ABSTRACT
Traditionally, circular RAN hsa_circ_0008035 was proven to function as a tumor inhibitor in gastric cancer. Nevertheless, much less was known about hsa_circ_0008035 in osteosarcoma (OSA). This project was undertaken to assess the role of hsa_circ_0008035 in OSA.
Hsa_circ_0008035 level in serum of OSA patients, OSA tissues and cell lines were measured by reverse transcription-quantitative PCR. After downregulation or overexpression of hsa_circ_0008035, cell proliferation, apoptosis and migration were tested in MG63, SAOS-2 or hFOB1.19 cells. Meanwhile, the expression level of miR-375 was analyzed. The binding between hsa_circ_0008035 and miR-375 was confirmed using bioinformatics and luciferase assay. Subsequently, the effects of miR-375 inhibition on MG63 cell growth and migratory potential were reevaluated. Eventually, the activating status of Notch pathway was assessed by Western blot.
Our results demonstrated that hsa_circ_0008035 was overexpressed in serum of OSA patients, OSA tissues and cells. Silencing hsa_circRNA_0008035 impeded OSA cell growth and migration, while hsa_circ_0008035 facilitated cell behaviors of hFOB1.19 cells. Additionally, hsa_circ_0008035 negatively modulated miR-375 expression. Meanwhile, miR-375 inhibition overturned the suppressive effects of silencing hsa_circRNA_0008035 on OSA cell behaviors. Furthermore, silencing hsa_circ_0008035 perturbed Notch pathway by adjusting miR-375 expression.
In conclusion, silencing hsa_circRNA_0008035 exerted repressive function on OSA cell growth and migration and Notch pathway by accelerating miR-375.
Introduction
Osteosarcoma (OSA), one of the most general primary malignant tumors, consists of mesenchymal cells and is characterized by forming bone-like tissues [Citation1]. OSA usually occurs around knee articulation, distal femur or proximal humerus [Citation2]. The annual incidence of OSA is approximately three per million, which is common in children and adolescents [Citation3]. Due to rapid growth, stronger invasiveness, potential early metastasis and resistance to chemotherapeutic drugs, the 5-y survival rate of recurrent or metastatic OSA is about 60% [Citation4]. Although surgical treatment of OSA is the most standard and basic method, including amputation surgery and limb survival surgery, the specific scope of resection is still controversial [Citation5]. With the extensive application of neoadjuvant chemotherapy and radiotherapy, personalized and precise treatments have received widespread attentions [Citation6]. However, these therapies cannot completely inhibit tumor growth or reduce the metastatic possibility of OSA, leading to a high level of OSA mortality [Citation7]. In order to break through the bottleneck period of OSA treatment, it is urgent to explore new mechanisms of OSA development and provide effective intervention strategies for early diagnosis and treatment of OSA.
Circular RNAs (circRNAs) are a category of competitive endogenous RNA molecules that exhibit pivotal activity in genetic regulation and tumor development [Citation8]. It was generally accepted that circRNAs were highly stable, of whose covalent ring structure could evade the decomposition of ribonuclease [Citation9]. Additionally, circRNAs participate in transcriptional regulation not only through circRNA-miRNA-mRNA axis but also via RNA-binding protein, affecting physiological processes [Citation8,Citation10,Citation11]. With reference to OSA, the enhancement of circRNA UBAP2 was observed in OSA specimens and cells, and evidently linked to OSA growth and poor prognosis of OSA patients [Citation12]. Besides, circRNA CDR1as modulated cellular vitality, migratory potential and epithelial-mesenchymal transition through targeting microRNA-7 (miR-7) in OSA cells [Citation13]. Equally important, hsa_circ_0008035 issued from exostosin glycosyltransferase 1 (EXT1) and located on chr8:118830673–118849440 with 18767 nt genomic length and 670 nt spliced length. Available data indicated that hsa_circ_0008035 facilitated the tumorigenesis of gastric carcinoma via regulating the miR-375/YBX1 axis [Citation14]. Nevertheless, no previous study provided information on the relative importance of hsa_circ_0008035 in OSA.
The purpose of the current study was to determine whether hsa_circ_0008035 participated in the occurrence of OSA and delved into the deep-rooted mechanism. This study made a major contribution to research on clinical OSA diagnosis and treatment by demonstrating the function of hsa_circ_0008035.
Materials and methods
Clinical samples and collection
The serum samples were obtained from 50 OSA patients (33 male and 17 female; 36 cases below 18 y old and 14 cases above 18 y old) diagnosed with OSA (11 cases of stage IIA and 39 cases of IIB/III) at China–Japan Union Hospital of Jilin University (Changchun, China) between June 2017 and June 2019. On the basis of the exclusion criteria (orthopedic diseases, cardiovascular diseases and nervous system-related diseases), 50 healthy volunteers (30 male and 20 female; 32 cases below 18 y old and 16 cases above 18 y old) were eligible for recruitment. Moreover, tissue samples of OSA participants were obtained when tumor tissues were resected. The resected OSA tissues were approximately more than 8 cm in diameter in 28 patients and less than 8 cm in 22 cases. The anatomic location of OSA tissues was different: 40 OSA tissues were located in tibia/femur. Additionally, 38 patients with OSA were absent from distant metastasis while 38 patients with OSA presented distant metastasis. From each patient, peritumor tissue samples were collected from resected. Our study was conducted in support of the Ethics Committee of the China–Japan Union Hospital of Jilin University, and all patients signed a consent form.
Cell culture
Human OSA cell lines (MG-63, SAOS-2, U-2OS, HOS and SW1353) and osteoblast cell line (hFOB1.19) were all purchased from American Type Culture Collection (Rockville, MD, USA). OSA cells and hFOB1.19 cells were respectively cultivated in Dulbecco’s Modified Eagle medium (Gibco, Rockville, MD, USA) or DF-12 medium (Gibco) that enriched with 10% fetal bovine serum (FBS, Gibco). The condition of cell culture was at 37°C and 5% CO2.
Cell transfection
For RNA interference, small interfering RNA (siRNA) duplexes targeting hsa_circ_0008035 (si-circ) were designed and chemically synthesized by GenePharma (Shanghai, China). The nucleotide sequence was as follows: si-circ, 5ʹ-AGG AGA GAG CAA GGT ATG ATT-3ʹ. Scrambled sequence was used as negative control of siRNA (si-NC). Overexpressing hsa_circ_0008035 plasmids (ex-circ) and control (NC) was constructed by GenePharma for our experiments. In addition, miR-375 inhibitor (5ʹ-UCA CGC GAG CCG AAC GAA CAA A-3ʹ) and NC inhibitor (5ʹ-UCU ACU CUU UCU AGG AGG UUG UGA-3ʹ) were utilized in this research. Lipofectamine RNAiMAX (Thermo Fisher, Carlsbad, CA, USA) was utilized to implement cell transfection. At 48 h after transfection, cells were collected for the following experiments.
Reverse transcription-quantitative PCR
TRIzol solutions (Invitrogen, Carlsbad, CA, USA) were employed to the extraction of RNA from serum samples, OSA tissues and cells. Next, reverse transcription was carried out by using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher). TaqMan Real-time PCR Master Mixes (Thermo Fisher) was used to perform qPCR. The primers used were as follows hsa_circ_0008035, forward, 5ʹ- CTA CCA GCC AAA CAC CGC T-3ʹ, reverse, 5ʹ-TCC AGG AAT CTG AAG GAC CCA-3ʹ; glyceraldehyde-3-phosphate dehydrogenase (GAPDH), forward, 5ʹ-AAG CCA CCC CAC TTC TCT CTA A-3ʹ, reverse, 5ʹ-AAT GCT ATC ACC TCC CCT GTG T-3ʹ; miR-375, forward, 5ʹ-TTT GTT CGT TCG GCT CGC-3ʹ, reverse, 5ʹ-GAT TGA ATC GAG CAC CAG TTA CG-3ʹ; U6, forward, 5ʹ-GAT GAC ACG CAA ATT CGT GAA-3ʹ, reverse, 5ʹ-GAT TGA ATC GAG CAC CAG TTA CG-3ʹ. The relative expression of hsa_circ_0008035 and miR-375 was normalized by GAPDH and U6 with 2–ΔΔCt method [Citation15].
Cell proliferation assay
Transfected cells were placed in 96-well plates (5 × 103/well). At 1–5 d, cells were collected and treated with 10 μL of cell counting kit (CCK, GlpBio, Montclair, CA, USA) solutions. After 4 h, the optical density of 450 nm was evaluated by utilizing a microtiter plate reader (Detie, Nanjing, China).
Cell apoptosis experiment
Annexin V-AbFluor TM 488 Apoptosis Detection Kit (Abbkine, Wuhan, China) was used to measure the percentage of apoptotic cells on a flow cytometer (BeamCyte, Changzhou, China). At 48 h after transfection, the transfected cells in culture flask that contained death or apoptotic cells were collected in a 5-mL eppendorf tube and centrifuged at 500 × g for 5 min. The pelleted cells were resuspended with 1 × annexin V-binding buffer to 1 × 106 cells/mL. The 5 μL of Annexin V-AbFluor TM 488 and 1 μL of propidium iodide working solution were added into each well. After 15 min of incubation, cells were washed twice and apoptotic cells were viewed under flow cytometer.
Cell migration experiment
Wound-healing experiment was executed to evaluate cell migratory capacity. Transfected or non-transfected cells were placed in 6-well plates and cultivated to 80% confluence. Artificial wound was produced on the cell monolayer, and then wound-healing time was measured. For every group, at least three wells of artificial wounds were formatted to detect cell migration. After the wound was formatted, the cell plate was maintained with a culture medium without FBS and then placed at 37°C for 48 h.
Luciferase assays
The hsa_circ_0008035 psi-CHECK-2 luciferase reporter vector was cloned by inserting oligo with wild type (wt) hsa_circ_0008035 into the BamHI/SalI site of pGL2 luciferase vector (Promega, Madison, WI, USA), and the recombinant was referred to hsa_circ_0008035-wt. The seed sites of hsa_circ_0008035 were mutated by several nucleotides in the seed site using QuikChange site-directed mutagenesis kit (Stratagene). HEK-293T cells plated in a 96-well plate were transfected with hsa_circ_0008035-wt/mut (100 ng per well), phRL-TK Renilla luciferase vector (25 ng per well, Promega) together with miR-375 mimic/mimic NC. Firefly and Renilla luciferase activity was detected 48 h after transfection using Dual-Glo Luciferase Assay System (Promega). The relative luciferase activity (the percentage of firefly luminescence relative to the Renilla control) was calculated.
Western blot
After using RIPA (Biorbyt, Wuhan, China) to implement protein extraction, proteins were loaded into SDS-PAGE gel. Followed by transference into polyvinylidene difluoride membranes, the membranes carried on proteins were sealed with 5% BSA blocking buffer (Solarbio), and incubated in primary antibodies overnight at 4°C. Primary antibodies were listed as follows: Notch-1 (3608, Cell Signaling Technology, CST, Boston, MA, USA), Hes1 (11988, CST), DLL4 (96406, CST) and β-actin (4970, CST). Next, HRP-conjugated anti-rabbit IgG (7074, CST) was utilized to hatch the membranes for 2 h. Finally, after the treatment of enhanced chemiluminescence (ECL) solutions (Pierce, Minneapolis, MN, USA), the gray value of protein bands was analyzed by ImageJ software (NIH, Bethesda, MD, USA).
Statistical analysis
All data arrangement and analysis were performed by utilizing GraphPad Prism 6.0 software (GraphPad, San Diego, CA, USA). The statistical analysis results of Student’s t-test and one-way analysis of variance (ANOVA) was showed as mean + standard deviation. P < 0.05 was suggested as statistically significant.
Results
Expression of hsa_circ_0008035 in serum of OSA patients, OSA tissues and OSA cells
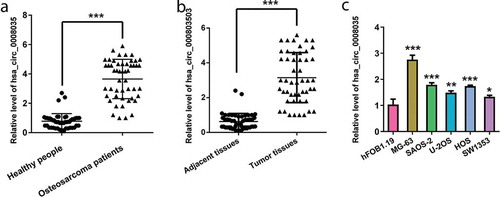
Initially, the expression of hsa_circ_0008035 in serum samples of healthy people and OSA patients was examined by reverse transcription-quantitative (RT-qPCR). ) presents that hsa_circ_0008035 was upregulated in serum samples of OSA patients, in comparison with healthy people (p < 0.001). Meanwhile, OSA tissues were collected from 50 patients to test the expression level of hsa_circ_0008035. As disclosed in ), overexpression of hsa_circ_0008035 was observed in OSA tissues, compared with the corresponding paracancerous specimens (p < 0.001). Beyond that, the expression of hsa_circ_0008035 in five types of OSA cell lines (MG-63, SAOA-2, U-2OS, HOS and SW1353) was also measured by RT-qPCR. It was clearly showed that hsa_circ_0008035 expression was significantly enhanced in MG-63, SAOA-2, U-2OS, HOS and SW1353 cells, by comparison with human osteoblast (hFOB1.19) (p < 0.05, p < 0.01, p < 0.001, )). These results proved that hsa_circ_0008035 was overexpressed in the serum of OSA patients, OSA tissues and OSA cells.
Figure 1. Expression of hsa_circ_0008035 in serum of OSA patients, OSA tissues and OSA cells. (a) The expression of hsa_circ_0008035 in serum of OSA patients (n = 50) and healthy people (n = 50) was examined by RT-qPCR. (b) The expression of hsa_circ_0008035 OSA tissues (n = 50) and the corresponding paracancerous specimens (n = 50) was tested by RT-qPCR. (c) The expression of hsa_circ_0008035 in OSA cell lines (MG-63, SAOA-2, U-2OS, HOS, SW1353) and human osteoblast (hFOB1.19) was analyzed by RT-qPCR. Data were shown as the mean + SD for three replications per group. * p < 0.05, ** p < 0.01, *** p < 0.001, t-test.
Effects of silencing hsa_circ_0008035 on cell growth and migration of MG63 and SAOS-2 cells
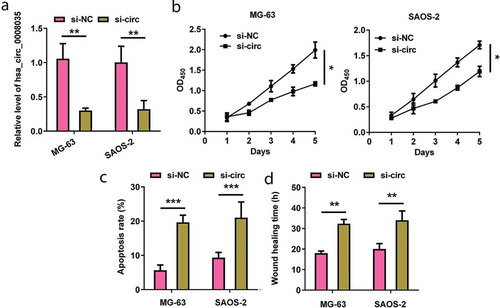
Among the five types of OSA cell lines, we elected MG-63 and SAOA-2 cell lines for the next experiments due to the overexpressed levels of hsa_circ_0008035. After successful transfection of si-hsa_circ_0008035 (p < 0.01, )), the impacts of silencing hsa_circ_0008035 on cell growth and migratory ability were examined. ) reveals that silencing hsa_circ_0008035 significantly declined cell proliferation after 2 d of cell transfection (p < 0.05). Quite the opposite, cell apoptotic potential was evidently enhanced by silencing hsa_circ_0008035 in MG-63 and SAOA-2 cells (p < 0.001, )). Beyond that, silencing hsa_circ_0008035 suppressed cell migratory ability by prolonging wound-healing time of MG-63 and SAOA-2 cells (p < 0.01, )). The above-mentioned data showcased that silencing hsa_circ_0008035 impeded cell growth and migration of MG-63 and SAOA-2 cells.
Figure 2. Effects of silencing hsa_circ_0008035 on cell growth and migration of MG63 and SAOS-2 cells. Si-hsa_circ_0008035 (si-circ) or si-NC was transfected into MG63 and SAOS-2 cells. (a) The expression of hsa_circ_0008035 was evaluated by RT-qPCR. (b) Cell proliferation, (c) apoptosis rate and (d) wound-healing time were assessed by utilizing CCK-8, flow cytometry and migration experiment. Data were shown as the mean + SD for three replications per group. * p < 0.05, ** p < 0.01, *** p < 0.001, ANOVA.
Effects of hsa_circ_0008035 on cell growth and migration of hFOB1.19 cells
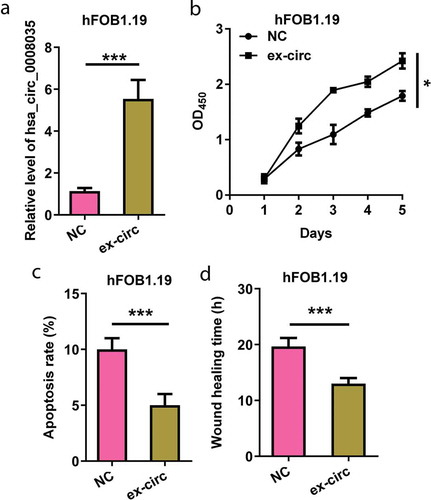
Next, we explored the effects of hsa_circ_0008035 on hFOB1.19 cell behaviors. Overexpressing hsa_circ_0008035 plasmid was utilized to upregulate hsa_circ_0008035 expression in hFOB1.19 cells (p < 0.001, )). As displayed in ), overexpression of hsa_circ_0008035 apparently accelerated cell proliferation of hFOB1.19 cells (p < 0.05, )). Instead, overexpression of hsa_circ_0008035 weakened cell apoptosis rate of hFOB1.19 cells (p < 0.001, )). Meanwhile, wound-healing time of hFOB1.19 cells was restrained by overexpression of hsa_circ_0008035 (p < 0.001, )). These consequences demonstrated that overexpression of hsa_circ_0008035 strengthened cell growth and migration in hFOB1.19 cells.
Figure 3. Effects of hsa_circ_0008035 on cell growth and migration of hFOB1.19 cells. Overexpressing hsa_circ_0008035 plasmid (ex-circ) or NC plasmid was transfected into hFOB1.19 cells. (a) The expression of hsa_circ_0008035 was evaluated by RT-qPCR. (b) Cell proliferation, (c) apoptosis rate and (d) wound-healing time were assessed by utilizing CCK-8, flow cytometry and migration experiment. Data were shown as the mean + SD for three replications per group. * p < 0.05, *** p < 0.001.
Regulation of hsa_circ_0008035 on miR-375 expression
Since miR-375 was reported as a crucial miRNA in the diagnosis and prognosis of OSA, RT-qPCR analysis was implemented to measure miR-375 expression. In ), we observed that the expression levels of miR-375 in OSA cell lines were markedly lower than that in hFOB1.19 cells (p < 0.05, p < 0.01 and p < 0.001). Additionally, ) shows that silencing hsa_circ_0008035 expedited miR-375 expression in MG-63 and SAOA-2 cells (p < 0.001). Moreover, overexpression of hsa_circ_0008035 cuts down the expression level of miR-375 in hFOB1.19 cells (p < 0.001, )). The aforementioned results suggested that hsa_circ_0008035 negatively regulated miR-375 expression.
Figure 4. Regulation of hsa_circ_0008035 on miR-375 expression. (a) The expression of miR-375 in OSA cell lines (MG-63, SAOA-2, U-2OS, HOS, SW1353) and human osteoblast (hFOB1.19) was estimated by RT-qPCR. (b) Si-hsa_circ_0008035 (si-circ) or si-NC was transfected into MG63 and SAOS-2 cells. The expression of miR-375 was examined by RT-qPCR. (c) Overexpressing hsa_circ_0008035 plasmid (ex-circ) or NC plasmid was transfected into hFOB1.19 cells. The expression of miR-375 was analyzed by RT-qPCR. (d) The complementary sites were indicated for the binding of hsa_circ_0008035 and miR-375. HEK-293T cells were transiently transfected with firefly luciferase reporter plasmids (hsa_circ_0008035-wt/mut), renilla luciferase reporter plasmids and miR-375 mimic/mimic NC. (e) The expression of miR-375 was analyzed by RT-qPCR to confirm transfection efficiency. (f) Binding analysis of hsa_circ_0008035 toward miR-375 using luciferase assay. Data were shown as the mean + SD for three replications per group. * p < 0.05, ** p < 0.01, *** p < 0.001.
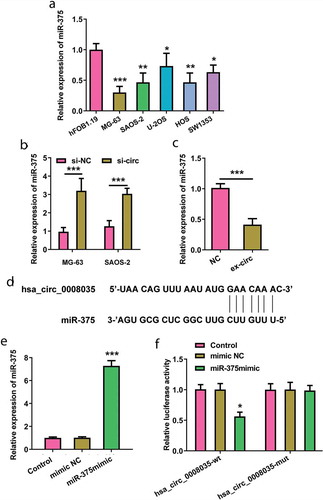
To identify hsa_circ_0008035 targeting miRNAs, we used RegRNA 2.0 (http://regrna2.mbc.nctu.edu.tw/). MiR-375 was selected as a candidate miRNA binding to hsa_circ_0008035 based on matching sites ()). To get insights into the direct regulation of miR-375 under hsa_circ_0008035, we experimentized luciferase reporter assay. The wt of hsa_circ_0008035 sequence complementary to miR-375-binding site was inserted into downstream of luciferase reporter gene vector and the recombinant was referred to hsa_circ_0008035-wt. Similarly, luciferase reporter gene vector containing mutant type (mut) of hsa_circ_0008035 sequence was named as hsa_circ_0008035-mut. HEK-293T cells were successfully co-transfected with hsa_circ_0008035-wt/mut and miR-375 mimic (p < 0.001, )). In the presence of miR-375 mimic, the expression of hsa_circ_0008035-wt reporters was reduced (p < 0.05, )). MiR-375-mediated transcriptional inhibition was disrupted by specific mutations within the hsa_circ_0008035 binding sequence ()). The result indicated that hsa_circ_000803 functioned as a sponge of miR-375.
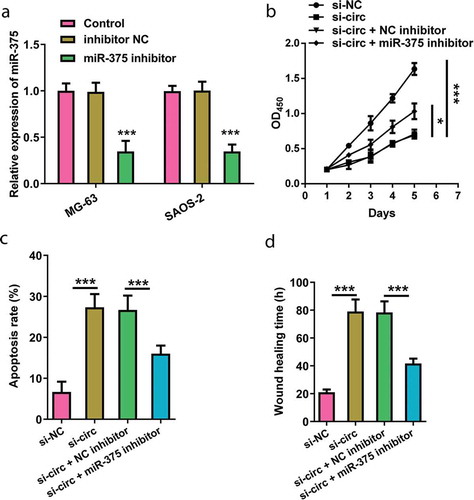
Involvement of miR-375 expression in the hsa_circ_0008035-modulated cell behaviors
MG-63 cells were co-transfected with si-hsa_circ_0008035 and miR-375 inhibitor (p < 0.001, )). Cell proliferation, apoptotic potential and migration were reevaluated. As presented in ), the declination of cell proliferation that was triggered by silencing hsa_circ_0008035 was reversed by miR-375 inhibition in MG-63 cells (p < 0.05). However, the enhancement of cell apoptosis in hsa_circ_0008035 silencing cells was overturned by miR-375 inhibition (p < 0.001, )). Besides, miR-375 inhibition strongly accelerated cell migration by shortening wound-healing time (p < 0.001, )). These data indicated that silencing hsa_circ_0008035 weakened cell growth and migration of MG-63 cells via upregulating miR-375 expression.
Figure 5. Involvement of miR-375 expression in the hsa_circ_0008035-modulated cell behaviors. Si-hsa_circ_0008035 (si-circ) and/or miR-375 inhibitor was transfected into MG-63 cells. (a) The expression of miR-375 was analyzed by RT-qPCR to confirm transfection efficiency. (b) Cell proliferation, (c) apoptosis rate and (d) wound-healing time were assessed by utilizing CCK-8, flow cytometry and migration experiment. Data were shown as the mean + SD for three replications per group. * p < 0.05, *** p < 0.001.
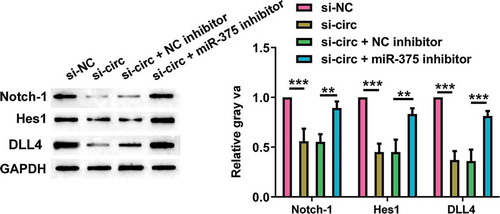
Effects of silencing hsa_circ_0008035 on Notch signaling pathway in MG-63 cells
Eventually, we explored the regulatory role of silencing hsa_circ_0008035 in Notch pathway in MG-63 cells. As shown in , silencing hsa_circ_0008035 noticeably inhibited the protein expression of Notch-1, Hes1 and DLL4 (p < 0.001). On the contrary, miR-375 inhibition reversed the declined levels of Notch-1, Hes1 and DLL4 in hsa_circ_0008035-silenced cells (p < 0.01, p < 0.001). These consequences corroborated that silencing hsa_circ_0008035 perturbed Notch signaling pathway in MG-63 cells through expediting miR-375 expression.
Figure 6. Effects of silencing hsa_circ_0008035 on Notch signaling pathway in MG-63 cells. Si-hsa_circ_0008035 (si-circ) and/or miR-375 inhibitor was transfected into MG-63 cells. The protein levels of Notch-1, Hes1 and DLL4 were evaluated by Western blot. Data were shown as the mean + SD for three replications per group. ** p < 0.01, *** p < 0.001.
Discussion
OSA is one of the highly harmful tumors, which originated from bone tissues and usually transferred to the lung through serum at the early stage, accompanied by a higher mortality rate [Citation16]. In our paper, overexpression of hsa_circ_0008035 was observed in serum of OSA patients, OSA tissues and OSA cells. Subsequently, bio-efficacies of hsa_circ_0008035 on cell proliferation, apoptotic potential and migration were detected in OSA cells and osteoblast. In addition, the expression and function of miR-375 in OSA cells were further explored. Finally, we verified that Notch signaling pathway was interrupted by silencing hsa_circ_0008035.
CircRNAs are novel non-coding RNA molecules that can be stable in serum or bodily fluids in the form of loop [Citation17]. Recently, based on microarray and bioinformatics analysis, the expression profile of circRNAs in OSA was unmasked [Citation18]. A multitude of studies disclosed that circRNAs were involved in the onset and progression of OSA by serving as oncogenetic molecules to accelerate tumor growth or as tumor suppressors to impede tumor progression [Citation19,Citation20]. For example, circRNA GLI2 exerted promoting function on OSA through targeting miR-125b-5p [Citation21]. Quite the opposite, it was demonstrated that hsa_circ_0002052 evidently weakened OSA cell proliferation and metastasis via miR-1205/APC2 axis [Citation22]. Hsa_circ_0008035 was reported to be upregulated in gastric cancer tissues and associated with malignant progression of gastric cancer [Citation14]. As similarly revealed, we proved that hsa_circ_0008035 was upregulated in the serum of OSA patients, OSA tissues and OSA cells. Besides, silencing hsa_circ_0008035 restrained cell proliferation, migratory ability and expedited cell apoptotic potential in OSA cells (MG-63 and SAOS). However, the opposite results were observed in hFOB1.19 cells which were overexpressed hsa_circ_0008035. The results were suggestive of repressive functions of silencing hsa_circ_0008035 in OSA cells, and hsa_circ_0008035 was expected to be a potential molecular biomarker for early diagnosis of OSA.
It is generally accepted that circRNAs act as miRNA sponges to play the role of competing endogenous RNA (ceRNA). The sequences of circRNAs contain potential miRNA binding sites that interfere with the binding of the miRNA to the target gene, thereby affecting gene regulation [Citation23,Citation24]. Previously report stated that hsa_circ_0008035 directly bound with miR-375. MiR-375, which was down-expressed in the serum of OSA patients, showed potential for reducing the proliferation of OSA cells [Citation25,Citation26]. Moreover, another study pointed out that it was by modulating miR-375 that silencing circFAT1 acted as an OSA inhibitor [Citation27]. The findings in our research were similar to those in a previous study that found that miR-375 expressed at a low level in OSA cells. Additionally, silencing hsa_circ_0008035 significantly triggered an enhancement of miR-375 expression in OSA cells. By contrast, overexpression of hsa_circ_0008035 reduced miR-375 level in hFOB1.19 cells. Moreover, online database and luciferase assay proved the direct binding between hsa_circ_0008035 and miR-375. Furthermore, suppressing miR-375 reversed the impacts of hsa_circ_0008035 inhibition on OSA cell behaviors. These results indicated that silencing hsa_circ_0008035 exhibited antitumor function via upgrading miR-375 in OSA cells.
Notch signaling pathway is a key pathway, which is closely related to invasion and metastasis of OSA cells [Citation28]. The classical Notch pathway goes through the following steps: binding between receptors and ligand, hydrolysis, activating the entire signaling pathway. Recent literature confirmed that the activation of Notch expression elicited OSA progression in a mice model [Citation29]. Moreover, a paper verified the involvement of Notch signaling network in miR-135b-regulated OSA recurrence and lung metastasis [Citation30]. Our data illustrated that Notch signaling pathway was perturbed by silencing hsa_circ_0008035 in OSA cells, while miR-375 inhibition abrogated the inactivation of Notch pathway. The findings indicated that Notch pathway might participate in hsa_circ_0008035/miR-375-modulated OSA cell growth and migration.
In spite of the narrow number of patients concluded in our present study, we proved the overexpression of hsa_circ_0008035 is significant in the serum and tissues of OSA patients. Further experiments explained the central importance of silencing hsa_circ_0008035 in OSA cell growth and migratory abilities, which provided a new strategy for the diagnosis and treatment of OSA. It is a limitation of our research that almost all results are observed from human OSA cell lines. On account of the difficulty to behave in vivo status, further researches like using primary OSA cells or animal model are needed.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92.
- Stitzlein RN, Wojcik J, Sebro RA, et al. Team approach: osteosarcoma of the distal part of the femur in adolescents. JBJS Rev. 2017;5:e5.
- Kager L, Tamamyan G, Bielack S. Novel insights and therapeutic interventions for pediatric osteosarcoma. Future Oncol. 2017;13:357–368.
- Friebele JC, Peck J, Pan X, et al. Osteosarcoma: a meta-analysis and review of the literature. Am J Orthop (Belle Mead NJ). 2015;44:547–553.
- Han G, Bi WZ, Xu M, et al. Amputation versus limb-salvage surgery in patients with osteosarcoma: a meta-analysis. World J Surg. 2016;40:2016–2027.
- Harrison DJ, Geller DS, Gill JD, et al. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18:39–50.
- Meazza C, Scanagatta P. Metastatic osteosarcoma: a challenging multidisciplinary treatment. Expert Rev Anticancer Ther. 2016;16:543–556.
- Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281.
- Dong R, Ma XK, Chen LL, et al. Increased complexity of circRNA expression during species evolution. RNA Biol. 2017;14:1064–1074.
- Du WW, Zhang C, Yang W, et al. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7:4183–4191.
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66.
- Zhang H, Wang G, Ding C, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017;8:61687–61697.
- Xu B, Yang T, Wang Z, et al. CircRNA CDR1as/miR-7 signals promote tumor growth of osteosarcoma with a potential therapeutic and diagnostic value. Cancer Manag Res. 2018;10:4871–4880.
- Huang S, Zhang X, Guan B, et al. A novel circular RNA hsa_circ_0008035 contributes to gastric cancer tumorigenesis through targeting the miR-375/YBX1 axis. Am J Transl Res. 2019;11:2455–2462.
- Hu M, Wang B, Qian D, et al. Human cytomegalovirus immediate-early protein promotes survival of glioma cells through interacting and acetylating ATF5. Oncotarget. 2017;8:32157–32170.
- Farfalli GL, Albergo JI, Lobos PA, et al. Osteosarcoma lung metastases. Survival after chemotherapy and surgery. Medicina (B Aires). 2015;75:87–90.
- Huang C, Shan G. What happens at or after transcription: insights into circRNA biogenesis and function. Transcription. 2015;6:61–64.
- Xi Y, Fowdur M, Liu Y, et al. Differential expression and bioinformatics analysis of circRNA in osteosarcoma. Biosci Rep. 2019;39. DOI:https://doi.org/10.1042/BSR20181514
- Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370.
- Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612.
- Li JF, Song YZ. Circular RNA GLI2 promotes osteosarcoma cell proliferation, migration, and invasion by targeting miR-125b-5p. Tumour Biol. 2017;39:1010428317709991.
- Wu Z, Shi W, Jiang C. Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/beta-catenin pathway by regulating miR-1205/APC2 axis. Biochem Biophys Res Commun. 2018;502:465–471.
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283.
- Mitra A, Pfeifer K, Park KS. Circular RNAs and competing endogenous RNA (ceRNA) networks. Transl Cancer Res. 2018;7:S624–s8.
- Shi ZC, Chu XR, Wu YG, et al. MicroRNA-375 functions as a tumor suppressor in osteosarcoma by targeting PIK3CA. Tumour Biol. 2015;36:8579–8584.
- Liu W, Zhao X, Zhang YJ, et al. MicroRNA-375 as a potential serum biomarker for the diagnosis, prognosis, and chemosensitivity prediction of osteosarcoma. J Int Med Res. 2018;46:975–983.
- Liu G, Huang K, Jie Z, et al. CircFAT1 sponges miR-375 to promote the expression of Yes-associated protein 1 in osteosarcoma cells. Mol Cancer. 2018;17:170.
- McManus MM, Weiss KR, Hughes DP. Understanding the role of Notch in osteosarcoma. Adv Exp Med Biol. 2014;804:67–92.
- Tao J, Jiang MM, Jiang L, et al. Notch activation as a driver of osteogenic sarcoma. Cancer Cell. 2014;26:390–401.
- Jin H, Luo S, Wang Y, et al. miR-135b stimulates osteosarcoma recurrence and lung metastasis via Notch and Wnt/beta-catenin signaling. Mol Ther Nucleic Acids. 2017;8:111–122.
