ABSTRACT
Circular RNA (circRNA) is currently considered to be a key regulatory molecule in cancer biology. In the present study, we aimed to explore the functional and clinical roles of circ-SLC7A6 (a circRNA derived from SLC7A6 gene) in non-small cell lung cancer (NSCLC). Circ-SLC7A6 was significantly downregulated in NSCLC tissues in comparison to para-carcinoma tissues. Low circ-SLC7A6 was closely associated with larger tumor size, lymph node metastasis, advanced clinical stage and adverse outcome. Exogenous expression of circ-SLC7A6 evidently inhibited the proliferation and invasion of NSCLC cells. Further investigations revealed that miR-21 was the direct functional target of circ-SLC7A6, in which circ-SLC7A6 abundantly sponged miR-21 and elevated a cohort of tumor suppressors, thus inhibiting NSCLC progression. Interestingly, QKI, elevated by circ-SLC7A6, could directly bind to the introns flanking circ-SLC7A6 to facilitate circ-SLC7A6 production. Importantly, in vivo xenograft tumor experiments showed that reintroduction of circ-SLC7A6 retarded tumor growth as well as decreased lung metastatic nodules. Overall, our study demonstrates that circ-SLC7A6 is a novel tumor suppressor in NSCLC, targeting circ-SLC7A6/miR-21 axis may be a promising treatment for NSCLC patients.
Introduction
Non-small cell lung cancer (NSCLC) is any kind of epithelial lung cancer except for small cell lung cancer (SCLC) [Citation1]. The most common types of NSCLC are squamous cell carcinoma, large cell carcinoma and adenocarcinoma, but there are several other rare types, all of which can occur in unusual histological variations [Citation2]. It is the leading cause of cancer-related mortality, representing close to 1 in 5 (18.4%) [Citation3]. Despite recent advances in screening, diagnosis and treatment of NSCLC, the 5-year relative survival rate is extremely low, less than 15%, especially advanced NSCLC cases [Citation4]. Continuous exploration of the pathogenesis of NSCLC will provide new therapeutic directions for NSCLC, thus prolonging the survival time.
Circular RNA (circRNA) is a kind of non-coding RNA molecules with closed loop structure, without 5ʹ-cap and 3ʹ-poly (A) structure [Citation5]. It is mainly located in the cytoplasm or stored in exosomes, not affected by RNA exonuclease, which enables it is highly stable and not easy to degrade [Citation6]. CircRNA has recently been proved to be widespread in various eukaryotes and expressed in a tissue- or cell-specific manner [Citation7]. CircRNA has a variety of functional patterns, among which its role as a “miRNA sponge” is well established [Citation8]. Specifically, circRNA is rich in a large number of miRNA response elements (MREs) that can sponge miRNAs and prevent miRNAs from interacting with mRNA in the 3ʹ-untranslated region (UTR), thus indirectly regulating the expression of miRNA downstream target genes [Citation9]. The most typical case is CDR1as/ciRS-7, which has been proved to have more than 70 conservative miR-7 binding sites and abundantly absorb miR-7, functioning as a miR-7 antagonist with a miRNA-binding capacity ten times higher than any other known transcript [Citation10,Citation11]. Similarly, the circRNA/miRNA interaction networks were also identified in a variety of human cancers that played crucial roles in tumorigenesis and aggressive progression, such as the circHIPK3/miR-7 axis in colorectal cancer [Citation12], the circ-BIRC6/miR-3918 axis in hepatocellular carcinoma [Citation13] and the circKDM4C/miR-548p axis in breast cancer [Citation14], etc.
In the present study, we described the functional and clinical implications of circ-SLC7A6, a circRNA originated from SLC7A6 gene (a protein coding gene that involves in amino acid transmembrane transporter activity [Citation15]), and further revealed the role of its involved circRNA/miRNA regulatory axis in the progression of NSCLC.
Materials and methods
Clinical samples, cell lines and nude mice
A total of 110 pairs of NSCLC and precancerous normal tissues were collected from Shengjing Hospital of China Medical University, which was immediately put into liquid nitrogen to ensure RNA quality for subsequent studies. We obtained the informed consent of all patients, and this study was conducted by the approval of medical ethics committee of Shengjing Hospital of China Medical University. We used two NSCLC cell lines including A549 and H460 to perform the functional experiments, they were routinely cultured in humidified chamber with DMEM medium. All nude mice used in our study were BLAB/c breed and maintained under specific conditions.
Reverse-transcription quantitative PCR (RT-qPCR)
Total RNA treated with or without 10 U RNase R (#R0301, Geneseed, Guangzhou, China) was extracted by TRIzol® reagent (Invitrogen, CA, USA). After that, two micrograms of RNA were reverse transcribed into cDNA using the PrimeScript RT Kit (Takara Bio, Kusatsu, Japan) as instructed by the manufacturer, followed by amplification and quantification by using SYBR Premix Taq™ Kit (Takara Bio). GAPDH was used to standardize gene expression levels, which was calculated by 2−ΔΔCt formula.
Generation of circ-SLC7A6-overexpressing cell lines
The full-length of circ-SLC7A6 was cloned into pLCDH-ciR vector (Geneseed), followed by infection into A549 and H460 cells in the presence with 8 μg/ml polybrene infection enhancer (Genechem, Shanghai, China). After that, cells were maintained in DMEM medium supplemented with 1.2 μg/ml puromycin (Genechem) for screening stable circ-SLC7A6-overexpressing cells.
Cell proliferation assay
CCK-8 assay was carried out by using CCK-8 solution (Liji, Shanghai, China) as per manufacturer’s instructions at the indicated time. The absorbance at 450 nm was recorded with a microplate reader. For colony formation assay, control and circ-SLC7A6-overexpressing A549 and H460 cells were plated on 96-well plates and grown for 10 days. Then, cells were stained by using crystal violet. For detecting DNA synthesis rate of A549 and H460 cells, the iFluor 647 EdU Staining Proliferation Kit (#ab222421, Abcam) was used in accordance with manufacturer’s protocol. In brief, cells were incubated with EdU solution for 3 h, followed by fixation for 15 min with methyl alcohol. Then, the permeabilization buffer and reaction mix were added into cells, the EdU positive cells were analyzed using a fluorescence microscope.
Transwell invasion assay
The chamber was pre-coated with 100 μl Matrigel for 2 h at 37°C and mounted on a 24-well plate. Then, 200 μl serum-free cell suspension was added onto the upper surface of the chamber, while 600 μl serum-containing DMEM medium was added into the lower surface. The chamber was plated into cell incubator and cultured for 18 h to allow the cells to invade the Matrigel, followed by staining with crystal violet.
Fluorescence in situ hybridization (FISH)
FISH assay was conducted by using FISH Detection Kit (GenePharma, Shanghai, China) with Cy5-labeled circ-SLC7A6 probe designed and synthesized by GenePharma. In short, the hybridization reaction mix was prepared with 70 μl hybridization buffer, 28 μl DEPC water and 2 μl circ-SLC7A6 probe, followed by addition into cells and incubation for 16 h. Then, cells were washed by SSC solution and PBS three times for 10 min each time, respectively. DAPI was added into cells for 5 min to stain nucleus. The staining results were observed under a fluorescence microscope.
RNA pull-down assay
A549 and H460 cell lysates were collected and incubated with biotinylated circ-SLC7A6 probe (GenePharma) for 3 h at room temperature. After that, the streptavidin-conjugated magnetic beads (Invitrogen) were added into the above probe-lysate complex and incubated for 30 min at room temperature. The complex enriched by circ-SLC7A6 was washed and eluted for qRT-PCR analysis. Besides, for miR-21 pulling down circ-SLC7A6, biotin-labeled miR-21 mimics were transfected into A549 and H460 cells by using Lipofectamine 3000 (Invitrogen) as per general protocol. Then, RNA pull-down was conducted as described above.
Luciferase reporter assay
The sequence of circ-SLC7A6 with wild-type or mutant miR-21 interaction site was inserted into the pMIR Report vector provided by Promega (Madison, WI, USA) and named as Luc-circ-SLC7A6-WT and Luc-circ-SLC7A6-Mut, respectively. After that, the vectors were co-transfected with control or miR-21 mimics into A549 and H460 cells by using Lipofectamine 3000 (Invitrogen). The luciferase activity in each well was measured by using the Dual Luciferase Reporter Assay Substrate Kit purchased from Promega.
RNA immunoprecipitation (RIP)
2 × 107 A549 and H460 cells were lysed in 100 μl RIP lysis buffer with 0.5 μl protease inhibitor cocktail and 0.25 μl RNase inhibitor, followed by collection of supernatants after centrifugation. For each reaction, 50 μl protein G magnetic beads with 5 μg anti-QKI (#ab126742, Abcam) or IgG were prepared in a microfuge tube as instructed. After that, 100 μl above supernatants were added into beads-antibody complex with 900 μl RIP immunoprecipitation buffer and incubated at 4°C overnight. Each well was re-suspended with 150 μl proteinase K buffer to digest protein at 55°C for 30 min. Samples were centrifuged and the supernatants were collected for RNA extraction and qRT-PCR.
Xenograft model
For subcutaneous tumor formation, 1 × 107 A549 cells in 0.2 ml PBS were injected subcutaneously into BALB/c nude mice, which were routinely grown in specific animal houses for 5 weeks. After that, the subcutaneous tumors were collected for immunohistochemistry (IHC) staining of Ki-67 (#ab16667, Abcam). For tail vein injection, 1 × 106 A549 cells in 0.1 ml PBS were injected into the lateral tail vein of BALB/c mice. After 6 weeks, the mice were killed by cervical dislocation and the lungs were collected to count surface metastases under a dissecting microscope, followed by hematoxylin & eosin (H&E) staining.
Statistical method
The relationships between circ-SLC7A6 and NSCLC clinical features were determined by Chi-square test, while the difference between two groups was compared by Student’s t-test. Kaplan-Meier plotter was used to measure the effect of circ-SLC7A6 on NSCLC patient survival, the difference was compared by Log-rank test. P < 0.05 was considered statistically significant.
Results
Circ-SLC7A6 is decreased in NSCLC
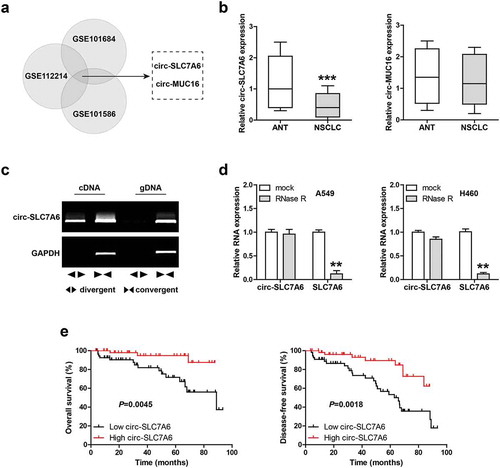
To identify the deregulated circRNAs in NSCLC, we analyzed three NSCLC GEO databases including GSE101684, GSE112214 and GSE101586. The overlapping results only contained two circRNAs, circ-SLC7A6 and circ-MUC16 ()). Then, we detected the expression of both in 110 paired NSCLC and adjacent normal tissues. The qRT-PCR results showed that circ-SLC7A6 was significantly downregulated in NSCLC, whereas circ-MUC16 had no significant change ()). We designed divergent and convergent primers to verify circ-SLC7A6, and the results displayed that circ-SLC7A6 was amplified by divergent primers only in cDNA, but not in gDNA (genomic DNA) ()). Besides, circ-SLC7A6 was highly resistant to RNase R, while linear SLC7A6 mRNA was digested in large quantities by RNase R ()), suggesting that circ-SLC7A6 is a bona fide circRNA. Importantly, we analyzed the relationships between circ-SLC7A6 and clinicopathological features of NSCLC patients, the results showed that low circ-SLC7A6 was closely associated with lymph node metastasis, larger tumor volume and higher TNM stage (). Moreover, patients with high circ-SLC7A6 had longer overall and disease-free survival time than those with low circ-SLC7A6 ()).
Table 1. The correlations between circ-SLC7A6 expression and clinicopathological characteristics in 110 NSCLC patients.
Figure 1. Low circ-SLC7A6 is observed in NSCLC. (a) Analysis of differentially expressed circRNA in NSCLC using the indicated three GEO databases. GSE112214 includes three pairs of NSCLC and matched adjacent normal tissues; GSE101684 includes tumor samples and paired adjacent normal tissues from four patients with early-stage lung adenocarcinoma; GSE101586 includes five paired lung adenocarcinoma and adjacent non-tumor tissues. (b) qRT-PCR analysis of circ-SLC7A6 and circ-MUC16 in 110 paired NSCLC and normal tissues. (c) RT-PCR analysis of circ-SLC7A6 and GAPDH expression using divergent and convergent primers. (d) qRT-PCR analysis of circ-SLC7A6/SLC7A6 expression in A549 and H460 cells treated with RNase R. (e) The survival curves of NSCLC patients based on median circ-SLC7A6 expression level. **p < 0.01, ***p < 0.001.
Overexpression of circ-SLC7A6 represses NSCLC cell proliferation and invasion
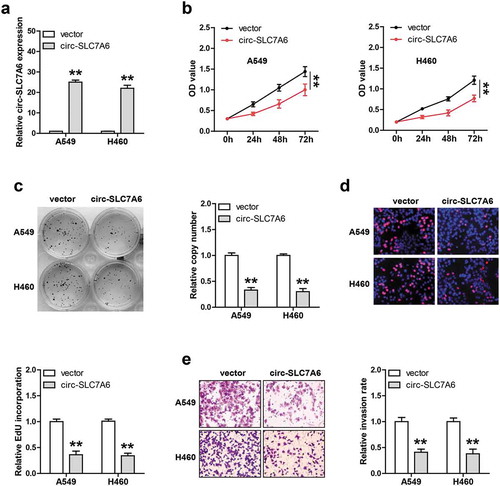
Next, we generated stable circ-SLC7A6-overexpressing A549 and H460 cell lines via using lentiviral vector ()). The CCK-8 results showed that cell viability was significantly weakened after circ-SLC7A6 overexpression ()). Similarly, less cell colony formation was observed in circ-SLC7A6-overexpressing group as compared with control group ()). The EdU staining results showed that exogenous circ-SLC7A6 expression evidently impaired DNA synthesis rate ()). Moreover, overexpression of circ-SLC7A6 significantly attenuated NSCLC cell invasion ability, as demonstrated by Transwell assay ()). These data indicate that circ-SLC7A6 is a tumor suppressor in NSCLC.
Figure 2. Exogenous circ-SLC7A6 expression inhibits NSCLC cell proliferation and invasion. (a) qRT-PCR analysis confirming lentivirus mediated overexpression of circ-SLC7A6 in A549 and H460 cells. (b–d) Cell viability, colony number and DNA synthesis rate of circ-SLC7A6-overexpressing A549 and H460 cells measured by CCK-8, colony formation and EdU assays, respectively. **p < 0.01, ***p < 0.001.
Circ-SLC7A6 sponges miR-21 in NSCLC
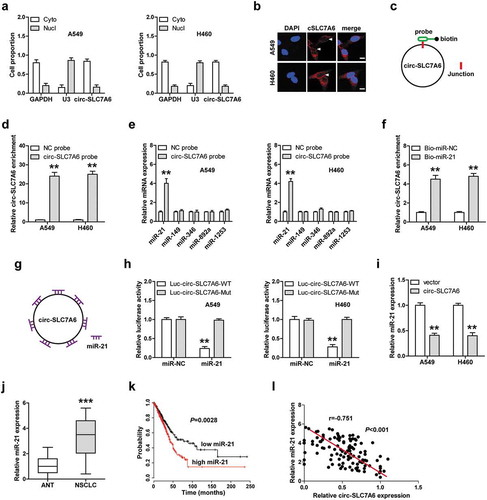
We tested the location of circ-SLC7A6, the qRT-PCR and FISH results both showed that circ-SLC7A6 was mainly located in the cytoplasm of A549 and H460 cells (,b)), implying that circ-SLC7A6 may function as a “miRNA sponge”. We analyzed the RegRNA V2.0 database (http://regrna2.mbc.nctu.edu.tw/detection.html) and the results showed that a large number of miRNAs were predicted to be complementary to circ-SLC7A6. According to alignment score, we selected the top five miRNAs (miR-21, miR-149, miR-346, miR-892a and miR-1253) for verification. The biotin-labeled circ-SLC7A6 probe was designed ()) and the qRT-PCR assay confirmed that it could effectively enrich circ-SLC7A6 ()). The RNA pull-down results showed that miR-21, but not other four miRNAs, was pulled down by circ-SLC7A6 probe in both A549 and H460 cells ()). Consistently, circ-SLC7A6 was also significantly enriched by biotinylated miR-21 probe in comparison to control probe ()). The sequence alignment results in the RegRNA V2.0 database showed that circ-SLC7A6 has seven miR-21 binding sites ()), we mutated them to perform luciferase reporter assay. As shown in ), miR-21 overexpression significantly reduced the luciferase activity of wild-type circ-SLC7A6 reporter, whereas this effect was disappeared after mutation of miR-21 binding sites. Moreover, reintroduction of circ-SLC7A6 notably decreased miR-21 expression ()), and high miR-21 level was observed in NSCLC tissues as compared with normal tissues ()). Besides, patients with high miR-21 had shorter survival time than patients with low miR-21 ()). Importantly, a strong negative correlation between circ-SLC7A6 and miR-21 expression was found in NSCLC tissues ()). These results suggest that circ-SLC7A6 acts as a sponge of miR-21 in NSCLC.
Figure 3. One circ-SLC7A6 sponges seven miR-21 in NSCLC cells. (a, b) qRT-PCR and FISH confirming the cytoplasmic location of circ-SLC7A6 in A549 and H460 cells. (c) The schematic diagram of designed biotin-labeled probe used for RNA pull-down assay. (d–f) RNA pull-down assay in A549 and H460 cells using biotin-labeled circ-SLC7A6 probe or miR-21 mimic probe, followed by qRT-PCR analysis. (g) The schematic diagram of one circ-SLC7A6 sponging seven miR-21. (h) Luciferase reporter assay in A549 and H460 cells co-transfected with miR-21 mimics and wild-type or mutant circ-SLC7A6 vector. (i, j) qRT-PCR analysis of miR-21 expression in circ-SLC7A6-overexpressing cells and in 110 paired NSCLC and normal tissues. (k) The survival curve of NSCLC patients with low or high miR-21 expression. (i) The correlation between circ-SLC7A6 and miR-21 expression in NSCLC tissues. **p < 0.01, ***p < 0.001.
Circ-SLC7A6 elevates a cohort of tumor suppressors
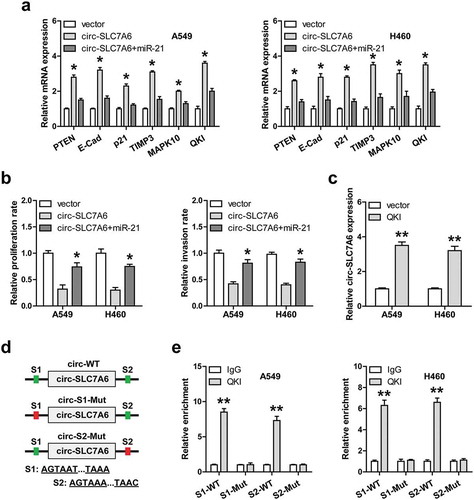
Given that circ-SLC7A6 could sponge and inhibit miR-21, we then tested whether circ-SLC7A6 affects the expression levels of the downstream targets of miR-21. A total of six well-validated miR-21 targets (PTEN, E-cadherin, p21, TIMP3, MAPK10 and QKI) were selected for qRT-PCR analysis. The results showed that overexpression of circ-SLC7A6 remarkably increased their expression levels, while these upregulation effects were significantly blocked by exogenous miR-21 expression in both A549 and H460 cells ()). Functionally, enforced expression of miR-21 could effectively rescue the weakened proliferation and invasion of NSCLC cells caused by circ-SLC7A6 overexpression ()). Of note, QKI, a RNA binding protein, was recently reported to be involved in circRNA formation [Citation16]. We then wondered whether circ-SLC7A6 was regulated by QKI. The qRT-PCR results showed that exogenous QKI expression markedly increased circ-SLC7A6 expression in both A549 and H460 cells ()). Importantly, one QKI binding site was respectively found in introns on both sides of circ-SLC7A6 ()), and the RIP results showed that QKI directly bound to both of them, whereas mutation of them blocked above effects ()).
Figure 4. QKI-induced circ-SLC7A6 elevates many tumor suppressors downstream of miR-21. (a) qRT-PCR analysis of six gene expression in circ-SLC7A6-overexpressing cells transfected with miR-21 mimics. (b) The proliferation and invasion of circ-SLC7A6-overexpressing A549 and H460 cells transfected with miR-21 mimics. (c) qRT-PCR analysis of circ-SLC7A6 expression in A549 and H460 cells after QKI overexpression. (d, e) The schematic diagram of QKI binding sites on introns flanking circ-SLC7A6, and RIP assay in A549 and H460 cells using the corresponding vectors. *p < 0.05, **p < 0.01.
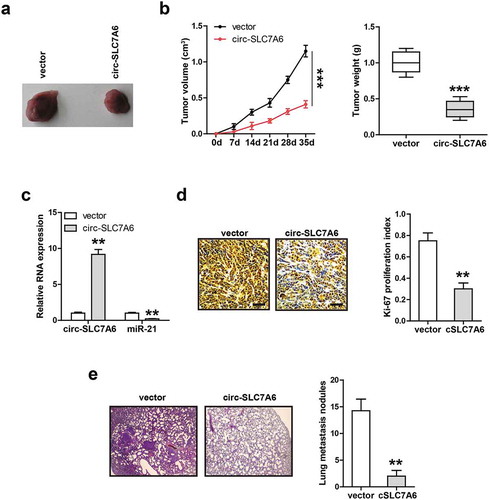
Restoration of circ-SLC7A6 inhibits tumor growth and metastasis in vivo
Ultimately, we explored the in vivo function of circ-SLC7A6. Through subcutaneous injection of circ-SLC7A6-overexpressing A549 cells into nude mice, we found that circ-SLC7A6 overexpression resulted in the reduction of tumor volume and weight (,b)). Importantly, miR-21 level was substantially decreased in circ-SLC7A6-overexpressing group as compared with control group ()). Similarly, overexpression of circ-SLC7A6 evidently decreased the number of Ki-67 positive cells ()). In addition, we established lung metastasis model via caudal vein injection of circ-SLC7A6-overexpressing A549 cells, an average of 13 lung metastatic nodules were observed in control group, while only 2.5 in circ-SLC7A6-overexpressing group ()). These data suggest that circ-SLC7A6 also functions as a tumor-inhibiting circRNA in vivo, which is in line with in vitro data.
Figure 5. Circ-SLC7A6 represses tumor growth and metastasis in vivo. (a, b) The volume and weight of tumor in control and circ-SLC7A6-overexpressing groups. (c) qRT-PCR analysis of miR-21 expression in circ-SLC7A6-overexpressing tumor tissues. (d) IHC staining of Ki-67 assessing cell proliferation in control and circ-SLC7A6-overexpressing groups. (e) H&E staining showing pulmonary metastasis nodule in control and circ-SLC7A6-overexpressing groups. **p < 0.01, ***p < 0.001.
Discussion
Recent research interest has been re-ignited in circRNA, a special type of non-coding RNA that functions as a crucial player in human life process. In the current study, we focused on the role of circ-SLC7A6 in NSCLC. We found that it was significantly downregulated in NSCLC, which was closely linked with unfavorable prognosis. Circ-SLC7A6 inhibited NSCLC cell aggressive behavior by abundantly sponging miR-21and upregulating a cohort of tumor suppressors. Importantly, we also verified the existence of circ-SLC7A6/miR-21 in vivo, and circ-SLC7A6 overexpression retarded the growth of transplanted tumor and decreased the formation of lung metastasis nodule. Thus, our results uncover the important role of circ-SLC7A6 in NSCLC, meanwhile providing new evidence for the “miRNA sponge” function of circRNA in cancer.
Due to its high stability, circRNA is proposed as an efficient biomarker of human diseases [Citation17,Citation18]. Recently, several circRNAs were identified to be significantly deregulated in cancer and used as biomarkers for diagnosis or prognosis. For instance, circ-FUT8 [Citation19], circ-CAMSAP1 [Citation20], circ-CLK3 [Citation21], circ-OSBPL10 [Citation22] and circ-BARD1 [Citation23] were shown as diagnostic or prognostic biomarkers for bladder cancer, colorectal cancer, cervical cancer, gastric cancer and breast cancer, respectively. Herein, we found that circ-SLC7A6 was highly resistant to RNase R, suggesting that it is extremely stable. And NSCLC patients with low circ-SLC7A6 had shorter survival time than patients with high circ-SLC7A6, implying its characteristics as a prognostic indicator of NSCLC. Of note, recent studies have shown that circRNA is also widely and stably present in urine, sweat, blood and even exosomes, and can be used as noninvasive biomarker for disease diagnosis [Citation24]. Whether circ-SLC7A6 can be utilized as a diagnostic biomarker for NSCLC is worthy of further study.
The “miRNA sponge” function of circRNA has been well proven on the premise that circRNA is mainly located in the cytoplasm [Citation25]. In our study, qRT-PCR and FISH assays showed that circ-SLC7A6 was a cytoplasmic circRNA, hinting that circ-SLC7A6 may act as a “miRNA sponge”. Through a series of screening and experimental confirmation, we found that circ-SLC7A6 could abundantly sponge and inhibit miR-21, a well-documented oncogene in different cancer types, including NSCLC [Citation26]. We also verified that miR-21 was significantly upregulated in NSCLC tissues and high miR-21 was positively correlated with adverse outcome. Of note, only a few studies have shown that circRNA has more than two binding sites of one miRNA, for instance, CDR1as contained more than 70 potential binding sites for miR-7 [Citation10,Citation11], and circ-SRY could simultaneously absorb 16 miR-138 [Citation27]. Here, we found and confirmed seven miR-21 binding sites on circ-SLC7A6, suggesting that circ-SLC7A6 is a powerful molecular sponge for miR-21. In addition, enforced expression of circ-SLC7A6 substantially reduced miR-21 expression both in vitro and in vivo, and exogenous miR-21 expression was able to rescue the attenuated malignant behavior of NSCLC cells caused by circ-SLC7A6 overexpression, indicating that miR-21 is a functional target of circ-SLC7A6. Most strikingly, a recent study showed that the synthetic circRNA was highly resistant to nuclease digestion and could competitively bind oncogenic miR-21 to suppress gastric cancer progression [Citation28]. Further study is needed to explore whether circ-SLC7A6 can be synthesized and whether this artificial circ-SLC7A6 is functional in vivo.
Conclusion
Our findings suggest that circ-SLC7A6 is a novel tumor suppressor that inhibits NSCLC tumorigenicity and metastasis through acting as an effective sponge for oncogenic miR-21. Targeting the regulatory axis of circ-SLC7A6/miR-21 implicates a promising therapeutic possibility for NSCLC patients.
Availability of data and materials
The analyzed data in this study are available from the corresponding author on reasonable request.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Heigener DF, Reck M. Lung cancer in 2017: giant steps and stumbling blocks. Nat Rev Clin Oncol. 2018;15(2):71–72.
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. NATURE. 2018;553(7689):446–454.
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Non-Small Cell Lung Cancer Treatment (PDQ(R)): Health Professional Version; 2002.
- Li HM, Ma XL, Li HG. Intriguing circles: conflicts and controversies in circular RNA research. Wiley Interdiscip Rev RNA. 2019;e1538. DOI:https://doi.org/10.1002/wrna.1538
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157.
- Li X, Yang L, Chen LL, et al. Functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442.
- Kristensen LS, Andersen MS, Stagsted L, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691.
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283.
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. NATURE. 2013;495(7441):333–338.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. NATURE. 2013;495(7441):384–388.
- Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Deaths 2018;9(4):417.
- Yang G, Wang X, Liu B, et al. circ-BIRC6, a circular RNA, promotes hepatocellular carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell Cycle. 2019;18(9):976–989.
- Liang Y, Song X, Li Y, et al. circKDM4C suppresses tumor progression and attenuates doxorubicin resistance by regulating miR-548p/PBLD axis in breast cancer. ONCOGENE. 2019;38(42):6850–6866.
- Bodoy S, Fotiadis D, Stoeger C, et al. The small SLC43 family: facilitator system l amino acid transporters and the orphan EEG1. Mol Aspects Med. 2013;34(2–3):638–645.
- Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. CELL. 2015;160(6):1125–1134.
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
- Lei B, Tian Z, Fan W, et al. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301.
- He Q, Yan D, Dong W, et al. circRNA circFUT8 upregulates krupple-like factor 10 to inhibit the metastasis of bladder cancer via sponging miR-570-3p. Mol Ther Oncolytics. 2020;16:172–187.
- Zhou C, Liu HS, Wang FW, et al. circCAMSAP1 promotes tumor growth in colorectal cancer via the miR-328-5p/E2F1 axis. Mol Ther. 2020;28(3):914–928.
- Hong H, Zhu H, Zhao S, et al. The novel circCLK3/miR-320a/FoxM1 axis promotes cervical cancer progression. Cell Death Dis. 2019;10(12):950.
- Wang S, Zhang X, Li Z, et al. Circular RNA profile identifies circOSBPL10 as an oncogenic factor and prognostic marker in gastric cancer. ONCOGENE. 2019;38(44):6985–7001.
- Zhao J, Zou H, Han C, et al. Circular RNA BARD1 (Hsa_circ_0001098) overexpression in breast cancer cells with TCDD treatment could promote cell apoptosis via miR-3942/BARD1 axis. Cell Cycle. 2018;17(24):2731–2744.
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984.
- Kristensen LS, Hansen TB, Veno MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. ONCOGENE. 2018;37(5):555–565.
- Bica-Pop C, Cojocneanu-Petric R, Magdo L, et al. Overview upon miR-21 in lung cancer: focus on NSCLC. Cell Mol Life Sci. 2018;75(19):3539–3551.
- Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409.
- Liu X, Abraham JM, Cheng Y, et al. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol Ther Nucleic Acids. 2018;13:312–321.
