. (d) Sphere formation of A431 cells and cSCCSCs as described in C (scale bar = 300μm).
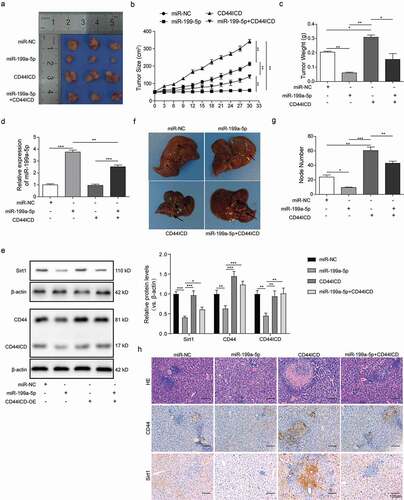
MiR-199a-5p suppressed cSCC xenograft tumor growth and metastasis by targeting CD44ICD activation
We transplanted the A431 cells into a xenograft mouse model and assessed the tumor formation after adenovirus injection, which carried miR-199a-5p or CD44ICD CDS. Notably, the mice injected with CD44ICD adenovirus displayed enhanced tumor growth as compared to the miR-NC group. In contrast, the tumors grew notably slower in the mice injected with miR-199a-5p adenovirus. The tumor growth was mostly restored by the co-injection of CD44ICD and miR-199a-5p adenoviruses ()). Furthermore, average tumor size and weight of the miR-199a-5p group were markedly lower than in miR-NC, and much higher in the CD44ICD group. The tumor growth in the miR-199a-5p group was largely enhanced by the CD44ICD adenovirus injection (,)). We collected tumor tissues from the four mice groups and performed biochemical analysis. Relative expression of miR-199a-3p was quantified by QPCR, and we observed that miR-199a-3p expression was completely abolished in CD44ICD group ()). MiR-199a-5p group displayed decreased protein levels of Sirt1, CD44, and CD44ICD, which were conversely restored by co-injection of CD44ICD adenovirus ()). In addition, we observed more abundant liver nodules in CD44ICD group and a far smaller quantity in the miR-199a-5p group (,)), as well as the HE staining and immunohistochemistry analysis for CD44 and Sirt1 expression in the metastatic nodular tissues ()). Interestingly, the miR-199a-5p group regained tumor migration ability by co-injecting the CD44ICD adenovirus (–)). All these reuslts showed that miR-199a-5p supressed the cSCC cells xenograft tumor growth and metastasis, which could reversed by overexpreesion of CD44ICD.
Figure 6. MiR-199a-5p suppressed cSCC xenograft tumor growth and metastasis by targeting CD44ICD activation. (a) The tumor sizes were measured every five days for A431 xenografts treated with miR-199a-5p or CD44ICD overexpression adenoviruses or combination with the two. (b) Observation of the xenograft tumors separated from nude mice after 30-days-treatment as described in A (n = 5). (c) Weights of these xenograft tumors in each group nude mice as described in B (n = 5). (d) Q-PCR analysis for the expression of miR-199a-5p in the tumor tissues as described in B. (e) Western blot analysis for the expression of Sirt1, CD44 and CD44ICD in the tumor tissues as described in B. (f) Observation of the metastatic livers separated from A431 nude mice after 30-days-treatment with miR-199a-5p or CD44ICD overexpression or combination with the two. The arrows showed the metastatic nodules on the surface of liver tissues. (g) Counting of metastatic nodules on the liver surface from the nude mice as described in E. (h) HE staining and CD44, Sirt1 immunohistochemistry analysis of the matastastic liver from the nude mice as described in E (scale bar = 100μm). * p < 0.05, ** p < 0.01, *** p < 0.001.
Discussion
Accumulating studies have demonstrated that CD44 plays essential roles in tumorigenesis, tumor cell growth and metastasis, and transduces a variety of intracellular signals via sequential proteolytic cleavages in the ectodomain and intracellular domain. CD44+ cancer cells in SCCs tumors have been reported to possess stem cell-like properties, resembling higher abilities to self-renew and to differentiate [Citation4,Citation5,Citation30], and are closely related to a generalized resistance to apoptosis and chemotherapy. Previously, it has been shown that the molecular expression of cancer stemness markers, such as Oct4, Sox2 and Nanog, was induced by CD44ICD overexpression [Citation31]. The coordinated stemness factor networks drive cancer cell stemness in gastric carcinoma [Citation32], breast cancer [Citation33] and HNSCC [Citation34]. In consistence, we revealed in this study that CD44ICD positively correlated with cSCCSCs stemness by promoting stemness factor expression, increasing cancer cell proliferation and sphere formation. Interestingly, we observed that DAPT, a γ-secretase inhibitor which inhibits CD44 intracellular cleavage, not only prevented the release of CD44ICD, but also tended to inhibit the CD44 expression, which might be attributed to the fact that the released CD44ICD may translocate into the nucleus and enhance the intact CD44 expression [Citation35,Citation36].
Differentially expressed miRNAs have been highlighted as important guide molecules in cancer development. MiR-199a-3p and miR-199a-5p are produced by the same miR-199a precursor and share a high degree of sequence similarity. We previously reported that miR-199a functioned as an anti-tumor molecule in cSCC by targeting the CD44 expression [Citation37]. There is evidence that miR-199a-3p was under-expressed in CD44+ prostate cancer cells [Citation38], and overexpression of miR-199a-3p inhibited the stem cell-like properties, including self-renewing and tumor-initiating by directly targeting CD44 [Citation39]. However, the interactive roles of miR-199a-5p and CD44 in tumor progression remains elusive. In the present study we focus on the anti-tumor function of miR-199a-5p and identified Sirt1 as a downstream target for miR-199a-5p in cSCCSCs. In contrast to miR-199a-5p, we detected a relative high expression of Sirt1 in cSCCSCs, which was efficiently inhibited by miR-199a-5p overexpression. Interestingly, miR-199a-5p overexpression or Sirt1 knockdown decreased stemness properties in cSCCSCs, and the cells displayed reduced expression of a set of stemness markers as well as impaired sphere-forming ability. We demonstrated that miR-199a-5p-Sirt1 interactions played a key role in cSCCSCs stemness regulation. On the other hand, our finding that Sirt1 was directly down-regulated by miR-199a-5p in cSCCSCs might help us understand, in part, how the cellular activity of Sirt1 was controlled at transcriptional level, which remained unclear for a long time [Citation40].
We also found that Sirt1 knockdown repressed CD44 cleavage and CD44ICD nuclear translocation, and potentially inhibited cSCCSCs stemness. Sirt1 is an NAD-dependent deacetylase and known to play important roles in human cancers by serving as either tumor suppressor or promoter under specific conditions [Citation41]. Particularly, a variety of transcriptional factors were reported to act as Sirt1 substrates, and their posttranslational acetylation was altered in various cancers. We assumed that the inhibitory effect of Sirt1 knockdown on CD44 cleavage and CD44ICD nuclear translocation could be attributed to its deacetylation function. Similarly, a previous study showed that internalized CD44 enhanced acetylation of the transcription factor STAT3, formed a complex with STAT3 and p300 (acetyltransferase), and co-migrated to the nucleus [Citation8]. The complex bound to the cyclin D1 promoter and increased cell proliferation by activating cyclin D1 expression. No association between CD44 and unacetylated STAT3 was observed, suggesting that the cellular function of CD44 could be greatly acetylase or deacetylase dependent. However, whether and how other molecules are interacting with Sirt1 and CD44 or CD44ICD in cSCCSCs remains an open question. More intriguingly, our in vitro and in vivo experiments demonstrated that Sirt1 expression was efficiently inhibited by miR-199a-5p overexpression, resulting in effectively reduced CD44ICD expression and stemness characteristics. Together, our results concluded that the miR-199a-5p directly targeted the deacetylase Sirt1 and played a key regulation role in the CD44 mediated signal transduction processes in cSCC.
In summary, we examined the expression and function of miR-199a-5p in CD44+ human cSCC, and unveiled for the first time a miR-199a-5p/Sirt1/CD44ICD signaling pathway. By clarifying the cellular interactions among these molecules and their regulation mechanisms, our study provided theory insights for CSCs properties in cSCC and shielded important light on therapeutic development homing in on this cancer subpopulation.
Ethics statement
Animal experiments were performed according to the institutional ethical guidelines from the Institutional Animal Care and Use Committee of Xiangya Hospital affiliated to Central South University.
Supplemental Material
Download TIFF Image (1.9 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
SUPPLEMENTAL DATA
Supplemental data for this article can be accessed at here.
Additional information
Funding
References
- Dotto GP, Rustgi AK. Squamous cell cancers: a unified perspective on biology and genetics. Cancer Cell. 2016;29(5):622–637.
- Nicolis SK. Cancer stem cells and “stemness” genes in neuro-oncology. Neurobiol Dis. 2007;25(2):217–229.
- Faber A, Goessler UR, Hoermann K, et al. SDF-1-CXCR4 axis: cell trafficking in the cancer stem cell niche of head and neck squamous cell carcinoma. Oncol Rep. 2013;29(6):2325–2331.
- Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104(3):973–978.
- Zhao J-S, Li W-J, Ge D, et al. Tumor initiating cells in esophageal squamous cell carcinomas express high levels of CD44. PLoS One. 2011;6(6):e21419.
- Kajita M, Itoh Y, Chiba T, et al. Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J Cell Biol. 2001;153(5):893–904.
- Murakami D, Okamoto I, Nagano O, et al. Presenilin-dependent gamma-secretase activity mediates the intramembranous cleavage of CD44. Oncogene. 2003;22(10):1511–1516.
- Lee JL, Wang MJ, Chen JY. Acetylation and activation of STAT3 mediated by nuclear translocation of CD44. J Cell Biol. 2009;185(6):949–957.
- Screaton GR, Bell MV, Jackson DG, et al. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc Natl Acad Sci U S A. 1992;89(24):12160–12164.
- Ponta H, Herrlich P. The CD44 protein family: roles in embryogenesis and tumor progression. Front Biosci. 1998;3:d650–656.
- Herold-Mende C, Seiter S, Born AI, et al. Expression of CD44 splice variants in squamous epithelia and squamous cell carcinomas of the head and neck. J Pathol. 1996;179(1):66–73.
- Thorne RF, Legg JW, Isacke CM. The role of the CD44 transmembrane and cytoplasmic domains in co-ordinating adhesive and signalling events. J Cell Sci. 2004;117(Pt(3)):373–380.
- Senbanjo LT, Chellaiah MA. CD44: A multifunctional cell surface adhesion receptor is a regulator of progression and metastasis of cancer cells. Front Cell Dev Biol. 2017;5:18.
- Okamoto I, Kawano Y, Murakami D, et al. Proteolytic release of CD44 intracellular domain and its role in the CD44 signaling pathway. J Cell Biol. 2001;155(5):755–762.
- Yanaihara N, Caplen N, Bowman E, et al. Unique MicroRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198.
- Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179.
- O’Donnell KA, Wentzel EA, Zeller KI, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843.
- Jiang J, Gusev Y, Aderca I, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14(2):419–427.
- Wang S-H, Zhou J-D, He Q-Y, et al. MiR-199a inhibits the ability of proliferation and migration by regulating CD44-Ezrin signaling in cutaneous squamous cell carcinoma cells. Int J Clin Exp Pathol. 2014;7(10):7131–7141.
- Shin AN, Han L, Dasgupta C, et al. SIRT1 increases cardiomyocyte binucleation in the heart development. Oncotarget. 2018;9(8):7996–8010.
- Lammich S, Okochi M, Takeda M, et al. Presenilin-dependent intramembrane proteolysis of CD44 leads to the liberation of its intracellular domain and the Secretion of an Aβ-like peptide. J Biol Chem. 2002;277(47):44754–44759.
- Jiang LY, Zhang XL, Du P, et al. gamma-secretase inhibitor, dapt inhibits self-renewal and stemness maintenance of ovarian cancer stem-like cells in vitro. Chin J Cancer Res. 2011;23(2):140–146.
- Li LC, Wang DL, Wu YZ, et al. Gastric tumor-initiating CD44(+) cells and epithelial-mesenchymal transition are inhibited by gamma-secretase inhibitor DAPT. Oncol Lett. 2015;10(5):3293–3299.
- Yu J, Auwerx J. Protein deacetylation by SIRT1: an emerging key post-translational modification in metabolic regulation. Pharmacol Res. 2010;62(1):35–41.
- Bradbury CA, Khanim FL, Hayden R, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19(10):1751–1759.
- Huffman DM, Grizzle WE, Bamman MM, et al. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67(14):6612–6618.
- Stunkel W, Peh BK, Tan YC, et al. Function of the SIRT1 protein deacetylase in cancer. Biotechnol J. 2007;2(11):1360–1368.
- Hida Y, Kubo Y, Murao K, et al. Strong expression of a longevity-related protein, SIRT1, in Bowen’s disease. Arch Dermatol Res. 2007;299(2):103–106.
- Rane S, He M, Sayed D, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104(7):879–886.
- Facompre N, Nakagawa H, Herlyn M, et al. Stem-like cells and therapy resistance in squamous cell carcinomas. Adv Pharmacol. 2012;65:235–265.
- Cho Y, Lee H-W, Kang H-G, et al. Cleaved CD44 intracellular domain supports activation of stemness factors and promotes tumorigenesis of breast cancer. Oncotarget. 2015;6(11):8709–8721.
- Matsuoka J, Yashiro M, Sakurai K, et al. Role of the stemness factors sox2, oct3/4, and nanog in gastric carcinoma. J Surg Res. 2012;174(1):130–135.
- Gwak JM, Kim M, Kim HJ, et al. Expression of embryonal stem cell transcription factors in breast cancer: oct4 as an indicator for poor clinical outcome and tamoxifen resistance. Oncotarget. 2017;8(22):36305–36318.
- Habu N, Imanishi Y, Kameyama K, et al. Expression of Oct3/4 and Nanog in the head and neck squamous carcinoma cells and its clinical implications for delayed neck metastasis in stage I/II oral tongue squamous cell carcinoma. BMC Cancer. 2015;15:730.
- Wang Z, Zhao K, Hackert T, et al. CD44/CD44v6 a reliable companion in cancer-initiating cell maintenance and tumor progression. Front Cell Dev Biol. 2018;6:97.
- Cui W, Ke JZ, Zhang Q, et al. The intracellular domain of CD44 promotes the fusion of macrophages. Blood. 2006;107(2):796–805.
- Wang S, CAO KE, He Q, et al. miR-199a-5p induces cell invasion by suppressing E-cadherin expression in cutaneous squamous cell carcinoma. Oncol Lett. 2016;12(1):97–101.
- Liu R, Liu C, Zhang D, et al. miR-199a-3p targets stemness-related and mitogenic signaling pathways to suppress the expansion and tumorigenic capabilities of prostate cancer stem cells. Oncotarget. 2016;7(35):56628–56642.
- Henry JC, Park J-K, Jiang J, et al. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2010;403(1):120–125.
- Yamakuchi M. MicroRNA regulation of SIRT1. Front Physiol. 2012;3:68.
- Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009;5(2):147–152.
