ABSTRACT
It is hard to supply satellite cells as a cell source for therapy of muscle degenerative disease since the sampling of muscle tissue is very invasive to a patient with muscular disease. Direct conversion allows us to get specific cell types by transduction of defined transcriptional factors. To induce myogenic direct conversion, we transduced five transcriptional factors including Pax3, Sox2, Klf4, c-Myc, and Esrrb into mouse embryonic fibroblasts. We found that the transduction of the five transcriptional factors induced myogenic direct conversion of fibroblast. We revealed that the transduced cells with the five transcriptional factors were converted to myogenic lineage cells through a paraxial mesoderm-like stage. The expression level of myogenic-related genes of the transduced cells gradually increased as the passage increased. The induced myogenic lineage cells differentiated into muscle fibers in virto and in vivo. The current study revealed that the five transcription factors generated myogenic lineage cells from fibroblast via a paraxial mesoderm stage. The induced myogenic lineage cells may have a potential being applied as cell source for degenerative muscle disease.
Introduction
Since Yamanaka et al. found induced pluripotent stem cells (iPSC) in 2006, it has been revealed that certain transcriptional factors can determine cell fate [Citation1]. Direct conversion indicates that the defined transcriptional factors determine cell fate without passing the pluripotent stage [Citation2,Citation3]. The induction of defined transcriptional factors leads to direct conversion to specific cell types including neuron, neuronal stem cells, Schwann cells, cardiomyocytes, hepatocytes, and blood progenitor [Citation2–Citation10]. Various combinations were applied to get the direct conversion to specific cell types. The transduction of three transcriptional factors (Ascl1, Brn2, and Myt1 l) in fibroblasts induced direct conversion into neurons [Citation2]. Giedre et al. revealed that the combination of Fezf2, Brn2, and Myt1 l induced functional excitatory cortical neurons [Citation3]. The induction of Tbx5, Gata4, and Mef2 c in dermal fibroblasts triggered direct conversion to cardiomyocytes which function in vivo [Citation7].
However, there is a paucity in the literature of myogenic direct conversion. There are only a few studies on direct conversion into myogenic stem cells [Citation11–Citation13]. The heterogeneity of myogenic stem cells makes it hard to define the combination of transcriptional factors which is needed in myogenic direct conversion. Paired-box 3 (Pax3) is a key transcription factor expressed in muscle precursor cells and drives the onset of myogenesis in the embryo [Citation14,Citation15]. Pax3 activates myogenic regulatory factor (MRF) genes [Citation16,Citation17], such as Myf5, MyoD, and myogenin, which are essential for muscle specification [Citation18–Citation20]. In a previous study, ectopic expression of Pax3 is sufficient to induce MRF genes in the embryo and activate myogenesis in the absence of myogenic developmental signals [Citation21]. Similarly, the expression of exogenous Pax3 leads to the expression of MRF genes in several stem cells including embryonic stem (ES), iPSC, and mesenchymal stem cells (MSC), which results in myogenic differentiation [Citation22–Citation24]. However, the overexpression of Pax3 in fully differentiated somatic cells, not stem cells, did not induce myogenic differentiation [Citation24]. Thus, Pax3 is essential, but not sufficient to convert somatic cells into myogenic lineage cells. MyoD is one of the most potent transcriptional factors that regulate myogenic differentiation. The single transduction of MyoD induces myogenic differentiation in somatic cells, unlike Pax3 does not induce myogenesis in somatic cells [Citation25,Citation26]. However, the transduced cells with MyoD alone do not proliferate and self-renew [Citation27].
Recent studies revealed that the combination of several transcriptional factors induces myogenic conversion from somatic cells [Citation11–Citation13]. Ito et al. found that the combination of Pax3, Mef2b, Pitx1, Pax7, and MyoD leads to muscle progenitor cells from embryonic or adult fibroblasts [Citation11]. Also, our previous study revealed that the transduction of Six1, Eya1, Pax3, and Esrrb triggered stably expandable myogenic stem cells which have both self-renewal and myogenic differentiation capacity [Citation12]. Ori et al. revealed that the transduction of MyoD in fibroblasts with supplying of small molecules induced myogenic progenitor cells [Citation13]. However, there is few research on how somatic cells become muscle stem cells with defined factors.
We hypothesized that the combination of Pax3 with pluripotent transcription factors could reprogram somatic cells into myogenic lineage cells. Here, we combined Pax3 with Sox2, Klf4 c-Myc, and Esrrb to enhance myogenesis in mouse embryonic fibroblasts (MEF). The current study found that the combination of Pax3, Sox2, Klf4, cMyc, and Esrrb induced the direct myogenic conversion via paraxial mesoderm-like stage.
Results
Ectopic expression of PSKME induce dmyogenic gene expressions in mouse embryonic fibroblasts (MEF)
In order to induce myogenic conversion of MEF, we transduced five transcriptional factors; Pax3, Sox2, Klf4, c-Myc, and Esrrb which are involved in myogenesis or pluripotency ()). Each factor were designated as P, S, K, M, and E, respectively. Five to eight wk after transduction, the cells showed morphological changes. The cells were distinguished from the MEF. They were smaller than MEF. The cells had a distinct nucleus with scant cytoplasm. The cells formed a cell cluster. The cell clusters were isolated from adjacent cells ()). Among the isolated cell clusters, Myf5 positive clusters were selected by RT-PCR. The isolated clusters expressed all the exogenous genes including Pax3, Sox2, Klf4, c-Myc, and Esrrb both in mRNA and protein levels ()). We named the clusters as PSKME which are positive for Myf5 and expressing all the exogenous genes, Pax3, Sox2, Klf4, c-Myc, and Esrrb.
Figure 1. Myogenic reprogramming of MEF by Pax3, Sox2, Klf4, cMyc, and Esrrb (a) The process of induction of Pax3, Sox2, Klf4, cMyc, and Esrrb. The PSKME cells indicates MEF cells which are transduced by Pax3, Sox2, Klf4, cMyc, and Esrrb. The PSKME cells represents morphological changes including formation of cell cluster. The morphological changes assessed by bright-field microscopy. MEF, mouse embryonic fibroblast. Scale bar = 200 µm. (b) RT-PCR of exogenous transcriptional factors. The PSKME cells expressed all the five exogenous transcriptional factors. (c) Western blotting of exogenous transcriptional factors and Myf5. The PSKME cells expressed all the five exogenous transcriptional factors and Myf5 in protein level. (d) RT-PCR of myogenic related factors in early passage (P5). The PSKME cells expressed only Myf5 among myogenic-related genes. P5, passage 5. (e) RT-PCR of myogenic related factors in middle (P10) and late passages (P20). P10, passage 10; P20, passage 20. (f) RT-PCR of myogenic development-related genes. The PSKME cells express myogenic-related genes. c-Met, receptor of hepatocyte growth factor; Mesp2, mesoderm posterior 2; Eomes, eomesodermin; Tbx4, T-box transcription factor 4. (g) Representative images of FACS analysis of PDGFR-α and FLK1. The PSKME cells are PDGFRα+/FLK-1−, whereas MEF cells are negative for both, suggesting that the PSKME cells have characteristics of paraxial mesoderm. P5. PDGFR-α, platelet-derived growth factor receptor-α; FLK1, receptor of vascular endothelial growth factor.
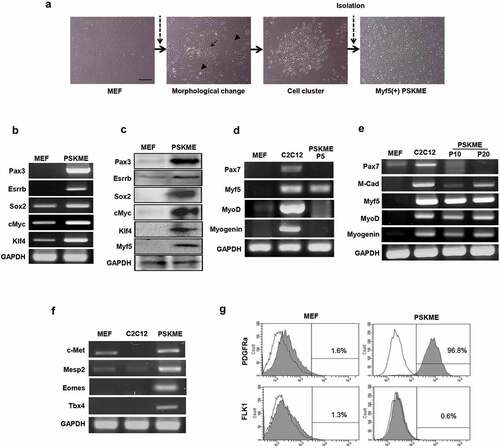
The early passage of PSKME expressed only Myf5, but not other muscle-related genes, such as Pax7, MyoD, and Myogenin ()). However, the PSKME started to express myogenic-related genes such as M-Cadherin, MyoD, and Myogenin when it reached to passage 10 (P10). Also, the expression levels of myogenic-related genes of PSKME increased in passage 20 (P20) when compared to it of P10. Interestingly, Pax7, which is one of the most important myogenic markers was not expressed even in late passaging in which other myogenic genes were expressed ()). In conclusion, the combination of transcriptional factors PSKME has converted MEF into the myogenic lineage.
The PSKME expressed paraxial mesoderm markers
We investigated gene expressions related to muscle development. PSKME cells expressed all the muscle development markers including c-Met, mesoderm posterior 2 (Mesp2), eomesodermin (Eomes), and T-box transcription factor 4 (Tbx4). Especially, PSKME cells exclusively expressed Tbx4, suggesting that the cells are myotome-like population, which are distinguished from MEF or C2C12 ()). FACS analysis indicates that most of the PSKME cells expressed platelet-derived growth factor receptor-α (PDGFR-α) (96.8%), while few MEF were positive for PDGFR-α (1.6%). In contrast, few PSKME expressed receptors of vascular endothelial growth factor (FLK-1) (0.6%). Thus, most of the PSKME cells were PDGFRα+/FLK-1− which are paraxial mesoderm markers ()). Therefore, these data suggested that the PSKME cells are distinct from MEF and have characteristics of muscle development, especially paraxial mesoderm stage.
iMLCs were established by clonal selection of PSKME cells
To establish induced myogenic lineage cell line from PSKME cells, we performed clonal selection using single-cell culture. For clonal selection, PSKME cells at passage 20 were stained with the antibody against the stage-specific embryonic antigen 3 (SSEA3) cell surface marker. SSEA3 antibody was used to reduce the heterogeneous PSKME and to isolate stem cell-like cells from PSKME cells. The PSKME cells had SSEA3 positive population (13.2%), and each SSEA3 positive cells were isolated into single cells by fluorescence-activated cell sorting (FACS). Each single cells were cultured in individual wells and monitored by live-cell images ()).
Figure 2. Clonal selection of PSKME cells and identification of iMLCs. (a) Representative images of sorted-single cells from SSEA3 positive PSKME cells (Blue line). The each single SSEA3 cell is plated on 96 well plate. (b) Phase-contrast images of single cells incubated in proliferation or myogenic differentiation media. Arrows indicate differentiated myofibers. Scale bar = 100 µm. (c) RT-PCR of myogenic-related genes of iMLCs incubated in proliferation or differentiation media. M, MEF; C, C2 C12; P, proliferation media; D, differentiation media. (d) Transgene silencing analysis. The PSKME expresses all the induced transcription factors. However iMLC show silencing of transduced genes. (e) Integration test. All the transgenes are integrated. SKM, Sox2, Klf4 and Esrrb.
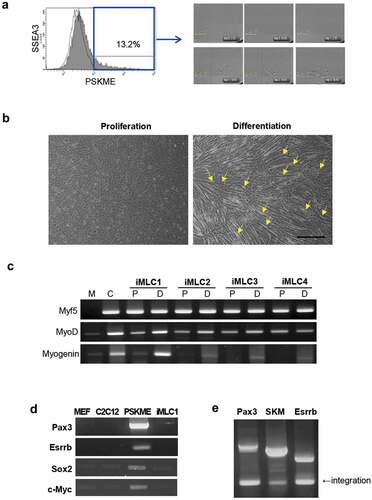
Finally, 17 clones were selected and propagated for further studies. The myogenic differentiation ability of these clones was evaluated by incubating them in myogenic differentiation media. Five to 10 d after incubation, the cells differentiated represented the characteristics of myogenic differentiation. On the observation under the bright-field microscope, the cells were elongated and fused together with other elongated cells. The differentiated cells had several nuclei in a row, which are called multinucleated fibers. The differentiated fibers showed an increase in width as well as length ()). The clones derived from PSKME cells have myogenic differentiation capacity named induced myogenic lineage cells (iMLCs).
In addition, we compared myogenic gene expression in these clones when cultured in proliferation or myogenic differentiation media. Myf5 and MyoD were expressed in all clones incubated in proliferation or differentiation medium. The expression of MyoD was increased in differentiation medium when compared that in proliferation medium, especially, in iMLC1, which represents prominent myotube formation. The expression of myogenin, a late myogenic differentiation marker, was increased in all the clones incubated in myogenic differentiation media, whereas the expression of myogenin was weak or absent in proliferation media ()).
The transgenes, including Pax3, Esrrb, Sox2, and c-Myc, were silenced in iMLCs, while all the transgenes were integrated into the genome, which indicate that the iMLC was reprogrammed completely. The integration test was performed on monocistronic vector expressing Pax3, polycistronic vector of Sox2, Klf4, and cMyc (SKM), and monocistronic vector expressing Esrrb (,e)). However, the PSKME cells expressed all the transgenes indicating that they are in the process of reprogramming ()). In addition, partial silencing was observed in some clones, named Non-iMLCs. The Non-iMLCs expressed some transgenes. The Non-iMLCs expressed less level of MyoD when compared to it of iMLCs showing complete silencing of transgenes (Fig S1).
These findings indicate that the PSKME cell is a mixture of completely reprogrammed myogenic cells and partially reprogrammed myogenic cells. For this reason, single-cell sorting should be performed to get the completely reprogrammed myogenic cells.
iMLCs represented the characteristics of muscle stem cells
To assess the ability of iMLC to proliferate as it continues to pass, we continued subculturing. The iMLC was stably expandable until it reached passage 40, and there was no difference between the growth rates in the early, middle, and late passages ()).
Figure 3. Characterization of iMLC. (a) Growth curve of iMLC according to passage. (b) Representative FACS profile of iMLC at passage 7. The white area indicates isotype control. The gray area indicate the percentage of positive cells against each cell surface antigens in iMLC. (c) qRT-PCR analysis of myogenic transcription factors of iMLC in proliferation (white bar) and differentiation (black bar) media. Error bars represent SEM from three replicates of three independent experiments. *p < 0.01. P, proliferating media; D, differentiation media. (d) Representative images of immunofluorescence staining of iMLCs. Scale bar = 250 µm. (e) The number of positive cells in immunofluorescence staining of i MLCs. The bar indicates the number of positive cells against each myogenesis markers per total cell number. Error bars represent SEM from three replicates of three independent experiments. P, proliferating media; D, differentiation media.
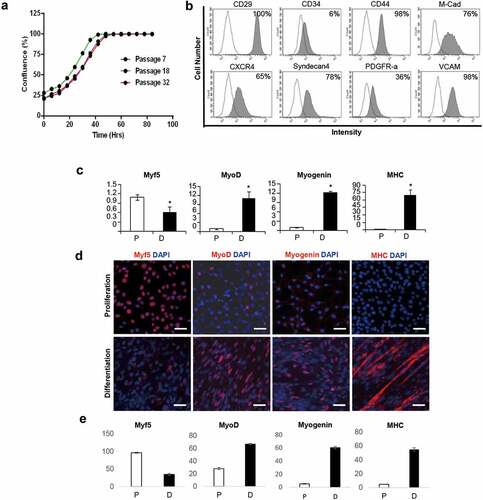
Most iMLCs were positive for CD29, CD44, M-cadherin, CXCR4, Syndecan-4, and VCAM, but a few cells also expressed CD34. And 36% of iMLC cells were positive for PDGFR-α ()). The cell surface markers of iMLC resembled that of Pax3-induced myogenic progenitor and satellite cells that express M-cadherin, syndecan-4, CD29, and CXCR4 [Citation22,Citation28–Citation30].
Next, we analyzed the expression levels of myogenic markers such as Myf5, MyoD, Myogenin, and myosin heavy chain (MHC) of the iMLCs which are incubated in proliferation media or differentiation media. In the proliferation media, the iMLC expressed a high level of Myf5 but low level of MyoD, Myogenin, and MHC. In the myogenic differentiation media, however, the expressions of MyoD, Myogenin, and MHC were significantly increased, which indicated that the iMLC can differentiate into muscle fibers in the myogenic differentiation media. In contrast, the expression of Myf5 was decreased in differentiation media ()).
The expressions of myogenic markers evaluated by immunofluorescence (IF) were the same as that by qRT-PCR. Most of the iMLCs incubated in proliferation media were positive for Myf5 (96.5 ± 1.6%), but only a small percentage of the cells were positive for middle to the terminal stage of myogenic differentiation markers including MyoD, Myogenin, and MHC (28.2 ± 1.6%, 5.4 ± 0.7% and 4.8 ± 0.3%). These expression patterns were totally adversed in iMLCs incubated in myogenic differentiation media. The iMLCs incubated in myogenic differentiation media expressed high levels of MyoD, Myogenin, and MHC (66.9 ± 1.1%, 60.5 ± 1.9%, and 54.9 ± 2.5%). The expression level of Myf5 was decreased in differentiation media (34.6 ± 3%) (,e)).
In conclusion, the proliferating iMLC expressed Myf5, but not myogenin and MHC. However, myogenin and MHC were strongly expressed in iMLC incubated in myogenic differentiation media.
iMLC differentiated into muscle fibers
We examined the capacity of iMLC to form myotubes in vitro and in vivo. The iMLCs were incubated in myogenic differentiation media for 12 d and stained against myogenin and troponin I which are the late myogenic differentiation markers. The iMLCs which were incubated in differentiation media showed all the characteristics of well-differentiated muscle fibers in vitro. The differentiated iMLCs had multinucleated fibers. The multinucleated fibers had 4 to 11 spindle-shaped nucleus, which were positive for myogenin in IF. The differentiated iMLCs represented elongation of cytoplasm, which were positive for Troponin I ()).
Figure 4. Myogenic differentiation potential of iMLC in vitro and in vivo. (a) Myogenic differentiation test in vitro. In differentiation media, iMLCs represented terminal stage of myogenic differentiation after incubated in myogenic differentiation media. The iMLCs are fused together and form multinucleated myotubes. Scale bar = 100 µm. (b) Myogenic differentiation test in vivo. The GFP-labeled iMLCs are transplanted into notexin-injured tibialis anterior muscles of nude (upper panel) or mdx mice (lower panel). The asterisks indicate codetection of GFP and dystrophin. Scale bars = 150 µm.
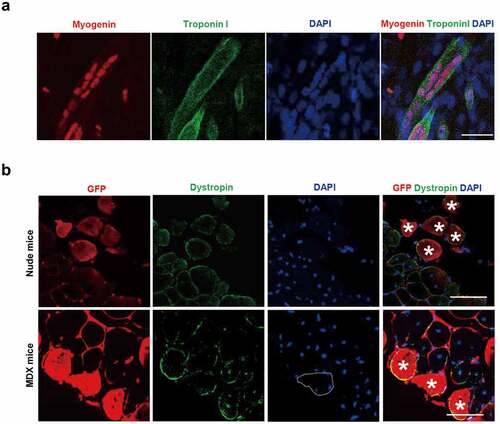
We then investigated the myogenic differentiation potential of iMLCs in vivo by transplanting GFP-tagged iMLCs into the tibialis anterior (TA) muscle of both nude mice and MDX mice. Four weeks after transplantation, the iMLC-transplanted TA muscles were analyzed to detect the co-expression of GFP and dystrophin. The GFP positive cells were observed in both nude and MDX mice indicating successful engraftment of iMLCs. Also, some of the GFP positive cells were positive for dystrophin, indicating that the transplanted iMLCs were differentiated into muscle fibers and showed terminal differentiation. Also, the cells which are positive for both GFP and dystrophin had peripheral nuclei, not a central nucleus, meaning that the cells were mature muscle fibers ()). These data demonstrated that iMLC has myogenic differentiation potential in vitro and in vivo.
Discussion
In this study, we revealed that the ectopic expression of Pax3, Sox2, Klf4, cMyc, and Esrrb induced myogenic lineage cells. The iMLCs by the ectopic expression of the five transcription factors can stably proliferate and differentiate into muscle fibers. The overexpression of the five transcriptional factors in MEF resulted in iMLCs after expressing paraxial mesoderm markers.
Myf5 is expressed in the major population of satellite cells and used for the identification and characterization of satellite cells. Myf5 has been known to control the myogenic differentiation of activated satellite cells [Citation31]. In addition, it plays an important role in myotome formation during embryonic myogenesis. In Myf5 mutant mice, myotome formation is delayed and initiates after MyoD expression [Citation32]. Thus, Myf5 is an early regulatory gene of myogenesis. Moreover, Myf5 is well known as a target of Pax3 [Citation22–Citation24]. The PSKME-transduced cells do not express Pax7 which plays an important role in early myogenesis (Fig, muscle development, and satellite-cell specification) [Citation33]. The overexpression of Pax3 induces Myf5 without going through Pax7 expression despite this important role of Pax7 in early myogenesis [Citation16]. That is the reason why PSKME transduced cell and iMLCs do not express Pax7, though Pax7 is an important early myogenic marker. From the perspective of expression of myogenic markers, the iMLCs are more like muscle progenitor cells such as myoblast, rather than muscle satellite cells. Pax3 has been known to induce Myf5 expression in stem cells such as induced pluripotent stem cells, embryonic cells, or mesenchymal stem cells [Citation22–Citation24]. Nevertheless, the overexpression of Pax3 alone cannot trigger the expression of Myf5 as much as that of myoblasts [Citation22]. It means that the ectopic expression of additional factors such as Sox2, KlF4, cMyc, and Esrrb is needed for MEF to express Myf5 as myogenic stem cells or myoblasts. As the subculture continues, the PSKME cells start to express M-cadherin, MyoD, and Myogenin, though the early passage of PSKME cells expresses only Myf5 (,e)). The expression of M-cadherin, MyoD, and Myogenin gradually increases as the number of passage increases which indicates that the reprogramming of MEF into myogenic lineage is a stochastic process ()). Interestingly, the cells transduced with the combinations of PSOKME (Pax3, Sox2, Oct4, Klf4, cMyc, Esrrb) or PSKOE (Pax3, Sox2, Klf4, Oct4, Esrrb) do not express MyoD and Myogenin though the number of passaging increases to passage 20 (Fig S2). PSOKME and PSKOE are the combinations which only add Oct4 or replacement of cMyc with Oct4 in PSKME which represent the expression of Myf5, MyoD, and myogenin. This implies that the ectopic expression of Oct4 may interfere with the reprogramming of MEF into paraxial mesoderm or myogenic lineage cells. The addition of Oct4 into PSKME or replacement of cMyc with Oct4 might lead MEF into pluripotent stem cells rather than myogenic lineage cells since the Oct4 is a very potent pluripotent transcriptional factor that even alone can reprogram adult neural stem cells into induced pluripotent stem cells [Citation34]. Thus, the combination of optimal transcriptional factors is necessary for direct conversion of MEF into myogenic lineage cells.
The expression of the myogenic transcriptional factors increases as passaging continues. Especially, PSKME cells do not express MyoD and myogenin in passage 5. However, it expresses the genes at late passage. The expression of the myogenic-related genes in PSKME increases gradually as passaging continues. These results indicate that the reprogramming of MEF into myogenic stem cells by the transduction of PSKME is a stochastic process. The balance between
The overexpression of five transcriptional factors, Pax3, Sox2, Klf4, cMyc, and Esrrb, gives MEF to be paraxial mesoderm-like cells which express PDGFRα+/FLK-1− ()). Ectopic expression of Pax3 makes ES cells PDGFRα+/FLK-1− cell population which are the surface markers of paraxial mesodermal progenitor cells [Citation35]. Indeed, paraxial mesoderm which is positive for PDGFRα generates functional muscle stem cells in vivo [Citation36]. Paraxial mesoderm cells transiently coalesce in the embryo to form somites [Citation37]. In turn, somites form the dermomyotome, in which cells expressing Pax3, patterning the epithelial sheet called the myotome [Citation15]. At this point, embryonic myogenesis occurs upon the activation of MRF genes [Citation32,Citation38]. The PSKME expresses genes that are related to muscle development ()). Eomes and Mesp2 are the markers of paraxial mesoderm and early somites [Citation39,Citation40]. Tbx4 is expressed in the developing myotome [Citation16]. Also, c-Met controls limb bud formation regulated by Pax3 [Citation41]. Thus, PSKME cells are paraxial mesoderm progenitor and could generate muscle stem cells.
To get homogeneous cells from PSKME cells, we performed single-cell sorting and evaluated its myogenic differentiation capacity. In this manner, we obtained cell lines, named as iMLCs that form myotubes and express terminal myogenic markers in myogenic differentiation media. The iMLC exhibits characteristics of activated satellite cells, which are precursors of muscle tissue, in terms of surface markers and gene expression, as well as the ability to form multinucleated myotubes in vitro and in vivo. Activated satellite cells eventually become committed to myogenesis, resulting in elevated myogenin and MHC [Citation31,Citation42]. We also found that the iMLC is muscle lineage-specific, which is not flexible, as the cells cannot survive in other media that promote adipogenesis, chondrogenesis, and osteogenesis (data not shown). The population of PDGFR-α (+) cells of iMLCs is smaller than it of PSKME at passage 5 () and )). It means that the PSKME and iMLCs have unique characteristics though the iMLCs are sorted from PSKME. The iMLCs loose the characteristics of paraxial mesoderm though the MEF undergoes the paraxial mesoderm stage in the early process of myogenic direct conversion. This phenomenon is similar to the ES cells with ectopic expression of Pax3, in which a number of PDGFRα+/FLK-1− cells gradually increased until day 5 and decreased thereafter [Citation22].
To evaluate the engraftment and myogenic differentiation capacity in vivo, we transplanted iMLCs in MDX mice without gamma irradiation. Most of the previous studies performed gamma or X-ray irradiation before transplantation of muscle stem cells to improve and maximize the engraftment efficiency [Citation43–Citation45]. The irradiation ablates the endogenous satellite cells and the transplanted cells do not compete with the recipient satellite cells which leads to an increase of engraftment efficiency [Citation43–Citation45]. However, we did not perform the gamma irradiation to reproduce the actual in vivo environment. This would be the reason that the engraftment efficiency is not that high when compared with the other transplantation experiment which performed gamma irradiation [Citation24]. Considering that the endogenous satellite cells impede the engraftment of exogenous stem cells, the detection of dystrophin positive cells in iMLC transplanted-MDX mice indicates that the iMLCs are engraftable and differentiate into muscle fiber in vivo.
In summary, we suggests that the ectopic expression of the defined factors including Pax3, Sox2, Klf4, c-Myc, and Esrrb can induce myogenic differentiation in fibroblasts via paraxial mesoderm stage. This study shows the new combination of transcriptional factors that trigger myogenic direct conversion in fibroblasts and provides an understanding of the process of the myogenic direct conversion.
Materials and methods
Reagents
The reagents were purchased from the following companies: culture media, trypsin, penicillin/streptomycin, reverse transcriptase, RNase out, TRITC-, and FITC-conjugated secondary antibodies from Thermo Fisher Scientific (Rockford, IL); bFGF-2, anti-Pax3 (Mab2459), anti-Esrrb (H6705) from R&D systems (Minneapolis, MN); FBS, HS from Hyclone (Logan, UT); PCR premix, Oligo (dT) from Bioneer (Daejeon, ROK); anti-Myf5 (C20), anti-MyoD (5.8A), anti-myogenin (F5D), anti-c-Myc (9E10), anti-Sox2 (H65), anti-MHC (A4.74), anti-troponin I (H170) from Santa Cruz Biotechnology (Santa Cruz, CA); anti-Klf4 (ab34814), anti-dytrophin (ab15277), anti-GFP (9F9) from Abcam (Cambridge, MA); anti-GAPDH (14C10), HRP-conjugated secondary antibodies from Cell signaling Technology (Danvers, MA); anti-M-cadherin (611,100) from BD Bioscience (Mississauga, Ontario, Canada); PE-conjugated anti-CD29, anti-CD34, anti-CD44, anti-CD56, anti-CXCR4, anti-FLK1, anti-PDGFRα, anti-Sca1 antibody, Alex flour 488-conjugated anti-SSEA3 antibody from eBioscience (San Diego, CA); PE-conjugated anti-Syndecan4, anti-CD31, from BD Bioscience (San Jose, CA); FITC-conjugated anti-VCAM from Bio-Rad (Hercules, CA); ECL detection kit from Thermo Fisher Scientific (Rockford, IL); all the other chemical from Sigma-Aldrich (St Louis, MO).
Plasmids
The Pax3 open reading frame of length 1440 bp was subcloned into the pLJM lentiviral vector from Addgene (19,319), which contains a selective marker for puromycin resistance. A plasmid containing Esrrb (40,798) was procured from Addgene, in addition to polycistronic vectors containing SOKM (20,325). The plasmid containing SKM was constructed by self-ligation after the excision of Oct4 from SOKM. Plasmids were amplified in E. coli STBL3 (Invitrogen) and purified with Qiagen MidiPrep Kits.
Lentivirus preparation
Lentiviral vectors were packaged in 293 FT cells (Invitrogen) that were 90% confluent in 5 mL fresh MEF media. Lipofectamine 2000 (Invitrogen) was used to deliver 5 μg lentiviral backbone plasmid and 15 μg packaging plasmid (Invitrogen) prepared in 60 μL OPTI-MEM (Invitrogen). Viral supernatant was collected 48 and 72 h post-transfection, filtered through a 0.45 μM filter (Millipore), aliquoted, and frozen at −80°C until use. We used only lentiviruses with a minimum titer of 5 × 105 IFU/mL, as determined using Lenti-X GoStix (Clontech).
Generation of iMLC
Mouse embryonic fibroblasts isolated at E13.5 were prepared as previously described [Citation37]. MEF was seeded at 1.5 ~ 2 × 105 cells for each 60 mm plate coated with 2% gelatin. Cultures were then infected with 500 μL virus suspended in 1.5 mL MEF media (10% FBS and 2 mM L-Glutamine in DMEM) the following day. Two days after transduction, the culture media was replaced with 2 mL fresh media, and cells were treated with 5 μg/mL puromycin for the next 10–12 d. After selection on puromycin, cells were cultured in satellite-cell media, which is H-DMEM containing 10% FBS, 10% horse serum, 1% penicillin/streptomycin, and 5 ng/mL basic FGF. Media was changed every 2–3 d until morphological changes were observed. Clusters of cells with altered morphology were dissociated from the bottom of the plate, moved into a multi-well plate and passaged. Cells at passage 5–7 were used for RT-PCR. Prior to the first passage, undifferentiated cells were removed from PSKME cultures by scraping.
FACS analysis
Cells were detached from the culture dish using 0.05% trypsin, pelleted, and washed with PBS. Samples were then incubated for 1 h on ice with PE-conjugated antibodies against FLK-1, PDGFR-α, CD29, CD44, CD34, CXCR4, and syndecan-4. Samples were also probed with an unlabeled mouse antibody against M-cadherin, and then stained with PE-conjugated goat anti-mouse IgG. Cells were analyzed on FACS Aria (Becton-Dickinson).
RT-PCR
RNA was isolated using Trizol and then used as a template to synthesize cDNA via reverse transcription, following the manufacturer’s protocol (Invitrogen). C2C12 cell line was used as a positive control for the expression of myogenic markers which is an immortalized murine myoblast. C2C12 was purchased from ATCC. The cDNA was then amplified using Taq polymerase and primers listed in Supplementary Table S1. Real-time PCR was performed using 2× SYBR Green PCR mix with the Roche primers QT00199507, QT00101983, QT00112378, QT0106850, and QT01658692, which target, respectively, Myf5, MyoD, myogenin, myosin heavy chain, and glyceraldehyde-3-phosphate dehydrogenase.
Immunoblot analysis
Cell lysates for western blots were prepared using 1× RIPA buffer (ThermoScientific) supplemented with Complete Protease Inhibitor Cocktail (Roche). Samples containing 50 μg total protein were resolved on SDS-PAGE, and transferred to PVDF membrane (Immobilon-P; Millipore). The blot was probed overnight at 4°C with primary antibodies against Pax3 (1:500), Esrrb (1:1000), Sox2 (1:500), c-Myc (1:500), Klf4 (1:400), Myf5 (1:1000), and GAPDH (1:2000). After incubation with secondary antibodies (1:1000) conjugated to horseradish peroxidase, blots were developed with Chemiluminescent Substrate (ThermoScientific) and visualized by Image Analyzer (UVP).
Immunofluorescence
Cells that had been maintained on gelatin-coated plates were fixed for 15 min at room temperature in 4% paraformaldehyde. After washing with PBS, samples were incubated for 10 min at −20°C in 100% methanol and then washed three times with PBS. Cells were then blocked for 1 h at room temperature with 3% BSA and probed with primary antibodies against Myf5 (1:100), MyoD (1:100), myogenin (1:100), and MHC (1:100). FITC- or TRITC-conjugated secondary antibodies were used for staining.
The tibialis anterior muscle was harvested from mice 4 wk after cell transplantation. Double staining with GFP and dystrophin was performed according to a previous report [Citation46]. Briefly, frozen sections were blocked overnight at 4°C with 10% horse serum in PBS and then probed overnight at 4°C with mouse anti-GFP (1:100) and rabbit anti-dystrophin (1:100). Alexa 594-conjugated anti-mouse IgG and FITC-conjugated anti-rabbit IgG were used as secondary antibody. Images were obtained by confocal fluorescence microscopy (Zeiss).
Myogenic differentiation in vitro and in vivo
To induce myogenesis in vitro, cells were grown in high-glucose DMEM media containing 5% horse serum and 1% penicillin/streptomycin. Cells were cultured for 8–12 d and analyzed by RT-PCR and immunofluorescence. To examine myogenesis in vivo, cells were transduced with GFP and sorted by FACS. Cells expressing GFP were suspended in 20 μl PBS to a final density of 25,000 cells/μL and injected into the tibialis anterior of nude and MDX mice that had been injured with notexin at a dose of 40 μg/muscle. Myogenesis was assessed 4 wk later by co-imaging GFP and dystrophin. Animal experiments were conducted under a protocol approved by Kyungpook National University Animal Care and Use Committee (2014–0167).
Statistical analysis
Differences between samples were analyzed using Student’s two-tailed t-test for independent samples.
Supplemental Material
Download Zip (1.5 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data of this article can be accessed here.
Additional information
Funding
References
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006 25;126(4):663–676.
- Viebuchen T, Ostermeier A, Zhiping PP, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(2010):1035–1041.
- Miskinyte G, Devaraju K, Hansen MG. Direct conversion of human fibroblasts to functional excitatory cortical neurons integrating into human neural networks. Stem Cell Res Ther. 2017;8(1). DOI:10.1186/s13287-017-0658-3
- Hu W, Qiu B, Guan W, et al. Direct conversion of normal and alzheimer’s disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell. 2015;17(2):204–212.
- Cassady JP, D’Alessio AC, Sarkar S, et al. Direct lineage conversion of adult mouse liver cells and B lymphocytes to neural stem cells. Stem Cell Rep. 2014;3(6):948–956.
- Mazzara PG, Massimino L, Pellegatta M, et al. Two factor-based reprogramming of rodent and human fibroblasts into Schwann cells. Nature Commun. 2017. DOI:10.1038/ncomms14088.
- Ieda M, Ji-Dong F, Delgado-Olguin P, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–386.
- Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475(29):390–393.
- Ballester M, Bolonio M, Santamaria R, et al. Direct conversion of human fibroblast to hepatocytes using a single inducible polycistronic vector. Stem Cell Res Therapy. 2019;10(1):317.
- Szabo E, Rampalli S, Risueño RM, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468(25):521–526.
- Ito N, Kii I, Shimizu N, et al. Direct reprogramming of fibroblasts into skeletal muscle progenitor cells by transcription factors enriched in undifferentiated subpopulation of satellite cells. Sci Rep. 2017;7(1). DOI:10.1038/s41598-017-08232-2
- Lee EJ, Kim M, Kim YD, et al. Establishment of stably expandable induced myogenic stem cells by four transcription factors. Cell Death Dis. 2018;9:11.
- Bar-Nur O, Mattia FM, Gerli B, et al. Direct reprogramming of mouse fibroblasts into functional skeletal muscle progenitors. Stem Cell Reports. 2018;10(5):1505–1521.
- Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, et al. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005;19:1426–1431.
- Goulding M, Lumsden A, Paquette AJ. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development. 1994;120:957–971.
- Bajard L, Relaix F, Lagha M, et al. A novel genetic hierarchy functions during hypaxial myogenesis: pax3 directly activates Myf5 in muscle progenitor cells in the limb. Gene Dev. 2006;20:2450–2464.
- Lagha M, Sato T, Bajard L, et al. Regulation of skeletal muscle stem cell behavior by Pax3 and Pax7. Cold Spring Harb Symp Quant Biol. 2008;73:307–315.
- Relaix F, Rocancourt D, Mansouri A, et al. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953.
- Tremblay P, Dietrich S, Mericskay M, et al. A crucial role for Pax3 in the development of the hypaxial musculature and the long-range migration of muscle precursors. Dev Biol. 1998;203:49–61.
- Relaix F, Rocancourt D, Mansouri A, et al. Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes Dev. 2004;18:1088–1105.
- Maroto M, Reshef R, Münsterberg AE, et al. Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell. 1997;l 89:139–148.
- Darabi R, Gehlbach K, Bachoo RM, et al. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat Med. 2008;14:134–143.
- Filareto A, Parker S, Darabi R, et al. An ex vivo gene therapy approach to treat muscular dystrophy using inducible pluripotent stem cells. Nature Commun. See comment in PubMed Commons below 2013;4: 1549.
- Gang EJ, Bosnakovski D, Simsek T, et al. Pax3 activation promotes the differentiation of mesenchymal stem cells toward the myogenic lineage. Exp Cell Res. 2008;314:1721–1733.
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000.
- Kabadi AM, Thakore PI, Vockley CM. Enhanced MyoD-induced transdifferentiation to a myogenic lineage by fusion to potent transactivation domain. ACS Synth Biol. 2015;4(6):689–699.
- Sato T, Higashioka K, Sakurai H. Core transcription factors promotes induction of PAX3-positive skeletal muscle stem cells. Stem Cell Reports. 2019;13(2):352–365.
- Bober E, Franz T, Arnold HH, et al. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development. 1994;120:603–612.
- Mansouri A, Stoykova A, Torres M, et al. Dysgenesis of cephalic neural crest derivatives inPax7−/− mutant mice. Development. 1996;122:831–838.
- Mitchell KJ, Pannerec A, Cadot B, et al. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nat Cell Biol. 2010;12:257–266.
- Tedesco FS, Dellavalle A, Diaz-Manera J, et al. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. J Clin Investig. 2010;120:11–19.
- Ordahl CP, Le Douarin NM. Two myogenic lineages within the developing somite. Development. 1992;114:339–353.
- Seale P, Sabourin LA, Girgis-Gabardo A, et al. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102(6):777–786.
- Kim JB, Greber B, Araúzo-Bravo MJ, et al. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461(7264):649–653.
- Sakurai H, Era T, Jakt LM, et al. In vitro modeling of paraxial and lateral mesoderm differentiation reveals early reversibility. Stem Cells. 2006;24:575–586.
- Sakurai H, Okawa Y, Inami Y, et al. Paraxial mesodermal progenitors derived from mouse embryonic stem cells contribute to muscle regeneration via differentiation into muscle satellite cells. Stem Cells. 2008;26:1865–1873.
- Ordahl CP. Early stages of chick somite development. Anat Embryol (Berl). 1995;191:381–396.
- Denetclaw WFJ, Ordahl CP. Location and growth of epaxial myotome precursor cells. Development. 1997;124:1601–1610.
- Ciruna BG, Rossant J. Expression of the T-box gene Eomesodermin during early mouse development. Mech Dev. 1990;81:199–203.
- Saga Y, Hata N, Koseki H, et al. Mesp2: A novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev. 1997;11:1827–1839.
- Epstein JA, Shapiro DN, Cheng J, et al. Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc Natl Acad Sci USA. 1996;93:4213–4218.
- Beauchamp JR, Heslop L, Yu DS, et al. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biolol. 2000;151:1221–1234.
- Wakeford S, Watt DJ, Partridge TA. X-irradiation improves mdx mouse muscle as a model of myofiber loss in DMD. Muscle Nerve. 1991;14(1):42–50.
- Sampath SC, Sampath SC, Ho ATV. Induction of muscle stem cell quiescence by the secreted niche factor Oncostatin M. Nature Commun. 2018;9(1):1521.
- Cosgrove BD, Gilbert PM, Porpiglia E. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat Med. 2014;20(3):255–264.
- Liadaki K, Luth ES, Kunkel LM. Co-detection of GFP and dystrophin in skeletal muscle tissue sections. BioTechniques. 2007;42:699–700.
