ABSTRACT
Circular RNAs (circRNAs) correlate with cancer cell phenotypes. Particularly, circRNAs mediate the cancer process as microRNAs (miRNAs) sponges. This study was to ascertain the roles of hsa_circ_0009910 in phenotypic aspects of ovarian cancer cells. Mantel-Cox test was performed to analyze the correlation between hsa_circ_0009910 and survival outcomes of ovarian cancer. Minigene reporter was constructed and small interfering-RNA was designed for constructing hsa_circ_0009910-dysregulated and miR-145-upregulated cells identified by qRT-PCR. Proliferative and motile activities were monitored by CCK and Transwell. Western blot was applied for quantification of cyclin D1, CDK4, CDK6, MMP-2, MMP-9, IκBα, p65, Notch1, Hes1, and Hes5. miRNAs targets were predicted using a bioinformatics tool and confirmed using qRT-PCR and Dual-Luciferase reporter assay. Hsa_circ_0009910 was correlated with the poor prognosis of ovarian cancer patients. The ovarian cancer cell phenotypes were promoted by hsa_circ_0009910 while repressed by silencing hsa_circ_0009910. Hsa_circ_0009910 silence was responsible for the upregulation of the predicted miRNAs targets. Thereinto, miR-145 was confirmed as a miRNA target and negatively regulated by hsa_circ_0009910. miR-145 nullified the biological function of hsa_circ_0009910 in the proliferative and motile phenotypes, and the active status of NF-κB and Notch. Hsa_circ_0009910, representing unfavorable prognosis, induced proliferative and motile phenotypes by suppressing miR-145 in ovarian cancer cells.
Introduction
Ovarian cancer is the most lethal gynecological cancer, originating from germ cells, stromal cells or epithelial cells. According to the statistics from the World Health Organization, there will be appropriately 295,414 newly diagnosed ovarian cancer cases, and 184.799 deaths are estimated to happen in 2018 [Citation1]. The improvement in prevention and early screening manifests the promise in translating ovarian cancer into a controllable malignancy [Citation2]. Currently, two prominent extrapolations merge to interpret the pathogenesis of ovarian cancer: the incessant ovulation hypothesis [Citation3] and the gonadotropin hypothesis [Citation4]. However, to address the etiology of ovarian cancer, a molecular evaluation of the effects of non-protein coding RNA is required to account for the proliferative and motile phenotypes which are unmanageable in cancer process [Citation5,Citation6,].
Compelling data suggested that the pathological alteration of several circular RNAs (circRNAs) observed in patients diagnosed as ovarian cancer might be implicated in the development of this malignancy [Citation7,Citation8,]. The research for the functional underpinnings has revealed circRNAs as dynamic scaffolding structures for protein interactions [Citation9] and endogenous sponges for microRNAs (miRNAs) [Citation10]. What’s more, the biogenetic process of circRNAs per se is an approach to modulate gene expression through competing with pre-mRNA splicing [Citation11]. The disorder is confirmed to be different between exome and whole genome [Citation12]. Hsa_circ_0009910, derived from MFN2, locates on chr1:12049221–12052747 with 3526 nt genomic length and 315 nt spliced sequence length (its bioinformatics are available on http://circrna.org/cgi-bin/simplesearch.cgi). Here, we focused on its role as a miRNA sponge in cellular phenotypes.
The oncogenic potency of hsa_circ_0009910 has been validated in acute myeloid leukemia, gastric cancer, and osteosarcoma [Citation13–Citation15], and the studies confirmed that hsa_circ_0009910 regulates the cellular phenotype aspects through soaking up miR-20a-5p and miR-449a [Citation13,Citation15,]. It remains enigmatic about the possibility that hsa_circ_0009910 possesses the miRNA-binding element for the specific miRNA or the universal miRNAs. Definitely, it is necessary to assess this feasibility because the ovarian cancer cell phenotypes are regulated by a host of miRNAs [Citation16–Citation18]. Nowadays, bioinformatics tools have been developed for functional predictions and association analysis, such as circBase, circInteractome, circNet, etc., which are elaborately summarized by Dori and Bicciato [Citation19]. In our study, we predicted miRNAs targets of hsa_circ_0009910 using the bioinformatics tools.
Our study aimed to ascertain the role of hsa_circ_0009910 in the phenotypes of ovarian cancer cells, as well as a potential mechanism related to miR-145. The significant role of hsa_circ_0009910 in synergy with miR-145 proved in this study might be applied for the treatment of ovarian cancer. However, further clinical confirmation is required.
Materials and methods
Collection of human ovarian cancer tissues
To investigate the expression of circ_0009910 in human ovarian cancer, 50 pairs of ovarian cancer tissues and corresponding normal ovarian tissues were obtained from patients in Shengli Oilfield Central Hospital (Dongying, China). None of the patient received surgical treatment and therapies before this study. The normal tissues were excised far from the center of the tumor. After surgery, the tissue samples were freshly snap-frozen in liquid nitrogen and stored at −80°C for qRT-PCR analysis. Clinicopathological parameters of the patients are shown in . The present study was approved by the Medical Ethics Committee of Shengli Oilfield Central Hospital. All patients signed the informed consents before tissue collection.
Table 1. The clinicopathological parameters of patients with ovarian cancer enrolled in this study.
Cell culture
SKOV3 human ovarian carcinoma cells (ATCC HTB-77, Rockville, MD, USA) were seed on Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (FBS), 0.1 mg/mL streptomycin, 100 U/mL penicillin (all from Sigma-Aldrich, St. Louis, MO, USA). The cells were maintained in a humidified condition with 5% CO2 at 37°C.
Cell transfection
To induce the overexpression of hsa_circ_0009910, 5ʹ-flanking genomic sequence (~1 kb), sequence forming circRNA, and reverse complementary sequences of 5ʹ-flanking genomic sequence (~1 kb) were constructed into pcDNA3.1 plasmid (Invitrogen, Carlsbad, CA, USA) through enzyme digestion and ligation. To silence hsa_circ_0009910, hsa_circ_0009910 siRNA (si-circ_0009910) was transfected into SKOV3 cells. Scramble served as a control siRNA. miR-145 upregulated transfectants were obtained by incubating SKOV3 cells with miRNA mimics for miR-145 and miRNA negative control (miR-NC) (Dharmacon, Lafayette, CO, USA). The sequences of siRNA for hsa_circ_0009910, miR-145 mimic, and the corresponding negative control was suggested as follows: 5ʹ-GGCTTTTTTTGGCCGCGCAAT-3ʹ for si-circ_0009910, 5ʹ-GGACCGCGCCCAGTCTTAACT-3ʹ for si-NC, mimic sense 5ʹ-GUCCAGUUUUCCCAGGAAUCCCU-3ʹ and mimic antisense 5ʹ-GGAUUCCUGGGAAAACUGGACUU-3ʹ for miR-145 mimic, and mimic sense 5ʹ-AUGGAGUCCAGGGUUUACACUGU-3ʹ and mimic antisense 5ʹ-GCAAGGGAAUCCUUCUGGACGGA-3ʹ for miR-NC. All transfection was performed using Lipofectamine 2000 (Invitrogen) as transfection reagent according to the supplier’s protocol. Transfection efficiency at 72 h after transfection was confirmed by qRT-PCR with U6 or GAPDH genomic DNA as endogenous loading reference for miRNA or circRNA. All the sequences in this study have been provided in supplementary materials.
Viability assay
After transfection, the cells were collected and 5,000 cells were suspended in 100 μL medium followed by inoculation into a hole of 96-well plates. The cells were cultured and adhered to the wall for 48 h. The culture was added with 10 μL of CCK solution (Abmole Bioscience, Houston, TX, USA) according to the manufacturer’s specifications, and the reaction system was maintained for 3 h. The absorbance was detected using a microplate reader (Molecular devices, San Jose, CA, USA) at 450 nm. The viability from three independent examinations was expressed as percentage of the viability of transfected cells to control cells.
Transwell assay
The transfected cells were collected in serum-free culture medium and placed onto the upper chamber of 24-well HST Transwell Permeable Support with 0.4 μm Pore Polycarbonate Membrane (Corning, Cambridge, NY, USA). The lower side of the chamber was supplemented with 600 μL of culture medium containing 10% FBS. At 48 h after incubation, the culture medium was removed from both compartments, and the cells were fixed in methanol. The cells in the upper side were wiped away using cotton swab. The migrated cells in the lower side were stained using crystal violet. The cells were stained by 0.1% crystal violet for 30 min at room temperature. The stained cells were counted under a microscope (Olympus, Tokyo, Japan). To examine the invasive activity, Matrigel medium-coated membrane was applied.
Total RNA isolation and qRT-PCR analysis
Total RNA in tissues or cells was isolated using TRIzol reagent according to the product’s manual (Invitrogen). The content of RNA was detected using Nanodrop (Thermo Scientific Fisher, Waltham, MA, USA). The obtained RNA was purified using miRNEasy spin columns (QIAGEN, Hilden, Germany) according to the product’s manual. For miRNAs, the purified RNA was subjected to reverse transcription using SuperScript Master Mix (Invitrogen). qRT-PCR was performed using SYBR Green qPCR Master Mix (Thermo Fisher Scientific) on 7900 HT sequence detection system (Applied Biosystems, Foster City, CA, USA). For hsa_circ_0009910, the RNA extract was incubated with RNase R (Epicenter, Madison, WI, USA) before RNA purification, followed by reverse transcription and qRT-PCR analysis. U6 and GAPDH spike-in molecules were supplemented in reaction system for miRNAs and hsa_circ_0009910 normalization. Triplicates of each experiment were performed.
Dual-luciferase reporter assay
To construct hsa_circ_0009910-firefly luciferase plasmid, the sequence of spliced sequence of hsa_circ_0009910 was amplified using genomic DNA from SKOV3 cells. The wild type PCR product (WT) was constructed into the pGL4.20[luc2/Puro] vector (Promega, Madison, WI, USA) (pGL-luc2/circ-WT). To establish the mutant of hsa_circ_0009910 (circ-MUT), site-directed mutagenesis was performed using QuickChange II XL Site-Directed Mutagenesis Kit (Agilent, Palo Alto, CA, USA). For reporter assay, HEK-293 cells (ATCC CRL-1573) were co-transfected with pGL-luc2/circ-WT or pGL-luc2/circ-MUT and pGL4.79[hRluc/Neo] vector (Promega) as a co-reporter. After selection, miR-145 mimic or miR-NC was introduced into the stable transfectants using Lipofectamine 2000 (Invitrogen). After incubation, the bioluminescence was detected using dual-luciferase assay system (Promega) under a GloMax20/20 luminometer (Promega).
Protein extraction and western blot assay
After transfection, the cells were washed in cold phosphate-buffered saline, followed by lysed in RIPA lysis buffer (Beyotime, Shanghai, China). Equal amounts of proteins quantified by BCA protein assay kit (Pierce, Appleton, WI, USA) were electrophoresed to separate interest proteins. Polyvinylidene difluoride (GE Healthcare, Pittsburg, PA, USA) membrane carrying protein blots were incubated with 5% bovine serum albumin (BSA) (Thermo Scientific Fisher) for 1 h at room temperature. The primary antibodies used contained anti-cyclin D1 (1:1,000) (Millipore #04-221, Bedford, MA, USA), anti-CDK4 (1:1,000) (Abcam #ab68266, Cambridge, MA, USA), anti-CDK6 (1:1,000) (CST #3136, Danvers, MA, USA), anti-MMP-2 (1:1,000) (CST #87809), anti-MMP-9 (1:1,000) (Abcam #ab38898), anti-phospho-IκBα (Ser32) (1:1,000) (CST #2859), anti-IκBα (1:1,000) (CST #4814), anti-phospho-p65 (Ser536) (1:1,000) (CST #3031), anti-p65 (1:1,000) (CST #8242), anti-Notch1 (1:1,000) (Santa Cruz Biotechnologies #sc-6014, Santa Cruz, CA, USA), anti-Hes1 (1:1,000) (CST #11988), anti-Hes5 (1:1,000) (Abcam #ab25374) and anti-β-actin (1:1,000) (CST #4967). The incubation with the abovementioned primary antibodies was carried out at 4°C overnight. The antibodies were prepared in blocking buffer. Goat anti-rabbit IgG marked by horseradish peroxidase (1:5,000) (Abcam #ab6721) was used to probe primary antibodies. The incubation continued for 1 h. ImageJ software system (National Institutes of Health, Bethesda, MD, USA) was used to scan and signalize the protein bands.
Statistical analysis
All data were expressed as mean ± standard deviation. Comparison was carried out using GraphPad Prism 8.0 software (GraphPad, San Diego, CA, USA) with analytical methods Student’s t test and one-way analysis of variance followed by Bonferroni’s test. Statistical significance was indicated by the P-values (*P < 0.05, **P < 0.01, and ***P < 0.001). According to the level of hsa_circ_0009910, the specimens were divided into two groups (Youden’s index): low level (n = 23) and high level (n = 27). The comparison between the survival curves was carried out using Mantel-Cox test.
Results
Hsa_circ_0009910 and prognosis analysis of patients with ovarian cancer
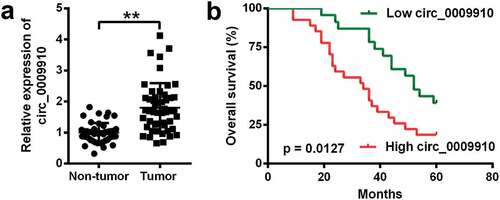
The ovarian cancer tissues were acquired from 50 patients with ovarian cancer to assess the level of hsa_circ_0009910; the detection of hsa_circ_0009910 was done using qRT-PCR analysis. We observed hsa_circ_0009910 was universally deposited in ovarian cancer tissues relative to the corresponding normal tissues (P < 0.01) ()). Based on the expression of hsa_circ_0009910 in tumor tissues, we described the survival curves and compared the difference of prognosis. Results from Mantel-Cox test unraveled that the difference in hsa_circ_0009910 expression was conspicuous between ovarian cancer tissues and para-carcinoma tissues (P = 0.0127) ()). Thus, hsa_circ_0009910, which was accumulated in ovarian cancer tissues, represented the unfavorable prognosis.
Figure 1. Human ovarian tissues accumulated hsa_circ_0009910 and hsa_circ_0009910 indicated the unfavorable prognosis. (a) Expression of hsa_circ_0009910 in 50 pairs of ovarian cancer tissues and adjacent normal tissues by qRT-PCR. (b) Kaplan–Meier Survival curves were depicted according to hsa_circ_0009910 expression and the significant difference of the curves was tested by Mantel-Cox method. **P < 0.01.
Effects of hsa_circ_0009910 on cellular phenotypes associated with proliferation and motility
To detect the functional effects of hsa_circ_0009910 in the cellular phenotypes of ovarian cancer cells, episomal hsa_circ_0009910 was artificially upregulated in SKOV3 cells (P < 0.001) ()); genomic hsa_circ_0009910 was silenced by siRNA (P < 0.01) ()). The transfectants were thereafter subjected to cascades of analysis at cellular and molecular levels. The results showed hsa_circ_0009910 overexpression increased the viability of SKOV3 cells (P < 0.01) ()), whereas there was a decrease in the viability when hsa_circ_0009910 expression was blocked by its siRNA (P < 0.01) ()). At the molecular level, the protein expression of proliferative markers such as cyclin D1 (P < 0.001), CDK4 (P < 0.001) and CDK6 (P < 0.001) were notably enhanced in the cells overexpressing hsa_circ_0009910 after transfected with pcDNA3.1 harboring hsa_circ_0009910 relative to empty vector (), while the proliferative markers were observed to be conspicuously decreased in the cells transfected with hsa_circ_0009910 siRNA compared with scramble (P < 0.05, P < 0.01) ().
Figure 2. Overexpression of hsa_circ_0009910 caused changes in cellular phenotypes associated with proliferation and motility. (a) qRT-PCR analysis of hsa_circ_0009910 levels in SKOV3 cells transfected with pcDNA3.1 carrying genomic sequence coding hsa_circ_0009910 or hsa_circ_0009910 siRNA. Vector and scramble served as control. GAPDH was used for qRT-PCR normalization. (b) CCK-8 assay examined the percentage of viable cells after transfection. (c–d) Western blot analysis was carried out for molecular change in cyclin D1, CDK4 and CDK6. (e–f) Migratory and invasive cells were counted under flow cytometry after crystal violet staining. (g–h) Western blot analysis was used for the detection of MMP-2 and MMP-9. β-Actin was used for protein normalization. Error bars were standard deviation from three independent experiments. P-values were calculated using student’s t test or one-way analysis of variance. *P < 0.05, **P < 0.01, ***P < 0.001.
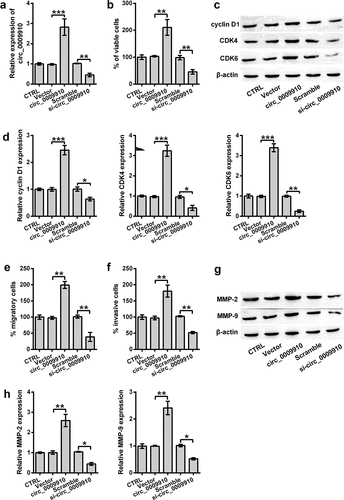
In addition, we further ascertained the involvement of hsa_circ_0009910 in migration and invasion. At the cellular level, hsa_circ_0009910 overexpressed cells showed an activity in migration and invasion (P < 0.01) (), while hsa_circ_0009910 deficient cells were repressed to migrate or invade (P < 0.01) (). At the protein level, the intracellular levels of MMP-2 and MMP-9 were incremented in hsa_circ_0009910 overexpressed cells (P < 0.01) while decremented in hsa_circ_0009910 silenced cells (P < 0.05) (). Consequently, we concluded that the upregulation of hsa_circ_0009910 was associated with the proliferative and motional capacities of ovarian cancer cells.
Identification and expression of miRNAs targeted by hsa_circ_0009910
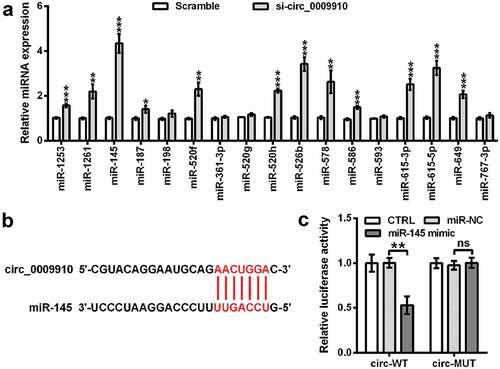
Next, the bioinformatics tool circInteractome (https://circinteractome.nia.nih.gov/) was used to predict the target miRNAs of hsa_circ_0009910. The analyzer identified a multitude of miRNAs as the major targets of hsa_circ_0009910, including miR-1253, miR-1261, miR-145, miR-187, miR-198, miR-520 f, miR-361-3p, miR-520 g, miR-520 h, miR-526b, miR-578, miR-586, miR-593, miR-615-3p, miR-615-5p, miR-649, and miR-767-3p. To validate the modulatory role of hsa_circ_0009910, SKOV3 cells were enforced to underexpress hsa_circ_0009910, followed by qRT-PCR analysis for the level of these target miRNAs. We found hsa_circ_0009910 silence obviously resulted in the accumulation of miR-1253, miR-1261, miR-145, miR-187, miR-520 f, miR-520 h, miR-526b, miR-578, miR-586, miR-615-3p, miR-615-5p, miR-649, and miR-767-3p in SKOV3 cells (P < 0.05, P < 0.01, P < 0.001) ()). Since it was evident that miR-145 was enhanced in hsa_circ_0009910 silenced cells, we experimentally validated the complementary relationship between hsa_circ_0009910 and miR-145 based on the predicted sequence ()) using a dual luciferase reporter assay system. The results suggested that there was a notable decrease in luciferase activity in SKOV3 cells which were cotransfected with miR-145 mimic and pGL containing spliced sequence of hsa_circ_0009910 (P < 0.01) ()). The activity was not significantly affected in SKOV3 cells after transfection with miR-145 mimic and pGL containing mutant spliced sequence of hsa_circ_0009910 (P > 0.01) ()). Hence, the conclusive evidence was provided here that hsa_circ_0009910 directly targeted miR-145 to negatively mediate its expression.
Figure 3. Hsa_circ_0009910 silence mediated the expression of miRNAs and complementarily targeted miR-145. (a) qRT-PCR analysis of miRNAs levels in SKOV3 cells transfected with hsa_circ_0009910 siRNA with scramble as control. U6 was used for qRT-PCR normalization. (b) Sequence base-pairing between hsa_circ_0009910 and miR-145. (c) Dual-luciferase reporter assay for ascertaining the binding relationship between linear spliced hsa_circ_0009910 and miR-145. Error bars were standard deviation from three independent experiments. P-values were calculated using student’s t test or one-way analysis of variance. nsP > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001.
Antagonistic effects of miR-145 against hsa_circ_0009910 on the phenotypes of ovarian cancer cells
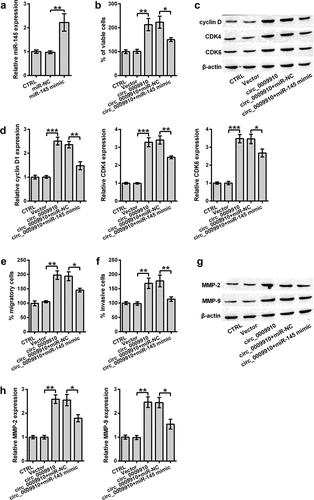
Although miR-145 was confirmed as a target miRNA of hsa_circ_0009910, studies were required to understand the role of miR-145 in hsa_circ_0009910-mediated phenotypes of ovarian cancer cells. We firstly determined the expression of miR-145 in SKOV3 cells transfected with miR-145 mimic; this was carried out using qRT-PCR analysis. miR-145 was apparently deposited in the transfectants (P < 0.01) ()). Continually, the increment in the viability of hsa_circ_0009910-overexpressed SKOV3 cells was significantly counteracted due to miR-145 mimic transfection (P < 0.05) ()). Meanwhile, the results suggested that cyclin D1, CDK4 and CDK6 protein expression in hsa_circ_0009910 transfected cells were distinctly decreased after transfection with miR-145 mimic (P < 0.05, P < 0.01) (). Additionally, the cellular abilities of migration and invasion were inhibited by miR-145 irrespective of hsa_circ_0009910 overexpression; this result was further confirmed by the downregulation of MMP-2 and MMP-9 in the cells transfected with hsa_circ_0009910 and miR-145 (P < 0.05) (). Thus, we observed an antagonistic relationship between miR-145 and hsa_circ_0009910; hsa_circ_0009910 might induce the phenotypes of ovarian cancer cells through downregulating miR-145.
Figure 4. miR-145 antagonized hsa_circ_0009910 in modulating cellular phenotypes associated with proliferation and motility of SKOV3 cells. (a) qRT-PCR analysis of miR-145 levels in SKOV3 cells transfected with miR-145 mimic or miR-NC. U6 served as an internal control. (b) CCK-8 assay examined the percentage of viable cells after co-transfection with hsa_circ_0009910 and miR-145 mimic. Vector and miR-NC served as control. (c–d) Western blot analysis was carried out for molecular change in cyclin D1, CDK4 and CDK6. (e–f) Migratory and invasive cells were counted under flow cytometry after crystal violet staining. (g–h) Western blot analysis was used for the detection of MMP-2 and MMP-9. β-Actin was used for protein normalization. Error bars were standard deviation from three independent experiments. P-values were calculated using student’s t test or one-way analysis of variance. *P < 0.05, **P < 0.01, ***P < 0.001.
Modulation of NF-κB and notch pathways by hsa_circ_0009910 through managing miR-145
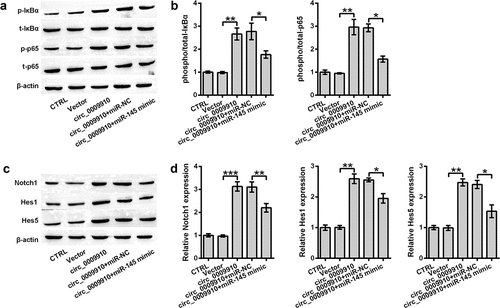
To assess whether NF-κB and Notch are responsive to hsa_circ_0009910, we examined the activation of IκBα, p65 and change in Notch1, Hes1 and Hes5; this was done using Western blot analysis. Hsa_circ_0009910 facilitated significant phosphorylation of IκBα and p65 (P < 0.01) () as well induced the abundance of Notch1, Hes1, and Hes5 (P < 0.01, P < 0.001) (). Reversely, the phosphorylation of IκBα and p65 or the activation of IκBα and p65 was blocked in the cells transfected with hsa_circ_0009910 and miR-145 mimic (P < 0.05) (); the same results were noticed in the expression of Notch1, Hes1 and Hes5 (P < 0.05, P < 0.01) (). Therefore, it was possible that hsa_circ_0009910 was responsible for signaling triggering or transduction in NF-κB and Notch pathways through downregulating miR-145.
Figure 5. miR-145 upregulation buffered the inhibitory role of hsa_circ_0009910 in signaling transduction of NF-κB and Notch pathways. Western blot analysis for signaling transducers of (a–b) NF-κB and (c–d) Notch pathways in SKOV3 cells co-transfected with miR-145 mimic and hsa_circ_0009910. β-Actin was used for protein normalization. Error bars were standard deviation from three independent experiments. P-values were calculated using student’s t test or one-way analysis of variance. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
The potential role of hsa_circ_0009910 has been identified in myeloid leukemia [Citation13], osteosarcoma [Citation15], and gastric cancer [Citation14] while its biological significance is not completely understood in ovarian cancer cells. In this study, the functional effects of hsa_circ_0009910 were confirmed in SKOV3 ovarian cancer cells. Furthermore, we investigated the involvement of miR-145 in the cellular phenotypes modulated by hsa_circ_0009910.
The accumulation of hsa_circ_0009910 has been reported in several types of cancers, including myeloid leukemia [Citation13], osteosarcoma [Citation15], and gastric cancer [Citation14]. These results have associated the accumulation of hsa_circ_0009910 with the poor prognosis of patients. Consistently, we noticed the enrichment of hsa_circ_0009910 in the tumor specimens from 50 patients with ovarian cancer, and the upregulation of hsa_circ_0009910 represented the unfavorable prognosis. These reports including our results showed that the changes of hsa_circ_0009910 expression in patients with ovarian cancer may be a reflection of pathological information. Pointedly, a cohort of circRNAs has been found to be dysregulated in tumor sites, and the individual expression of circRNAs indicates a distinctiveness of metastatic lesions and primary tumors [Citation20]. Thus, we hazarded a guess that hsa_circ_0009910 might pose advantages for cellular phenotypes associated with proliferation and motility.
As extrapolated, our results illustrated the physiological importance of hsa_circ_0009910 in proliferation as well as migration and invasion of ovarian cancer cells at cellular and molecular dimensions. Additionally, it has been presented that hsa_circ_0009910 knockdown arrested the myeloid leukemia cells at S stage and caused the progress of apoptosis [Citation13]. Knockdown of hsa_circ_0009910 reduced the proliferative activity and colony formation of gastric cancer cells [Citation14]. Silencing hsa_circ_0009910 caused the decrease in cyclin D1 and Bcl-2 while resulted in the upregulation of Bax in osteosarcoma cells [Citation15]. Besides, hsa_circ_0009910 silence was responsible for the switch in N-cadherin and E-cadherin expression as well as the downregulation of snail [Citation14]. The pathological changes of these markers might be accredited to circRNA-mediated dysregulation of miRNAs [Citation21–Citation24]. Thereafter, we predicted the target miRNAs of hsa_circ_0009910 on circular RNA interactome.
We observed a modulatory potency of hsa_circ_0009910 on miR-1253, miR-1261, miR-145, miR-187, miR-520 f, miR-520 h, miR-526b, miR-578, miR-586, miR-615-3p, miR-615-5p, miR-649, and miR-767-3p through associating with the binding elements. Particularly, the downregulation of miR-145 has been detected in serous and clear cell ovarian cancer specimens [Citation25]. The decrease in miR-145 reflects the unfavorable prognosis of patients with high-grade ovarian serous carcinomas [Citation26]. miR-145 possesses mRNA-binding elements of mRNAs coding for p70S6K1, MUC1 [Citation25], high-mobility group A2 [Citation26], metadherin [Citation27], Sp1, and CDK6 [Citation28]; these proteins are indispensable for proliferative and motile phenotypes of ovarian cancer cells. It has to be emphasized that hsa_circ_0009910 was firstly identified as a miRNA sponge soaking up miR-145 for mediating cellular phenotypes implicated in ovarian cancer process.
A cohort of studies has been carried out to ascertain the molecular mechanisms whereby circRNA endogenously interacts with cellular phenotypes which might be associated with the status of signaling cascades; constitutive active status of NF-κB contributes to the establishment of immune-evasive environment and induces tumor-promoting phenotypes [Citation29,Citation30,]; Notch activation promotes epithelial–mesenchymal transition and drove motile phenotypes of ovarian cancers [Citation31,Citation32,]. Here we found hsa_circ_0009910 silence caused the bluntness of signaling transduction in NF-κB and Notch pathways. Strikingly, the modulatory role of hsa_circ_0009910 has been validated in JAK/STAT signaling pathway; specifically, hsa_circ_0009910 silence fortified the phosphorylation of signaling transducers including JAK1 and STAT3 [Citation15]. Hsa_circ_0009910 emerges as a post-transcriptional mediator of miR-449a that operates through base pairing with the target sites; miR-499a downstream binds to 3ʹ-UTR of interleukin-6 receptor (IL-6 R) [Citation15]. IL6 R signaling participates in inducing the occurrence of active status of STAT3 during the peritoneal spread of high-grade ovarian carcinoma [Citation33].
The finding of this study unraveled that hsa_circ_0009910 deposited in ovarian cancer tissues represented the unfavorable outcome. Hsa_circ_0009910 induced tumor-promoting phenotypes through soaking up the target miRNAs such as miR-145. Hsa_circ_0009910-mediated miR-145 underexpression exhibited a therapy target for ovarian cancer therapy. This study proved the biological role of hsa_circ_0009910 in ovarian cancer cells. However, further studies are required to elaborate on how miR-145 is sponged by hsa_circ_0009910 and to ascertain whether the experimental results would reappear in vivo.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This research was ratified via the Medical Ethics Committee of the Shengli Oilfield Central Hospital (Dongying, China).
Supplemental Material
Download MS Word (18.7 KB)Disclosure statement
The authors declare that they have no conflict of interest.
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
- Cortez AJ, Tudrej P, Kujawa KA, et al. Advances in ovarian cancer therapy. Cancer Chemother Pharmacol. 2018;81:17–38.
- Casagrande JT, Louie EW, Pike MC, et al. “Incessant ovulation” and ovarian cancer. Lancet. 1979;2:170–173.
- Cramer DW, Welch WR. Determinants of ovarian cancer risk. II. Inferences regarding pathogenesis. J Natl Cancer Inst. 1983;71:717–721.
- Pandey R, Woo HH, Varghese F, et al. Circulating miRNA profiling of women at high risk for ovarian cancer. Transl Oncol. 2019;12:714–725.
- Marton E, Lukacs J, Penyige A, et al. Circulating epithelial-mesenchymal transition-associated miRNAs are promising biomarkers in ovarian cancer. J Biotechnol. 2019;297:58–65.
- Karedath T, Ahmed I, Al Ameri W, et al. Silencing of ANKRD12 circRNA induces molecular and functional changes associated with invasive phenotypes. BMC Cancer. 2019;19:565–581.
- Bao L, Zhong J, Pang L. Upregulation of circular RNA VPS13C-hsa-circ-001567 promotes ovarian cancer cell proliferation and invasion. Cancer Biother Radiopharm. 2019;34:110–118.
- Du WW, Zhang C, Yang W, et al. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7:4183–4191.
- Zhao Y, Alexandrov PN, Jaber V, et al. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer’s Disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes (Basel). 2016;7:116–124.
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66.
- Paramasivam N, Granzow M, Evers C, et al. Identification and prioritization of causal variants of human genetic disorders from exome or whole genome sequencing data. OBM Genet. 2018;2:017.
- Ping L, Jian-Jun C, Chu-Shu L, et al. Silencing of circ_0009910 inhibits acute myeloid leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol Dis. 2019;75:41–47.
- Liu M, Liu KD, Zhang L, et al. Circ_0009910 regulates growth and metastasis and is associated with poor prognosis in gastric cancer. Eur Rev Med Pharmacol Sci. 2018;22:8248–8256.
- Deng N, Li L, Gao J, et al. Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem Biophys Res Commun. 2018;495:189–196.
- Majem B, Parrilla A, Jimenez C, et al. MicroRNA-654-5p suppresses ovarian cancer development impacting on MYC, WNT and AKT pathways. Oncogene. 2019;38:6035–6050.
- Salem M, Shan Y, Bernaudo S, et al. miR-590-3p targets cyclin G2 and FOXO3 to promote ovarian cancer cell proliferation, invasion, and spheroid formation. Int J Mol Sci. 2019;20:1810–1826.
- Biamonte F, Santamaria G, Sacco A, et al. MicroRNA let-7g acts as tumor suppressor and predictive biomarker for chemoresistance in human epithelial ovarian cancer. Sci Rep. 2019;9:5668.
- Dori M, Bicciato S. Integration of bioinformatic predictions and experimental data to identify circRNA-miRNA associations. Genes (Basel). 2019;10:642–655.
- Ahmed I, Karedath T, Andrews SS, et al. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget. 2016;7:36366–36381.
- Caiment F, Gaj S, Claessen S, et al. High-throughput data integration of RNA-miRNA-circRNA reveals novel insights into mechanisms of benzo[a]pyrene-induced carcinogenicity. Nucleic Acids Res. 2015;43:2525–2534.
- Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun. 2018;495:2369–2375.
- Li B, Xie F, Zheng FX, et al. Overexpression of CircRNA BCRC4 regulates cell apoptosis and MicroRNA-101/EZH2 signaling in bladder cancer. J Huazhong Univ Sci Technolog Med Sci. 2017;37:886–890.
- Yuan Y, Liu W, Zhang Y, et al. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun. 2018;503:870–875.
- Wu H, Xiao Z, Wang K, et al. MiR-145 is downregulated in human ovarian cancer and modulates cell growth and invasion by targeting p70S6K1 and MUC1. Biochem Biophys Res Commun. 2013;441:693–700.
- Kim TH, Song JY, Park H, et al. miR-145, targeting high-mobility group A2, is a powerful predictor of patient outcome in ovarian carcinoma. Cancer Lett. 2015;356:937–945.
- Dong R, Liu X, Zhang Q, et al. miR-145 inhibits tumor growth and metastasis by targeting metadherin in high-grade serous ovarian carcinoma. Oncotarget. 2014;5:10816–10829.
- Zhu X, Li Y, Xie C, et al. miR-145 sensitizes ovarian cancer cells to paclitaxel by targeting Sp1 and Cdk6. Int J Cancer. 2014;135:1286–1296.
- Harrington BS, Annunziata CM. NF-kappaB signaling in ovarian cancer. Cancers (Basel). 2019;11:1182.
- Momeny M, Yousefi H, Eyvani H, et al. Blockade of nuclear factor-kappaB (NF-kappaB) pathway inhibits growth and induces apoptosis in chemoresistant ovarian carcinoma cells. Int J Biochem Cell Biol. 2018;99:1–9.
- Zhou J, Jain S, Azad AK, et al. Notch and TGFbeta form a positive regulatory loop and regulate EMT in epithelial ovarian cancer cells. Cell Signal. 2016;28:838–849.
- Chiaramonte R, Colombo M, Bulfamante G, et al. Notch pathway promotes ovarian cancer growth and migration via CXCR4/SDF1alpha chemokine system. Int J Biochem Cell Biol. 2015;66:134–140.
- Colomiere M, Ward AC, Riley C, et al. Cross talk of signals between EGFR and IL-6R through JAK2/STAT3 mediate epithelial-mesenchymal transition in ovarian carcinomas. Br J Cancer. 2009;100:134–144.
