ABSTRACT
The early embryonic development is important for the subsequent embryo implantation, and any defects in this process can lead to embryonic aneuploidy, which causes miscarriage and birth defects. Survivin is the member of inhibitor of apoptosis protein (IAP) family, and it is also an essential subunit of chromosomal passenger complex (CPC), which regulates both apoptosis and cell cycle control in many models. However, the roles of survivin in mouse early embryos remain unclear. In the present study, we showed that survivin activity was essential for mouse early embryo development. Our results showed that survivin mainly accumulated at chromosomes at metaphase stage and located at the spindle midzone at anaphase and telophase stages during the first cleavage. Loss of survivin activity led to the failure of cleavage in early mouse embryos. Further analysis indicated that survivin involved into spindle organization and chromosome alignment. Moreover, inhibition of survivin induced oxidative stress and DNA damage, showing with the increase of ROS level, the positive γH2A signal, and the increase of Rad51 level. We also observed the occurrence of autophagy and apoptosis in the survivin-inhibited embryos. In summary, our study suggested that survivin was a critical regulator for early embryo development through its regulation on spindle organization, chromosome alignment, and DNA damage.
Introduction
The early embryo development begins with the zygote formation. After fertilization, the zygotes complete the first cleavage and enter the two-cell embryonic stage followed by the mitotic spindle formation and chromosome separation [Citation1,Citation2]. Subsequently, the embryos undergo successive mitosis and develop to morulae [Citation3]. Next, unequal division leads to the formation of two different cell groups: the cells located inside the morulae form the ectoderm and endoderm, namely inner cell mass (ICM); while the cells outside of the morulae form the trophectoderm (TE) [Citation4]. At about the 30-cell stage, the morulae develop into blastulas after coelomation [Citation5]. Accurate DNA replication and chromosome segregation are important for the embryo to maintain integrity of genome [Citation6]. And DNA damage could cause the blocking of cell cycle, which further affects the cleavage and embryo quality [Citation7]. Therefore, during early embryonic development, any mistake in any link will cause the failure of embryo implantation and embryo death.
Survivin is known as a bifunctional protein that related with apoptosis and cell cycle control [Citation8,Citation9]. Apoptosis inhibition may be a general feature of neoplasia, and the BIR domain (Baculovirus IAP Repeats) of survivin is supposed to be important for anti-apoptotic regulation [Citation10]. As the member of inhibitor of apoptosis protein family (IAP), survivin could inhibit cell death by physically interacting with caspases. Thus, survivin is a potential target for apoptosis-based therapy in cancer [Citation9,Citation11,Citation12]. Meanwhile, survivin is also identified as the composition of the chromosomal passenger complex (CPC). CPC contains four subunits: the inner centromere protein (INCENP), survivin, Aurora-B kinase, and Borealin/Dasra-B [Citation13,Citation14]. CPC participates in several key mitotic events, such as the correct attachment of chromosome-microtubule, activation of the spindle assembly checkpoint (SAC), and construction/regulation of the contractile apparatus that drives cytokinesis [Citation15]. SAC is important to guarantee precise chromosome segregation by inhibiting the activity of the anaphase-promoting complex/cyclosome (APC/C) [Citation16]. In human cells, survivin is crucial to stable association of BubR1 to kinetochores and sustained SAC signaling [Citation17]. During mouse and porcine oocytes meiosis, survivin is an important factor to regulate SAC activity [Citation18–Citation20]. During embryonic development, survivin has been shown to be an essential inhibiting apoptotic factor. Moreover, it is shown that survivin is expressed in preimplantation embryos and could protect the embryos from apoptosis by inhibiting an apoptotic pathway [Citation21], and survivin prevents insect midgut from cell death during postembryonic development [Citation22].
Although survivin has been showed as an important inhibitor of apoptosis in early embryos; however, it is still unclear whether survivin has other regulatory functions as the composition of CPC during embryonic development. In the present study, we used the survivin inhibitor YM-155 to investigate the functions of survivin during mouse early embryo development. Our results showed that survivin regulated spindle morphology, chromosome alignment, and DNA damage repair, which was essential for embryonic development.
Materials and methods
Antibodies and chemicals
The survivin inhibitor YM-155 was from Selleck (Shanghai). Rabbit monoclonal anti-survivin antibody was from Cell Signaling Technology (Beverly, MA, USA), rabbit monoclonal anti-γ-H2A.X antibody, rabbit monoclonal anti-MAP1LC3A antibody, and cytochalasin B were from Abcam (Cambridge, UK), rabbit polyclonal anti-RAD51 antibody was purchased from Proteintech (Proteintech, CHI, USA). Mouse monoclonal anti-α-tubulin-FITC antibody and Hoechst 33,342 were from Sigma-Aldrich Corp (St. Louis, MO, USA). Alexa Fluor 488 goat anti-rabbit antibody and Alexa Fluor 594 goat anti-rabbit antibody were from Invitrogen (Carlsbad, CA, USA). EGTA was from Solarbio (Beijing). All other reagents not mentioned were from Sigma.
Parthenogenetic activation and embryo culture
We followed the guidelines of the Animal Research Institute Committee of Nanjing Agricultural University, and the animal facility had license authorized by the experimental animal committee of Jiangsu Province (SYXK-Su-20,170,007). Female mice (8 wk) were used to super-ovulated. After 48 h of intraperitoneal injection with 5 IU pregnant mare serum gonadotrophin (PMSG), the mice were then injected with 5 IU human chorionic gonadotrophin (HCG). Finally, after 14 h the cumulus-oocyte complexes (COCs) were collected from the ampullae of oviducts. To get the exposed MII oocytes, COCs were treated with 10 mg/mL hyaluronidase for 5 min. For production of parthenogenetic embryos, MII oocytes were activated by parthenogenetic activation medium, which includes 5 μg/mL cytochalasin B, 2 mM EGTA and 5 mM SrCl2 in M16 culture medium. Embryos were cultured in M16 culture medium, under mineral oil at 37°C in 5% CO2 atmosphere.
YM-155 treatment
YM-155 was dissolved in DMSO to 15 mM for reserve solution. We chose 15 nM and 20 nM (M16 diluted) working concentration for pre-experiment, and we chose 15 nM for the following experiments. The embryos were cultured in the M16 medium for different times and analyzed by immunofluorescence staining.
Immunofluorescence staining and confocal microscopy
Embryos were fixed 30 min in 4% paraformaldehyde (in phosphate-buffered saline (PBS)), then permeabilized for 20 min with 0.5% Triton X-100 in PBS, and blocked for 1 h in blocking buffer (1% BSA-supplemented PBS) at room temperature. For survivin, tubulin, γ-H2A.X and LC3A staining, the embryos were incubated with primary antibodies (survivin 1:200; α-tubulin-FITC 1:100; γ-H2A.X 1:200; MAP1LC3A 1:200) at 4°C overnight, then embryos were washed 3 times (5 min each time) by wash buffer (0.1% Tween 20 and 0.01% Triton X-100 in PBS). For secondary antibody staining, Alexa Fluor 488 goat anti-rabbit or Alexa Fluor 594 goat anti-rabbit antibody were incubated (1:200) for 1 h at room temperature. Finally, embryos were incubated with Hoechst 33,342 at room temperature for 10–20 min. After staining, samples were mounted on glass slides and observed with a confocal laser-scanning microscope (Zeiss LSM 800 META, Germany).
Reactive oxygen species (ROS) detection
Reactive Oxygen Species Assay Kit (DCFH-DA, Beyotime Institute of Biotechnology, China) was used to analyze the ROS levels of in 2-cell embryos. The 2-cell-embryos were transferred from M16 to DCFH-DA (1:800, with M16 medium dilution) for 30 min at 37°C. Then, we washed the embryos with fresh M16 for three times. Confocal fluorescent microscope (OLYMPUS CKX53, Japan) was used to detect the ROS fluorescent signals.
Real-time quantitative PCR analysis
Total RNA was extracted from 30 2-cell embryos using the Dynabeads mRNA DIRECT kit (Invitrogen Dynal AS, Norway). The first-strand cDNA was generated with PrimeScript RT Master Mix (Takara, Japan), under the following reaction conditions: 37°C for 15 min, 85°C for 5 s, hold at 4°C. The following primers were used to amplify the full-length coding sequence of target genes by RT-PCR: Sod1-F: 5′-AAC CAG TTG TGT TGT CAG GAC-3′, Sod1-R: 5′- CCA CCA TGT TTC TTA GAG TGA GG-3′; Sod2-F: 5′-CAG ACC TGC CTT ACG ACT ATG G-3′, Sod2-R: 5′-CTC GGT GGC GTT GAG ATT GTT-3′ Rad51-F: 5′-AAG TTT TGG TCC ACA GCC TAT TT-3′, Rad51-R: 5′-CGG TGC ATA AGC AAC AGC C-3′; Rad54-F: 5′-GAC AGT AAC TCC TAA GAA ACG CA-3′, Rad54-R: 5′-GCC GGT TGA GTA GCT GAG TC-3′; P62-F: 5′-ATG TGG AAC ATG GAG GGA AGA-3′, P62-R: 5′-GGA GTT CAC CTG TAG ATG GGT-3′; Bax-F: 5′-TGA AGA CAG GGG CCT TTT TG-3′, Bax-R: 5′-AAT TCG CCG GAG ACA CTC G-3′; Bak-F: 5′-GTG ACC TGC TTT TTG GCT GAT-3′, Bak-R: 5′-GGT CTC TAC GCA AAT TCA GGG-3′; mTOR-F: 5′- ACC GGC ACA CAT TTG AAG AAG −3′, mTOR-R: 5′- CTC GTT GAG GAT CAG CAA GG −3′; beclin-1-F: 5′- ATG GAG GGG TCT AAG GCG TC −3′, beclin-1-R: 5′- TCC TCT CCT GAG TTA GCC TCT −3′; gapdh-F: 5′- AGG TCG GTG TGA ACG GAT TTG −3′, gapdh-R: 5′- TGT AGA CCA TGT AGT TGA GGT CA −3′.
Real time-PCR reaction system (20 µL) was used: 10xFaste Universal SYBR Green Master (ROX) 10 µL; forward primer and reverse primer 0.8 µL, respectively; cDNA template 2 µL; ddH20 6.4 µL. Real time-PCR was conducted with a fast real-time PCR system (ABI Step One Plus). And the reaction conditions as following: Denaturation for 30 s at 95°C; 40 cycles of PCR for the quantitative analysis (95°C for 5 s and 60°C for 30 s), hold at 4°C. The relative expression of each gene was calculated using the 2−ΔΔCt method and performed three times per sample.
Western blot analysis
One hundred mice 2-cell embryos were placed in Laemmli sample buffer and heated at 100°C for 10 min. Proteins were separated by SDS-PAGE at 165 V for 80 min and then electrophoretically transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA) at 20 V for 75 min. After transfer, the membranes were then blocked with TBST (TBS containing 0.1% Tween 20) containing 5% nonfat milk at room temperature for 1 h. After blocking, the membranes were incubated with rabbit monoclonal anti-γ-H2A.X antibody (1:500), rabbit polyclonal anti-RAD51 antibody (1:500), and rabbit monoclonal anti-tubulin antibody (1:2000) at 4°C overnight. After washing 5 times in TBST (5 min each time), membranes were incubated for 1 h at room temperature with HRP-conjugated Pierce Goat anti-Rabbit IgG (1:5000). After washing for 5 times, the membranes were visualized using chemiluminescence reagent (Millipore, Billerica, MA). The experiment was repeated 3 times with different samples.
Statistical analysis
At least three biological replicates were performed for each analysis. Means ± SEM were used to express the results of group. All analyses were performed using GraphPad Prism7.00 software (GraphPad, CA, USA). Results of P < 0.05 were considered statistically significant (differences P < 0.05 denoted by *, P < 0.01 denoted by **, differences P < 0.001 denoted by *** and P < 0.00001 denoted by ****).
Results
Localization of survivin during mouse embryonic development
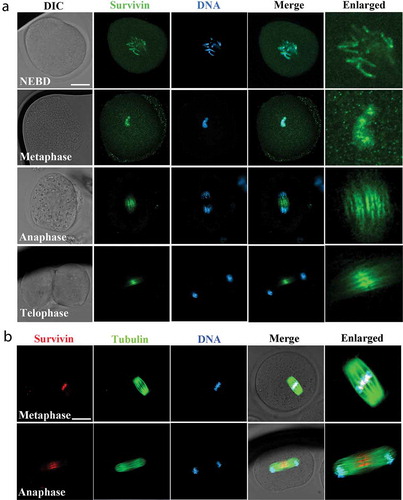
We first examined subcellular localization of survivin at different stages of embryo first cleavage by immunofluorescent staining. Our results showed that survivin was mainly accumulated at chromosomes after nuclear membrane breakdown (NEBD) and at metaphase stages; when chromosomes segregated and the zygote entered anaphase and telophase stages, survivin was mainly located at the midzone and midbody (). We also co-stained survivin with microtubules, as shown in , survivin accumulated at the central spindle at the anaphase in the zygotes. The localization pattern of survivin indicated that survivin might interact with chromosomes-related functions during embryonic development.
Figure 1. The localization of survivin during mouse early embryonic development. (a) Subcellular localization of survivin during mouse first cleavage of embryos. Survivin was enriched at the chromosomes (NEBD and metaphase stages) and spindle midzone (anaphase and telophase stages). Green, survivin; blue, DNA. Bar = 20 μm. (b) Co-stain of survivin and tubulin during mouse first cleavage of embryos. Survivin was enriched at the chromosomes (metaphase stage) and spindle midzone (anaphase stage). Red, survivin; green: tubulin; blue, DNA. Bar = 20 μm.
Inhibition of survivin activity affects the first cleavage of mouse early embryonic development
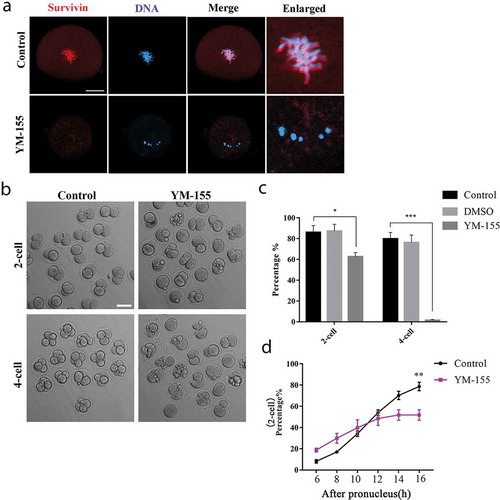
To investigate the function of survivin, we used survivin inhibitor YM-155 to discover the prospective role during embryonic development. We treated embryos with 15 nM concentration of YM-155 for subsequent experiments. After treated the embryos with YM-155, the specific localization of survivin disappeared (). The cleavage rate of control group and DMSO group showed no significant difference (the rate of 2-cell: control group vs DMSO group: 86.00 ± 6.51%, n = 284 vs. 87.23 ± 6.53%, n = 158, p > 0.05; the rate of 4-cell: control group vs DMSO group: 79.84 ± 6.11%, n = 203 vs. 76.20 ± 7.24%, n = 132, p > 0.05. ); we then compared control group and YM-155 treatment group in the following experiments. Our results showed that after the YM-155 treatment, most embryos could not complete first cleavage and failed to form the 2-cell embryos; moreover, almost all embryos were unable to develop to 4-cell embryos (). Statistical analysis data also confirmed this result (the rate of 2-cell: control group vs YM-155 group: 86.00 ± 6.51%, n = 284 vs. 62.67 ± 3.84%, n = 257, p < 0.05; the rate of 4-cell: control group vs YM-155 group: 79.84 ± 6.11%, n = 203 vs. 1.55 ± 0.78%, n = 179, p < 0.001) (). Moreover, we analyzed the 2-cell rate at different time points to check the cell cycle progression, and the results showed that at the starting of the embryos cleavage, there was a higher rate of 2-cell embryos in YM-155 treatment group; however, after 16 h culture, the 2-cell rate was significantly lower compared with the control group (). Our results indicated that inhibition of survivin might result in precocious anaphase and induce the decrease of cleavage rate.
Figure 2. Disrupting survivin activity inhibits cleavage during mouse early embryonic development. (a) Subcellular localization of survivin after YM-155 treatment. The specific localization of survivin was disappeared. Red, survivin; blue, DNA. Bar = 20 μm. (b) DIC images of 2-cell and 4-cell cleavage rate of control group and YM-155 treatment group. Bar = 100 μm. (c) The rate of 2-cell and 4-cell for early embryos of control group, DMSO group and YM-155 treatment group. There was no difference between control group and DMSO group during embryonic development (Control group: n = 284; DMSO group: n = 158). The rate of 2-cell and 4-cell was decreased in YM-155 group compared with control group (2-cell: Control group: n = 284; YM-155 group: n = 257); (4-cell: Control group: n = 203; YM-155 group: n = 179). *Significant difference (p < 0.05); ***Significant difference (p < 0.001). (d) Statistical data for 2-cell rate at different time points. **Significant difference (p < 0.01) (Control group: n = 195; YM-155 group: n = 201).
Inhibition of survivin affects spindle morphology and chromosome alignment at the first cleavage of mouse embryos
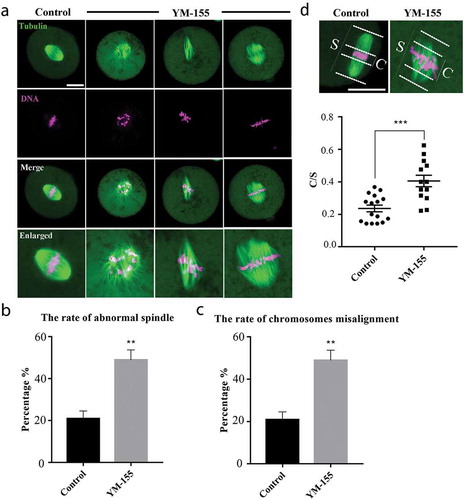
To clarify the causes of survivin on the cleavage of mouse embryos, based on the localization of survivin we examined the spindle morphology and chromosome alignment at metaphase of first cleavage embryos. As shown in , in the control group, 1-cell embryos exhibited complete barrel-shaped spindles, with the well-arranged chromosomes at the equatorial plate of spindle. By contrast, after the YM-155 treatment, the spindles of the embryos showed a variety of defects, such as the multipolar and nonpolar spindle, and the chromosomes in embryos of the YM-155 treatment group were severely misaligned. This could be supported by the statistical analysis data (the rate of abnormal spindle: control group vs YM-155 group: 20.86 ± 3.72%, n = 151 vs. 48.46 ± 7.88%, n = 139, p < 0.01) (); (the rate of chromosome misalignment: control group vs YM-155 group: 20.86 ± 3.72%, n = 151 vs. 44.06 ± 5.92%, n = 139, p < 0.01) (). Moreover, we measured the width of chromosome plate (C) and the spindle (S) at metaphase stage, we calculated the C/S and found that it was remarkably lower in control group compared with the YM-155 treatment group (0.24 ± 0.02, n = 16 vs. 0.41 ± 0.04, n = 13, p < 0.001. ). These results indicated that survivin regulated the development of mouse embryos by affecting spindle morphology and chromosome alignment at metaphase of first cleavage.
Figure 3. Survivin inhibition affects spindle morphology and chromosome alignment during mouse early embryonic development. (a) Representative images of spindle morphology in the control group and YM-155 treatment group. Compared with the control group, the spindles morphology of the embryos in YM-155 treatment group showed a variety of defects, and chromosomes were severely misaligned. Green: tubulin; magenta, DNA. Bar = 20 μm. (b) The proportion of abnormal spindle morphology in the control group and YM-155 treatment group. **Significant difference (p < 0.01) (Control group: n = 151; YM-155 group: n = 139). (c) The proportion of chromosome misalignment in the control group and YM-155 treatment group. *Significant difference (p < 0.05) (Control group: n = 151; YM-155 group: n = 139) (d) Representative images and statistic of chromosomes width/spindle (C/S). ***Significant difference (p < 0.001). Green: tubulin; magenta, DNA. Bar = 20 μm.
Inhibition of survivin affects oxidative stress and DNA damage during mouse embryo development
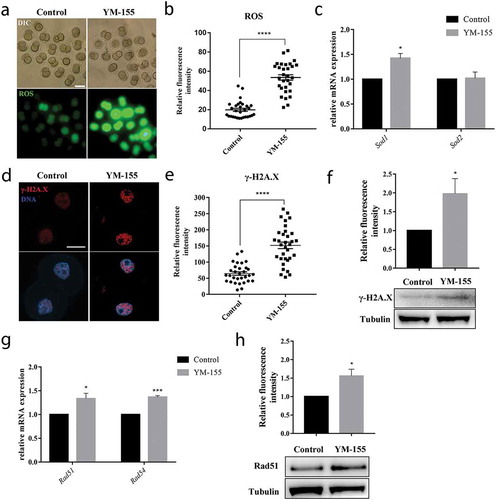
Previous studies showed that survivin was related with DNA damage response, and the oxidative stress could lead to DNA damage. In order to further explore the potential regulatory mechanism of survivin during embryo development, we explored the levels of ROS in control group and YM-155 treatment group in 2-cell embryos. Our results showed that YM-155 treatment caused the increased ROS level compared with the control group (), which was also supported by the statistical analysis (the ROS signal fluorescence intensity: control group vs YM-155 group: 19.89 ± 1.61, n = 30 vs. 53.54 ± 2.87, n = 30, p < 0.0001) (). The mRNA level of sod1 also significantly increased in YM-155 treatment group: Control group vs YM-155 group: 1 vs. 1.43 ± 0.10, p < 0.05 (). These results suggested that survivin induced oxidative stress in mouse early embryos. Next, we employed γ-H2A.X staining to detect the DNA damage in 2-cell embryos. As shown in , the γ-H2A.X signal fluorescence intensity was highly increased in the YM-155 treatment group embryos. This also could be supported by the statistical analysis (the γ-H2A.X signal fluorescence intensity: control group vs YM-155 group: 64.66 ± 5.22, n = 31 vs. 151.9 ± 10.08, n = 33, p < 0.0001) (). A significant increase of γ-H2A.X level was shown in YM-155 treatment group compared to control group by western blotting (1.98 ± 0.40 vs.1, p < 0.05, ). Furthermore, the expression of DNA repaired-related genes Rad51 and Rad54 were increased after YM-155 treatment (Rad51: 1 vs. 1.34 ± 0.11, p < 0.05; Rad54: 1 vs. 1.37 ± 0.03, p < 0.001, ). A significant increase of Rad51 level was shown in YM-155 treatment group compared to control group by western blotting (1.56 ± 0.18 vs.1, p < 0.05, ). These results showed that loss of survivin activity induced DNA damage during embryonic development.
Figure 4. Survivin inhibition affects oxidative stress and DNA damage during mouse early embryonic development. (a) Representative images of ROS fluorescence intensity in the control group and YM-155 treatment group. Green: ROS. Bar = 100 μm. (b) The fluorescent intensities of ROS in the control group and YM-155 treatment group. ****Significant difference (p < 0.0001). (c) The mRNA expression of Sod1 and Sod2 in the control group and YM-155 treatment group. Sod1 expression was significant increased, *Significant difference (p < 0.05). (d) Representative images of γ-H2A.X fluorescence intensity in the control group and YM-155 treatment group. Red: γ-H2A.X; blue, DNA. Bar = 20 μm. (e) The fluorescent intensities of γ-H2A.X in the control group and YM-155 treatment group. ****Significant difference (p < 0.0001). (f) Western blot analysis for γ-H2A.X expression in the YM-155 group and control group. Relative intensities of γ-H2A.X and tubulin were assessed by densitometry. *, significant difference (p < 0.05). (g) The mRNA expression of Rad51 and Rad54 in the control group and YM-155 treatment group. *Significant difference (p < 0.05); ***Significant difference (p < 0.001). (h) Western blot analysis for Rad51 expression in the YM-155 group and control group. Relative intensities of Rad51 and tubulin were assessed by densitometry. *, significant difference (p < 0.05).
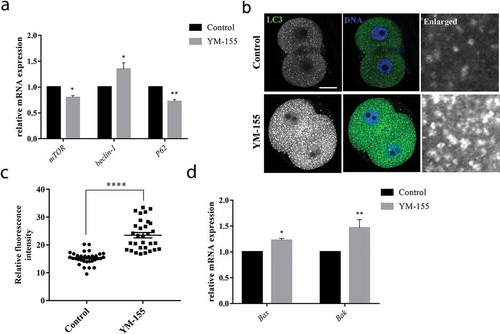
Inhibition of survivin affects autophagy level during mouse embryo development
The abnormal levels of oxidative stress could further induce autophagy and apoptosis. Next, we examined the autophagy level in the control group and YM-155 treatment group. We examined the autophagy relative genes mTOR, Beclin-1 and P62 mRNA level after YM-155 treatment. The results showed that mTOR, Beclin-1 and P62 mRNA level changed in YM-155 treatment group. As shown in , the mTOR and P62 mRNA expression significantly decreased in YM-155 treatment group: mTOR: 1 vs. 0.809 ± 0.04, p < 0.01; P62: 1 vs. 0.72 ± 0.04, p < 0.05. The Beclin-1 mRNA expression significantly increased in YM-155 treatment group: 1 vs. 1.35 ± 0.12, p < 0.05. Moreover, by immunofluorescence staining of LC3 at 2-cell embryos, we found that more dots of LC3 signals in the YM-155 treatment group (). Statistical analysis showed that the LC3 signal fluorescence intensity significantly increased in the YM-155 treatment group (control group vs YM-155 group: 15.31 ± 0.40, n = 32 vs. 23.52 ± 0.96, n = 31, p < 0.0001, ). Since survivin is an important apoptosis-related protein, we examined the apoptosis-relative genes Bax and Bak mRNA level after YM-155 treatment. The results showed that Bax and Bak mRNA level expression significantly increased in YM-155 treatment group. As shown in : Bax: 1 vs. 1.23 ± 0.04, p < 0.05; Bak: 1 vs. 1.47 ± 0.16, p < 0.01. These results indicated that survivin induced autophagy and apoptosis during mouse embryonic development.
Figure 5. Inhibition of survivin induces autophagy and apoptosis during mouse early embryonic development. (a) The mRNA expression of mTOR, beclin-1 and P62 in the control group and YM-155 treatment group. mTOR and beclin-1, *Significant difference (p < 0.05); P62, **Significant difference (p < 0.01). (b) Representative images of LC3 fluorescence intensity in the control group and YM-155 treatment group. Green: LC3; blue, DNA. Bar = 20 μm. (c) The fluorescent intensities of LC3 in the control group and YM-155 treatment group. ****Significant difference (p < 0.0001). (d) The mRNA expression of Bax and Bak in the control group and YM-155 treatment group. *Significant difference (p < 0.05); **Significant difference (p < 0.01).
Discussion
This study was designed to explore the functions of survivin during mouse early embryonic development. Our results indicated that survivin regulated spindle morphology and chromosome alignment in the metaphase stage of first cleavage in mouse embryos. Moreover, inhibition of survivin also induced oxidative stress, which further led to DNA damage, and caused autophagy and apoptosis. Our results provided evidence that survivin played critical roles for mouse early embryonic development.
Survivin, also known as BIRC5, is reported to be essential for cell division and can inhibit cell death, which is a well-known cancer therapeutic target gene [Citation23,Citation24]. During embryonic development, survivin has been shown to be an essential inhibiting apoptotic factor [Citation21,Citation22]. However, it is unclear whether survivin has other regulatory functions during embryonic development. In this study, we showed that survivin expressed in mouse zygotes and it mainly accumulated at chromosomes after NEBD and at metaphase stage, when chromosomes segregated and the embryo entered anaphase and telophase stages, survivin was located at the midzone of the spindle. This localization pattern is similar to its localization in proliferating cells [Citation25–Citation27]. In porcine oocytes, survivin was mainly co-localized with chromosomes after germinal vesicle breakdown (GVBD) and at the metaphase stage [Citation19]. During mouse oocyte meiotic maturation, our previous study also showed that survivin was mainly associated with centromeres in metaphase stage, and distributed at the midbody of spindle in anaphase and telophase stages [Citation20].
Previous studies indicate that survivin could also affect mitotic spindle assembly by dampening microtubule dynamics and help to ensure chromosomes alignment [Citation28,Citation29]. In Xenopus egg extracts, survivin is important for spindle assembly and it is a necessary component of the mitotic spindle [Citation30]. Meanwhile, survivin is reported to regulate spindle organization and chromosome segregation during rat oocyte meiosis [Citation31]. Based on the specific localization of survivin and the previous studies, we wondered whether the function of survivin in embryos was similar with other cells. We used survivin inhibitor YM-155 to explore the roles of survivin in mouse embryos. Our results showed that after the treatment of YM-155, the cleavage of the embryos was disturbed. It is shown that survivin-depleted oocytes failed to complete cytokinesis [Citation19]. Our results also showed that inhibition of survivin caused abnormal of spindle morphology and chromosomes arrangement at metaphase in mouse early embryos. These results were consistent with previous reports, revealing the conserved function of survivin in different cells and species.
In human breast tumors, survivin shows close relationship with oxidative stress [Citation32]. Similarly, our results showed that inhibition of survivin caused the increase of reactive oxygen species (ROS) level; therefore, high levels of ROS might be one of the causes that contribute to early embryonic dysplasia after inhibition of survivin activity. It is well known that ROS could induce DNA damage and consequently, a DNA damage response (DDR) [Citation33,Citation34]. ROS could induce double-strand DNA breaks (DSBs) and oxidative clustered DNA lesions (OCDLs) [Citation35]. During the repair process, OCDLs could convert to DSBs such as active the base excision repair pathway, and the DSBs are one of the most serious DNA damage [Citation36]. Previous studies also showed that survivin inhibition could induce DNA damage [Citation37,Citation38], in cancer cell survivin-ΔEx3 could be phosphorylated by the checkpoint kinase Chk2 during DNA damage [Citation39]; In Bcl-xL silenced glioma cell lines, YM-155 treatment led to the mitochondrial dysfunction and DNA damage [Citation38]. To verify whether survivin has the similar functions during embryonic development, we analyzed the DNA damage and DNA repair factors. Our results showed that inhibition of survivin caused the increase of DNA damage and DNA repair in early mouse early embryonic development. Therefore, the inhibition of survivin could lead to the oxidative stress, the increase of ROS, and DNA damage during the embryonic development.
It is widely known that survivin is an anti-apoptosis protein, and survivin expresses in preimplantation embryos and could protect the embryos from apoptosis by inhibiting an apoptotic pathway [Citation21]. Autophagy can constitute the stress adaptation to avoid apoptosis, whereas sometimes it leads to apoptosis and cell death [Citation40]. Furthermore, autophagy could be activated by DNA damage and oxidative stress [Citation41]. Our results showed that inhibition of survivin increased the autophagy and apoptosis level during mouse early embryonic development. Similarly, survivin is shown to have a novel role in cancer cells to directly regulate autophagy [Citation42]. In prostate cancer cells, YM155-induced autophagy plays a pro-apoptotic role [Citation43]. Therefore, we suggested that survivin activated autophagy due to its effects on DNA damage, and further led to apoptosis during early embryonic development.
In conclusion, our results indicate that survivin is essential for mouse early embryo development through its regulation on spindle organization, chromosome alignment, and DNA damage.
Authors contribution
Meng-Hao Pan, Shao-Chen Sun conceived the study; Meng-Hao Pan performed the majority of the experiments; Jia-Qian Ju, Xiao-Han Li, Yi Xu, Jie-Dong Wang, Yan-Ping Ren, Xiang Lu contributed to the materials and methods; Meng-Hao Pan wrote the manuscript; Shao-Chen Sun revised the manuscript; all the authors approved the submission of the manuscript.
Acknowledgments
We thank Yuehua Ma for guiding use of the confocal microscope (Central Laboratory, College of Horticulture, Nanjing Agriculture University, Nanjing, China).
Data availability statement
All the data can be found in the manuscript.
Disclosure statement
The authors have no conflict of interest to declare.
Additional information
Funding
References
- Schatten G, Simerly C, Schatten H. Microtubule configurations during fertilization, mitosis, and early development in the mouse and the requirement for egg microtubule-mediated motility during mammalian fertilization. Proc Natl Acad Sci U S A. 1985;82:4152–4156.
- Yurttas P, Morency E, Coonrod SA. Use of proteomics to identify highly abundant maternal factors that drive the egg-to-embryo transition. Reproduction. 2010;139:809–823.
- Ziomek CA, Johnson MH. Cell surface interaction induces polarization of mouse 8-cell blastomeres at compaction. Cell. 1980;21:935–942.
- Marikawa Y, Alarcon VB. Establishment of trophectoderm and inner cell mass lineages in the mouse embryo. Mol Reprod Dev. 2009;76:1019–1032.
- McLaren A, Smith R. Functional test of tight junctions in the mouse blastocyst. Nature. 1977;267:351–353.
- Barnum KJ, O’Connell MJ. Cell cycle regulation by checkpoints. Methods Mol Biol. 2014;1170:29–40.
- O’Connell MJ, Walworth NC, Carr AM. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 2000;10:296–303.
- Kallio MJ, Nieminen M, Eriksson JE. Human inhibitor of apoptosis protein (IAP) survivin participates in regulation of chromosome segregation and mitotic exit. The FASEB Journal. 2001;15(14):1–19.
- Jaiswal PK, Goel A, Mittal RD. Survivin: A molecular biomarker in cancer. Indian J Med Res. 2015;141:389–397.
- Wheatley SP, McNeish IA. Survivin: a protein with dual roles in mitosis and apoptosis. Int Rev Cytol. 2005;247:35–88.
- Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54.
- Tamm I, Wang Y, Sausville E, et al. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998;58:5315–5320.
- Vader G, Kauw JJ, Medema RH, et al. Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Rep. 2006;7:85–92.
- Jeyaprakash AA, Klein UR, Lindner D, et al. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007;131:271–285.
- Carmena M, Wheelock M, Funabiki H, et al. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13:789–803.
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393.
- Lens SM, Wolthuis RM, Klompmaker R, et al. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. Embo J. 2003;22:2934–2947.
- Sun SC, Wei L, Li M, et al. Perturbation of survivin expression affects chromosome alignment and spindle checkpoint in mouse oocyte meiotic maturation. Cell Cycle. 2009;8:3365–3372.
- Chen L, Yin T, Nie ZW, et al. Survivin regulates chromosome segregation by modulating the phosphorylation of Aurora B during porcine oocyte meiosis. Cell Cycle. 2018;17:2436–2446.
- Sun SC, Liu HL, Sun QY. Survivin regulates Plk1 localization to kinetochore in mouse oocyte meiosis. Biochem Biophys Res Commun. 2012;421:797–800.
- Kawamura K, Sato N, Fukuda J, et al. Survivin acts as an antiapoptotic factor during the development of mouse preimplantation embryos. Dev Biol. 2003;256:331–341.
- He HJ, Hou L, Wang JX, et al. The apoptosis inhibitor survivin prevents insect midgut from cell death during postembryonic development. Mol Biol Rep. 2012;39:1691–1699.
- Wheatley SP, Altieri DC. Survivin at a glance. J Cell Sci. 2019;132:7.
- Li F, Aljahdali I, Ling X. Cancer therapeutics using survivin BIRC5 as a target: what can we do after over two decades of study? J Exp Clin Cancer Res. 2019;38:368.
- Jeyaprakash AA, Basquin C, Jayachandran U, et al. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure. 2011;19:1625–1634.
- Yamagishi Y, Honda T, Tanno Y, et al. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–243.
- Uren AG, Wong L, Pakusch M, et al. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol. 2000;10:1319–1328.
- Rosa J, Canovas P, Islam A, et al. Survivin modulates microtubule dynamics and nucleation throughout the cell cycle. Mol Biol Cell. 2006;17:1483–1493.
- Cheung CH, Chen HH, Kuo CC, et al. Survivin counteracts the therapeutic effect of microtubule de-stabilizers by stabilizing tubulin polymers. Mol Cancer. 2009;8:43.
- Canovas PM, Guadagno TM. Functional analysis of Survivin in spindle assembly in Xenopus egg extracts. J Cell Biochem. 2007;100:217–229.
- Wang K, Jiang GJ, Wei L, et al. Survivin is a critical regulator of spindle organization and chromosome segregation during rat oocyte meiotic maturation. Zygote. 2011;19:307–313.
- Pervin S, Tran L, Urman R, et al. Oxidative stress specifically downregulates survivin to promote breast tumour formation. Br J Cancer. 2013;108:848–858.
- Srinivas US, Tan BWQ, Vellayappan BA, et al. ROS and the DNA damage response in cancer. Redox Biol. 2019;25:101084.
- Wojtczyk-Miaskowska A, Schlichtholz B. DNA damage and oxidative stress in long-lived aquatic organisms. DNA Repair (Amst). 2018;69:14–23.
- Venkatachalam G, Surana U, MV C. Replication stress-induced endogenous DNA damage drives cellular senescence induced by a sub-lethal oxidative stress. Nucleic Acids Res. 2017;45:10564–10582.
- Sharma V, Collins LB, Chen TH, et al. Oxidative stress at low levels can induce clustered DNA lesions leading to NHEJ mediated mutations. Oncotarget. 2016;7:25377–25390.
- Glaros TG, Stockwin LH, Mullendore ME, et al. The “survivin suppressants” NSC 80467 and YM155 induce a DNA damage response. Cancer Chemother Pharmacol. 2012;70:207–212.
- Jane EP, Premkumar DR, Sutera PA, et al. Survivin inhibitor YM155 induces mitochondrial dysfunction, autophagy, DNA damage and apoptosis in Bcl-xL silenced glioma cell lines. Mol Carcinog. 2017;56:1251–1265.
- Lopergolo A, Tavecchio M, Lisanti S, et al. Chk2 phosphorylation of survivin-DeltaEx3 contributes to a DNA damage-sensing checkpoint in cancer. Cancer Res. 2012;72:3251–3259.
- Maiuri MC, Zalckvar E, Kimchi A, et al. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752.
- Galati S, Boni C, Gerra MC, et al. Autophagy: A Player in response to Oxidative Stress and DNA Damage. Oxid Med Cell Longev. 2019;2019:5692958.
- Lin TY, Chan HH, Chen SH, et al. BIRC5/Survivin is a novel ATG12-ATG5 conjugate interactor and an autophagy-induced DNA damage suppressor in human cancer and mouse embryonic fibroblast cells. Autophagy. 2020;16(7):1296–1313.
- Wang Q, Chen Z, Diao X, et al. Induction of autophagy-dependent apoptosis by the survivin suppressant YM155 in prostate cancer cells. Cancer Lett. 2011;302:29–36.
