ABSTRACT
The poor prognosis of late gastric carcinomas (GC) underscores the necessity to identify novel biomarkers for earlier diagnosis and effective therapeutic targets. MiRNA-324-5p has been shown to be over-expressed in GC, however the biological function of miRNA-324-5p implicated in gastric cancer and its downstream targets were not well understood. Wnt/β-catenin signaling pathway is aberrantly regulated in GC. We sought to explore if miRNA-324-5p promotes oncogenesis through modulating Wnt signaling and EMT. MiRNA-324-5p is highly expressed in GC based on qRT-PCR and TCGA data. In addition, in vitro cell proliferation, cell migration assays and in vivo animal exenograft were executed to show that miRNA-324-5p is an oncogenic miRNA in GC. MiRNA-324-5p activates Wnt signaling and induces EMT in GC. Further, SUFU was identified as a target of miRNA-324-5p confirmed by western blotting and luciferase assays. Spearson analysis and TCGA data indicate that the expression of SUFU is negatively associated with the expression of miRNA-324-5p. Rescue experiments were performed to determine if SUFU mediates the Wnt activation, EMT and oncogenic function of miRNA-324-5p. MiRNA-324-5p inhibitors plus SUFU siRNAs rescue partially the inhibitory effect on Wnt signaling and EMT caused by miRNA-324-5p inhibitors. Finally, the suppression of cell proliferation, migration, and colony formation ability induced by miRNA-324-5p inhibitors is alleviated by addition of SUFU siRNAs. In summary, miRNA-324-5p is overexpressed in vivo and exerts cell growth and migration-promoting effects through activating Wnt signaling and EMT by targeting SUFU in GC. It represents a potential miRNA with an oncogenic role in human gastric cancer.
Introduction
China has the largest population of GC patients. The prognosis of late GC patients is very dismal. This necessitates novel biomarkers for earlier diagnosis and effective therapeutic targets [Citation1]. Deeper understanding the GC oncogenesis mechanism will be helpful to identify biomarkers for earlier detection and more effective intervention to improve the overall therapeutic outcome of GC patients.
Mounting evidence has shown that miRNAs are implicated in all stages of tumor development from onset, progression to metastasis [Citation2,Citation3]. Previous studies have shown that miRNA-324-5p is up-regulated in a number of cancers such as gastric cancer [Citation4], lung cancer [Citation5], malignant melanoma [Citation6], HCC [Citation7], and acute myeloid leukemia[Citation8] but down-regulated in CRC [Citation9], medulloblastoma [Citation10], suggesting it is an important miRNA involved in tumor development [Citation11]. However, the biological effect induced by miRNA-324-5p in GC and underlying mechanism of its role were largely unknown.
Wnt/β-catenin signaling has been implicated in gastric carncinogenesis and tumor progression [Citation12–16]. Emerging data have shown that miRNAs are involved in Wnt/β-catenin signaling pathway by targeting Wnt associated genes at different regulation layer. We have shown that miRNA-194 activates Wnt/β-catenin signaling pathway by targeting Wnt negative regulator SUFU [Citation17].
In the present study, we identified that miRNA-324-5p is significantly overexpressed in GC compared to NS (non-neoplastic gastric tissues). MiRNA-324-5p activates cell proliferation, migration, and tumor formation in GC cells. In addition, miRNA-324-5p activates the Wnt/β-catenin signaling pathway and induces EMT (epithelial to mesenchymal transition). SUFU was discovered as a target of miRNA-324-5p. Finally, SUFU appears to mediate the Wnt activation, EMT and oncogenic function of miRNA-324-5p. In summary, we conclude that miRNA-324-5p enhances cell proliferative, migratory property, and induces EMT in GC, at least in part, by activating Wnt/β-catenin signaling via targeting SUFU.
Materials and methods
Tissue samples and cell lines
In total, we have examined 60 cases of gastric cancers (GC) and paired adjacent non-neoplastic gastric tissues (NS). The tissue samples were obtained from Shenzhen University first affiliated hospital and examined by two independent pathologists. Under an institutionally-approved protocol, all patients signed the consent forms. The tissue biopsies were immediately stored in liquid nitrogen after resection until needed.
The gastric cancer cells AGS, MKN28 were from the American Type Culture Collection and China Infrastructure of Cell line Resources, respectively. The gastric cancer cells BGC-823, SGC7901 were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and control HFE-145 (human normal gastric epithelial cells) were obtained from Dr. Duane T. Smoot at Meharry Medical College. MKN28, AGS and SGC7901 cells were cultured in RPMI1640 medium (Thermo Fisher Scientific, USA) supplemented with 10% FBS, (Gibco) 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen Life Technologies, USA). BGC-823 and HFE-145 were cultured in DMEM medium (Hyclone, Logan, Utah) supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. All cells were cultured in a 5% CO2 incubator at 37°C.
Quantitative reverse-transcriptase PCR (qRT–PCR)
For the detection of miRNA-324-5p and SUFU mRNA expression, total RNA from tissues or cultured cells was isolated using mirVana miRNA Isolation Kit (Invitrogen, Carlsbad, CA, USA). Frozen gastric tissues were ground to powder using a grinding machine. Cultured cells or tissues lysate were harvest using Lysis/Binding Solution with 1/10 volume of miRNA Homogenate Additive. The lysate was then mixed well by vortexing and incubated on ice for 10 min. Then equal volume of chloroform was added to the lysate. Collect the aqueous phase, add 1.25 volumes of ethanol, and mix thoroughly. The lysate/ethanol mixture was transferred into a filter cartridge which was washed twice. Finally, the RNA was eluted with nuclease-free water and stored it at −80 °C until used. MiRNA-324-5p expression level was determined using TaqMan® Universal Master MixⅡ(Applied Biosystems, Foster City, CA, USA) . RNU6B TaqMan RT–PCR amplicon (RNU6B TaqMan microRNA Assay kit, Applied Biosystems) was used as an internal control. PrimeScript™ RT reagent Kit and SYBR® Green Master Mix (Takara, Dalian) were used to synthesize cDNA and quantify the expression of SUFU. The primers of SUFU is: sense 5’-GACCCTGGTTACAAATTCTGTTGA-3’, antisense 5’-AGGCAGGATGGAGACCTTCAG-3’. The expression was normalized to the expression of GAPDH.
Transfection of miRNAs and siRNAs
Cholesterol-conjugated miRNA-324-5p inhibitors, their corresponding NCs and SUFU siRNAs (5’‑CGG CCT GAG TGA TCT CTA T‑3’, 5’‑GATCCA CAC CTG CAA GAG A‑3’ and 5’‑GCA GCT TGA GAGCGT ACA T‑3’), were obtained from Ribobio (Guangzhou, China). MiRNA-324-5p mimics, inhibitors and their respective NCs were chemically synthesized by Dharmacon (Lafayette, CO, USA).A mixture of three SUFU siRNAs, which successfully down-regulated SUFU expression [Citation17], was used. Lipofectamine RNAiMAX (Invitrogen) was used for transfection of cells with 30–50% confluence. The cells were transfected with either miRNA-324-5p mimics/inhibitors (60 nM), or co-transfected with miRNA-324-5p inhibitors (60 nM) and of SUFU-siRNAs(60 nM). Their respective NCs were used as the negative controls. 48 hrs after miRNAs/siRNAs transfection, RNA, and protein were harvested according to the established protocols as described in the qRT-PCR section and western blotting section, respectively.
Cell proliferation assay
CCK-8 assay (Dojindo, Japan) was performed to assess cell proliferation ability. 30–50% confluent cells were transfected with miRNA-324-5p inhibitors and subsequently cultured for 48 hours. Followed by cells were collected and inoculated into 96-well plates at 1000 cells/well. The transfected cells were cultured in a 37 °C incubator for 7 days and the absorbance at 450 nm was measured every 2 days using CCK-8 cell counting kits (Dojindo, Japan) with a Microplate Reader (Molecular Devices, USA).
Cell migration assay
According to the established procedure [Citation18], cell migration ability was assessed using Transwell chambers (24-well format, BD Biosciences, St Louis, MO, USA). 5 × 104 tumor cells were inoculated on the upper chamber with 8 μm pores 48 h post transfection, cultured with serum-free medium. 20% FBS growth medium (800 μl) was added to the lower chambers and served as a chemoattractant. Following 24 h incubation at 37°C, cells that migrated to the pores were fixed with 4% paraformaldehyde for 20 min and then stained with hematoxylin for 10 min. Then five random fields of view under the microscope were photographed and cells were counted and summarized for statistical analysis.
Wound healing assay
The wound-healing assay was performed to evaluate migratory ability. 30–50% confluent cells were inoculated in the six-well plate and tranfected with miRNA-324-5p inhibitors. Forty-eight hours later when cells reach a confluence of 80–90%, a 200 µl micropipette tip was used to create a scratch in the middle. At 0, 24, 48 h after wounding, the progression of migration was pictured using Olympus 1 × 71 camera system.
Western blotting
Laemmli sample buffer (Bio-Rad) with a protease inhibitor, complete EDTA-free (Roche) was used to extract total protein and protein concentration was measured by a BCA Protein Assay kit (Pierce, Rockford, MA, USA). Cytoplasmic protein and nucleoprotein were isolated by the NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Thermofisher Scientific, MA USA). Equal amounts of extracted protein were electrophoresed on 10% SDS-PAGE and transferred to a PVDF membrane (Millipore, USA). The membranes were blocked with 5% nonfat milk diluted with TBST(Tris-Buffered saline Tween-20) for 1 h and then incubated with the primary antibody at 4°C overnight. The primary antibodies used in this study included SUFU (#2522), Wnt/β-Catenin Activated Targets Antibody Sampler Kit (#8655) and Epithelial-Mesenchymal Transition (EMT) Antibody Sampler Kit (#9782) (1:1000, Cell Signaling, Danvers, MA, USA) and GAPDH monoclonal antibody (1:5000, Cell Signaling). After incubated with secondary antibody and washed with TBST, the membranes were subjected to exposure.
Immunofluorescence analysis
Forty-eight hours after transfection, washed cells were fixed with 4% formaldehyde for 15 min. Cells were then washed with PBS three times and permeated with 0.2% Triton X-100 (PBS) for 10 min at room temperature. Next, cells were incubated with an anti-β-catenin antibody (1:100; L54E2, Cell Signaling) overnight at 4°C. After washing with PBS, the cells were incubated with Alexa Fluor® 594-Conjugated anti‑mouse secondary antibody (1:1000, #8890; Cell Signaling) for 1 h at room temperature avoiding light. Nuclei were stained by DAPI II (Abbott Molecular, Abbott Park, Illinois). Images of cells under a Leica confocal microscope (Cellular Imaging Facility, Lausanne, Switzerland) were pictured and analyzed with Leica Application Suite (LAS) software.
Luciferase reporter assay
Topflash and Fopflash plasmids were purchased from Addgene (Cambridge, MA, USA). When cells reach a confluence of 30–50%, we transfected 60 nM of miRNA-324-5p inhibitors and 100 ng Topflash/Fopflash plasmids with 10 ng pTK-renilla via Lipofectamine 3000 (Invitrogen). Dual-Glo luciferase assay kit (Promega) was used to carry out the luciferase reporter assays. The luminescence intensity was normalized by dividing firefly luciferase activity to renilla luciferase activity.
SUFU 3’-UTR gene segment with predicted miRNA-324-5p binding sites and corresponding SUFU 3’-UTR mutant segment were cloned into a pDL-UTR vector (Promega, USA). Constructed reporter vectors were co-transfected with miRNA-324-5p mimics/inhibitors via Lipofectamine 3000 for 48 h according to the protocol (Invitrogen), luciferase activity was measured as described above.
Animals and subcutaneous tumor growth assays
Four-to-six-old BALB/c-nu female nude mice were purchased from the Shanghai Laboratory Animal Center (Shanghai, China). All the animal experiments were approved by Institutional Animal Care and Use Committee of Shenzhen University. The mice were raised in SPF Laboratory Animal Room and were free to obtain food and tap water during the experiment. 1 × 106 BGC-823 cells with transfection were suspended with 100 μl PBS and injected subcutaneously into the back of nude mice. Either 10 nmol nonspecific negative control (NSC-inh)(Ribobio Co. Guangzhou, China) or cholesterol-conjugated miRNA-324-5p inhibitors (324–5p-inh) was injected locally into the corresponding tumor location every 3 to 4 days for 32 days(n = 6 in each group). Tumor length and width were measured twice a week by caliper and tumor volume (mm3) was calculated according to the formula: volume (mm3 = 1/2× length×width2). Tumor-bearing mice were sacrificed on day 32.
TCGA analyses and statistical analyses
TCGA data for gene expression (polyA+ IlluminaHiSeq), miRNA expression (IlluminaHiseq) for GC was obtained from UCSC Xena (http://xena.ucsc.edu).We have analyzed 403 samples including 368 GCs and 35 adjacent normal tissues. Log2(x + 1) transformed RSEM normalized count gene expression data was used. The expression differences of miRNA-324-5p and SUFU between GCs and histologically normal gastric tissues adjacent to the tumor from TCGA were analyzed using Mann-Whitney U test.
Student’s t‑test was used for analysis of difference between two groups in the experiments. One‑way analysis of variances followed by Dunnett’s test was used for comparisons in datasets containing multiple groups. The correlation between the expression of miRNA-324-5p and SUFU in paired GC specimens was analyzed by Spearman’s correlation. Data are presented as mean ± SD from three independent experiments. A p value < 0.05 was considered significant.
Results
miRNA-324-5p is an oncogenic miRNA in GC
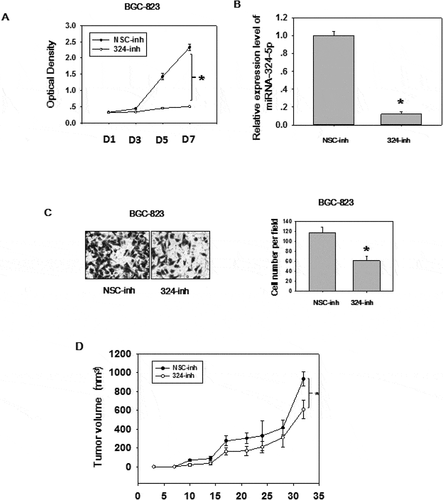
The expression level and biological effects of miRNA-324-5p in GC were not well studied, so we utilized qRT-PCR to test the expression level in 60 paired GC tissues ()). Significantly up-regulation (paired student t test, p < 0.0001) of miRNA-324-5p were observed in GC as compared to NS. We also analyzed GC data (368 GCs and 35 adjacent histologically normal samples) from TCGA to determine the expression level of miRNA-324-5p in GC. In agreement with our result, it showed that miRNA-324-5p was significantly overexpressed in GCs compared to the adjacent normal gastric tissues (p = 0.015, )). It should be mentioned that adjacent normal tissue is not real healthy tissue. It presents a special intermediate state between healthy tissue and tumor, however, it is relatively easy to access and good control for variation between patients and anatomic sites [Citation8,Citation19]. Thus, adjacent normal tissue is used as a control. Similarly, miRNA-324-5p is highly expressed in GC cell lines such as AGS, MKN28, SGC-7901, BGC-823 ()), suggesting it may be associated with gastric carcinogenesis. . To better understand the biological function of miRNA-324-5p, we transfect BGC-823 cells with miRNA-324-5p inhibitors. Inhibitor-transfected cells displayed decreased cell proliferation rate, suggesting miRNA-324-5p promotes cell proliferation ()). RT-PCR results demonstrate that miRNA-324-5p inhibitors successfully knockdown the expression level of miRNA-324-5p ()). Cell migration was also suppressed after transfection with miRNA-324-5p inhibitors, indicating that miRNA-324-5p plays a tumor-promoting role in gastric carcinogenesis ()). To further explore the biological effect induced by miRNA-324-5p in vivo, BGC-823 cells with transfection of miRNA-324-5p inhibitors or control were injected into flanks of SCID mice (N = 6/group) subcutaneously. Cholesterol-conjugated miRNA-324-5p inhibitor or control dissolved in PBS were injected locally into the corresponding tumor mass twice a week until sacrifice. Tumor volume was measured until day 32. As shown in ), tumor volume was significantly decreased in mice treated with miRNA-324-5p inhibitors than in those treated with NSC-inhibitors (p < 0.05). Altogether, these results revealed that miRNA-324-5p plays an oncogenic role in GC.
Figure 1. MiRNA-324-5p is highly expressed in GC
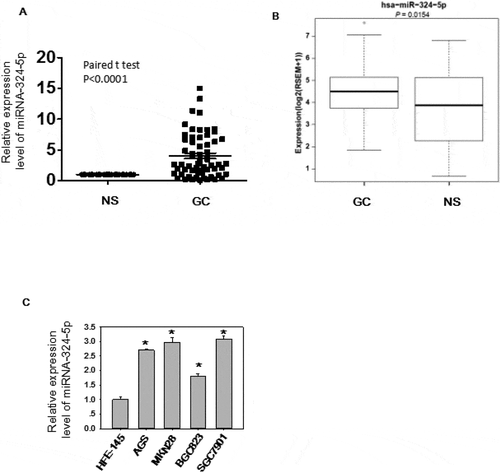
Figure 2. MiRNA-324-5p has an oncogenic effect in GC
miRNA-324-5p activates the Wnt/β-catenin signaling pathway and EMT
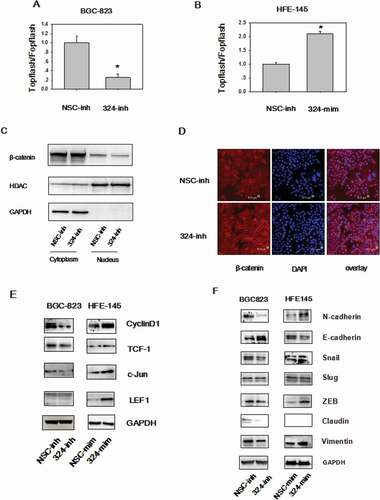
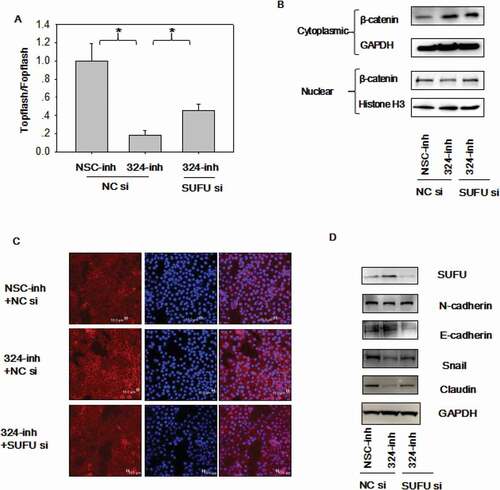
Wnt/β-catenin signaling is deregulated in 30% of GC [Citation12], highlighting the oncogenic property of this pathway. On the basis of the tumor promoting role of miRNA-324-5p, we investigate the relevance of miRNA-324-5p in regulating Wnt/β-catenin signaling. To this end, Topflash/Fopflash luciferase assays were performed in BGC-823 cells. There are 7 wild type TCF/LEF-binding sites in Topflash vector, while 6 mutated TCF/LEF binding sites upstream of a firefly luciferase reporter in Fopflash plasmid. Inhibition of miRNA-324-5p resulted in reduced β-catenin-dependent luciferase activities ()). Consistently, overexpression of miRNA-324-5p led to activation of Wnt/β-catenin signaling activity ()). It is well established that β-catenin nuclear translocation is required for Wnt/β-catenin signaling activation [Citation20]. Thus, cytoplasmic and nuclear localization of β-catenin was analyzed in BGC-823 cells. Western blotting after cytoplasmic and nuclear extraction illustrated that employment of miRNA-324-5p inhibitors resulted in decreased β-catenin translocation into the nucleus ()). Concordant with the Western blotting data, confocal immunofluorescence assay of β-catenin showed that use of miRNA-324-5p inhibitors led to β-catenin translocation to the cell membrane ()). This result was in line with the positive control result after cells incubated with XAV-939, a well-known Wnt signaling inhibitor [Citation17]. Moreover, Wnt-dependent gene expression was analyzed after miRNA-324-5p inhibition or activation. The expression level of Cyclin D, TCF-1, c-Jun was decreased dramatically in BGC-823 cells with treatment of miRNA-324-5p inhibitors ()). LEF-1 was expressed lowly at the basal level. Agreeably, increased expression of Cyclin D, TCF-1, c-Jun, LEF-1, and TCF1 resulted from miRNA-324-5p activation in HFE-145 cells. Together, it suggests that miRNA-324-5p activates Wnt signaling pathway.
During Epithelial-mesenchymal transition (EMT), an critical developmental process, the epithelial cells change into mesenchymal, fibroblast-like cells and harbor decreased intracellular adhesion and enhanced motility. EMT is the downstream output of activated Wnt signaling and EMT plays an essential role in all stages of cancer development. EMT was explored after up or down regulating miRNA-324-5p levels. A reduction of epithelial markers, such as E-cadherin and a gain of mesenchymal markers, such as Vimentin, Snail, N-cadherin, and ZEB were observed in HFE-145 cells with miRNA-324-5p mimics transfection. We suppose that miRNA-324-5p activates Wnt signaling which induces the expression of Wnt dependent EMT-TF (transcription factor) such as Snail and ZEB1 [Citation21]. Snail and ZEB1 in turn led to the overexpression of N-cadeherin, Vimentin and downregulation of E-cadherin. Similarly, EMT was repressed in BGC-823 cells with miRNA-324-5p inhibitors transfection ()). Of note, Slug expression level did not change, which is a good control for the expression differences observed in other EMT associated genes. In a word, these observations suggest that miRNA-324-5p activates the Wnt signaling pathway and induces EMT.
Figure 3. MiRNA-324-5p activates Wnt/β-catenin signaling pathway and EMT. (a)MiRNA-324-5p activated Topflash luciferase activity. BGC-823 cells were co-transfected with NSC-inh or miRNA-324-5p inhibitors, Topflash/Fopflash plasmids and pTK-renilla. *p < 0.05. (b) MiRNA-324-5p activated Topflash luciferase activity. HFE-145 cells were co-transfected with NSC-mim or miRNA-324-5p mimics, Topflash/Fopflash plasmids and pTK-renilla. *p < 0.05. (c) Suppression of miRNA-324-5p inhibited the translocation of β-catenin into the nucleus. Forty-eight hours after miRNA-324-5p inhibitors transfection, BGC-823 cells were lysed and immunoblotted with β-catenin antibody in cytoplasmic and nuclear portion, respectively. (d) Inhibition of miRNA-324-5p blocked the translocation of β-catenin into the nucleus. β-catenin localization was detected by confocal immunofluorescence 48 h after transfection of miRNA-324-5p inhibitors in BGC-823 cells. (e) MiRNA-324-5p activated Wnt/β-catenin downstream gene expression. Cell lysates were used to blot Wnt downstream gene antibody 48 h after transfection of miRNA-324-5p inhibitors or mimics in BGC-823 and HFE-145 cells, respectively. (f) MiRNA-324-5p induced EMT. Cell lysates were used to blot EMT markers 48 h after transfection of miRNA-324-5p inhibitors or mimics in BGC-823 and HFE-145 cells, respectively. Each experiment was repeated three times
SUFU is the target of miRNA-324-5p
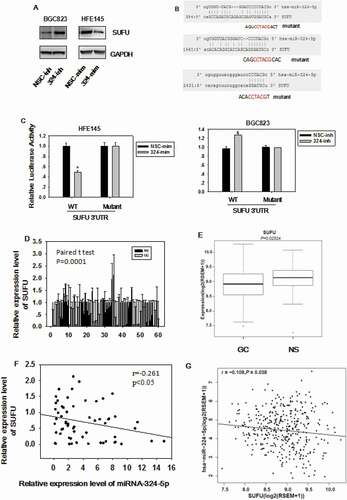
In order to elucidate which gene miRNA-324-5p targets to regulate Wnt/β-catenin signaling pathway and EMT, the target gene was predicted with miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/). We chose SUFU as a candidate gene based on the prediction and the fact that SUFU is a negative regulator of Wnt/β-catenin signaling by blocking β-catenin in the cytoplasm. As shown in ), miRNA-324-5p mimics transfection in HFE-145 which has low basal level of miRNA-324-5p expression resulted in reduced SUFU expression. On the other hand, miRNA-324-5p transfection in BGC-823 with relatively high basal level of miRNA-324-5p expression gave rise to the increased SUFU expression ()). To confirm that miRNA-324-5p directly targets SUFU mRNA through binding to its 3’UTR, three potential binding sites of miRNA-324-5p on 3’UTR of SUFU were analyzed as shown in ). A wild-type (pDL-SUFU-3’UTR-wt) or a mutant (pDL-SUFU-3’UTR-mut) reporter vector containing three mutated binding sites was co-transfected into HFE-145 cells with miRNA-324-5p mimics. Transfection of miRNA-324-5p mimics in HFE-145 cells significantly reduced SUFU 3’UTR dependent luciferase activity of the wild-type reporter, but not the mutant. Reciprocally, SUFU 3’UTR dependent luciferase activity of the wild-type reporter not the mutant was increased with the transfection of miRNA-324-5p inhibitors in BGC-823 cells ()), validating the specific interaction between miRNA-324-5p and the SUFU 3’UTR. Consistent with published literature, SUFU was down-regulated in 60 paired GC ()). TCGA data analysis also showed that SUFU was down-regulated in 386 GC tissues ()). More importantly, a negative correlation was found between miRNA-324-5p expression and SUFU expression in 60 cases of clinical-paired GC tissues (p < 0.05, R = −0.269, )), further strengthening the notion that SUFU was targeted by miRNA-324-5p. Furthermore, the inverse correlation was also found using TCGA database ()), suggesting that the negative correlation between miRNA-324-5p and SUFU is a common event and SUFU is a target of miRNA-324-5p. Admittedly, the correlation is somehow weak suggesting that there are other factors regulating the expression of SUFU.
Figure 4. SUFU is the direct target of miRNA-324-5p. (a) The expression level of SUFU was up-regulated by miRNA-324-5p inhibitors in BGC-823 and down-regulated by miRNA-324-5p mimics in HFE-145 cells. (b) The predicted binding sites of miRNA-324-5p on the 3’UTR region of SUFU and their corresponding mutant sites in luciferase reporters are shown. (c) HFE-145 and BGC-823 cells were co-transfected with miRNA-324-5p mimics/inhibitors and SUFU 3’UTR WT (wild-type)/Mutant reporter plasmids. SUFU 3’ UTR dependent luciferase activities were reduced or increased by miRNA-324-5p mimics or inhibitors, respectively,*p < 0.05. (d) SUFU expression level in 60 paired GC tissues was determined by qRT-PCR and SUFU was down expressed in GC. (e)RNA-seq analysis of SUFU mRNA levels in 368 GCs and 35 normal tissues queried from TCGA showed that SUFU was down-regulated in GC, *p < 0.05. (f) Spearman’s correlation analysis showed that there was a negative correlation between SUFU and miRNA-324-5p expression in 60 tested tissues, *p < 0.001. (g) Expression correlation analysis was performed between miRNA-324-5p and SUFU in GCs from TCGA database (n = 368), r = −0.108, p = 0.038. SUFU expression was negatively correlated with that of miRNA-324-5p
miRNA-324-5p activates Wnt/β-catenin signaling pathway and induces EMT via SUFU
To test if SUFU mediates the Wnt and EMT activation induced by miRNA-324-5p, the Topflash assay was performed in BGC-823 cells transfected with miRNA-324-5p inhibitors combined with SUFU siRNAs. As expected, β-catenin dependent luciferase activity was decreased in cells transfected with miRNA324-5p inhibitors and the inhibitory effect was alleviated by SUFU siRNAs partially ()). β-catenin was localized to the cytoplasm with miRNA-324-5p inhibition while SUFU siRNAs addition reversed the translocation ()). β-catenin was mostly localized to the cellular membrane when cells were treated with miRNA-324-5p inhibitors as illustrated by confocal immunofluorescence of β-catenin and more β-catenin moved to the cytoplasm after additional SUFU siRNAs transfection ()). Together, it suggests that SUFU mediates the Wnt activation induced by miRNA-324-5p. To explore if SUFU mediates the EMT induced by miRNA-324-5p, EMT markers were evaluated. Elevated expression of epithelial markers such as E-cadherin and a reduction of mesenchymal markers, such as N-cadherin, Snail, and Claudin were induced by miRNA-324-5p repression. They were attenuated partially in BGC-823 cells with combinational transfection of miRNA-324-5p inhibitor and SUFU siRNAs ()), suggesting that SUFU mediated EMT induced by miRNA-324-5p.
Figure 5. MiRNA-324-5p activates Wnt/β-catenin signaling pathway and EMT via SUFU. (a) Topflash luciferase activity was decreased by miRNA-324-5p inhibitors and SUFU siRNAs partially reversed this inhibitory effect. BGC-823 cells were transfected with miRNA-324-inh or co-transfected with miRNA-324-inh and SUFU siRNAs. (b) Interference of SUFU relieved the cytoplasmic retention of β-catenin induced by miRNA-324-5p blockage. (c) β-catenin translocation to nuclei induced by miRNA-324-5p inhibition was reversed by the coupled SUFU knockdown. Confocal immunofluorescence was performed to locate the β-catenin cytoplasmic or nuclear localization 48 h after transfection. (d) Suppression of SUFU alleviated the EMT induced by miRNA-324-5p inhibition. Each experiment was repeated three times
SUFU mediates miRNA-324-5p oncogenic function
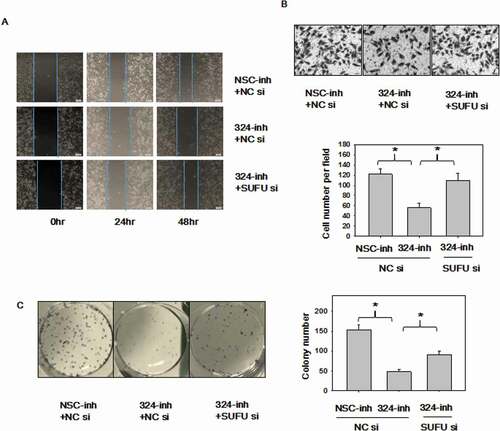
It is known that miRNA-324-5p, an oncogenic miRNA, activates Wnt pathway and induces EMT via SUFU. Finally, we sought to investigate if SUFU medicates the oncogenic function of miRNA-324-5p, rescue assays were performed with SUFU siRNAs in BGC-823 cells. Decreased ability to migrate observed in cells with miRNA-324-5p inhibition was reverted upon the silencing of SUFU in scratch assays ()) and transwell assays ()). Inhibition of miRNA-324-5p agreeably reduced colony-forming capacity in BGC-823 cells, while knockdown of SUFU rescued this inhibition ()). Overall, these findings implied that the oncogenic function of miRNA-324-5p is mediated by SUFU at least partially in GC.
Figure 6. MiRNA-324-5p promotes cell migration and colony formation ability by activating Wnt/β-catenin signaling pathway via SUFU. (a-b)Suppression of miRNA-324-5p weaken the migration capacity of BGC-823 cells while SUFU siRNAs can alleviate this inhibition. BGC-823 cells were transfected with miRNA-324-inh or co-transfected with miRNA-324-inh and SUFU siRNAs. Forty-eight hours after transfection, scratch assay and transwell assay were conducted. (c) MiRNA-324-5p inhibitors impaired colony forming capacity in BGC-823 cells while SUFU siRNAs reversed the depressive effect. 48 h after transfection with either miRNA-324-5p inhibitors or coupled SUFU siRNAs, 200 cells/well were inoculated into a 6-well plate. Cell colonies were stained and counted 14 days after plating. Each experiment was repeated three times. *p < 0.05
Discussion
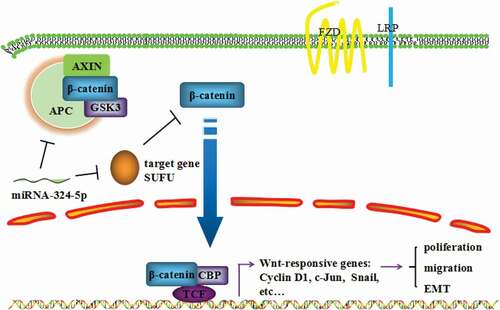
In the present study, we have explored the cancer-relevant function of miRNA-324-5p and underlying mechanism of its oncogenic behavior. We showed that GC tissues and cell lines harbor elevated expression level of miRNA-324-5p. GC cells displayed decreased cell proliferation rate, migration, and tumor formation ability when miRNA-324-5p were inhibited. In addition, Wnt/β-catenin signaling pathway was activated and EMT was induced by miRNA-324-5p. SUFU was identified to be a target of miRNA-324-5p. Finally, SUFU appears to mediate the Wnt activation, EMT, and oncogenic function of miRNA-324-5p. Together, we demonstrate that miRNA-324-5p enhances cell proliferative, migratory property, and induces EMT in GC, at least in part, through activating Wnt pathway by targeting SUFU ().
Figure 7. MiRNA-324-5p promotes cell proliferation, migration and EMT by activating Wnt/β-catenin signaling pathway via SUFU
The pre-miRNA-324 gave rise to two mature miRNA-324, miRNA-324-5p, and miRNA-324-3p during the maturation process. Previous studies have revealed miRNA-324-5p was up-regulated in lung cancer, malignant melanoma, HCC, acute myeloid leukemia, and GC but down-regulated in medulloblastoma and kidney cancer [Citation8,Citation11]. Until now, only a few studies have investigated the biological functions of miRNA-324-5p in human cancer. MiRNA-324-5p suppresses the proliferation of glioma cells through silencing GLI family zinc finger 1 (GLI1) [Citation22]. Up-regulation of miRNA-324-5p leads to inhibition of proliferation and invasion in colorectal cancer cells by targeting ELAVL1(Embryonic Lethal, Abnormal Vision, Drosophila-Like Protein 1) [Citation23]. In addition, miRNA-324-5p overexpression resulted in reduced migration and invasion by targeting matrix metallopeptidase 2 (MMP2), MMP9, ETS proto-oncogene 1 (ETS1), and Sp1 transcription factor gene expression in HCC [Citation24]. This is the first report describing the oncogenic function of miRNA-324-5p in GC. We have found that miRNA-324-5p is highly expressed in GC tissues as well as cell lines and inhibition of miRNA-324-5p resulted in reduced cell proliferation, migration and tumorigenicity capacity.
MiRNA-324-5p has the potential to be a prognostic factor for multiple cancers. A classifier of 19 miRNAs containing miRNA-324 has independent prognostic utility for pancreatic cancer. It determined the prognosis of three pancreatic ductal adenocarcinoma tumor subtypes which have significantly different survival time but the same clinical conditions [Citation25]. In addition, the expression levels of 6 miRNAs including miRNA-324 in PBMCs (peripheral blood mononuclear cells) of CHC (chronic hepatitis C) may act as significant risk biomarkers for the development of CHC. Since 75% CHC patients eventually develop HCC (Hepatocellular Carcinoma), the prognostic value of these miRNAs is high [Citation26]. We did not show miRNA-324-5p have potential prognostic value for GC, however, we found that miRNA-324-5p plays an oncogenic role for GC development, suggesting the necessity of future research on miRNA-324-5p prognostic capacity.
SUFU negatively regulates the Hedgehog and Wnt signaling pathways [Citation27]. It binds to GLI1 and inhibits its translocation to the nucleus. SUFU also suppresses Wnt signaling by forming a complex with β-catenin and enhancing β-catenin translocation to the cytoplasm [Citation20,Citation28,Citation29]. SUFU is identified as a very important tumor suppressor. It is down expressed in a number of tumors such as prostate cancer, lung cancer, glioblastoma, astrocytoma, glioma, and GC [Citation30–34]. In GC, the expression level of SUFU has been shown to be significantly and serially low in GC with different stages [Citation34,Citation35]. In our previous study, we found that SUFU was down expressed in GC tissues as well as cell lines and its expression was negatively related to tumor stage [Citation17]. However, the mechanism underlying its low expression in GC was poorly studied. Here, we showed that SUFU is targeted by miRNA-324-5p which is highly expressed in GC. The expression level of SUFU and miRNA-324-5p is negatively correlated with each other based on the RT-PCR results as well as TCGA analysis, providing a reason why SUFU is downregulated in GC.
EMT plays an important role in cancer advancement and metastasis. EMT regulation involves in a complicated network of transcriptional regulations engaged by many regulatory factors. A number of studies have demonstrated that miRNAs play a key role in the progression or suppression of EMT. MiRNAs target the master transcription factors of EMT and also regulate genes associated with signaling mediators, adhesion junction, and polarity complex proteins. For example, miRNA-200 family, which embraces miRNA-200a, miRNA-200b, miRNA-200 c, miRNA-141, and miRNA-429, negatively regulate EMT by targeting ZEB1/2 specifically [Citation36]. MiRNA-205, besides the miRNA-200 family, has been identified by microarray analyses as a negative regulator of EMT [Citation37]. Downregulation of miRNA-199a-5p in triple-negative breast cancer cells resulted in significantly elevated expression of EMT-associated genes such as ZEB1, CDH1, and TWIST [Citation38]. MiRNA-9 directly targets E-cadherin leading to EMT activation [Citation39].We found that miRNA-324-5p targets SUFU to induce EMT by activating Wnt signaling pathway, suggesting a novel mechanism underlying the potential significance of SUFU in gastric oncogenesis.
Conclusion
The present study revealed that miRNA-324-5p enhances cell proliferative, migratory property and induces EMT in GC, at least partially, through activating Wnt signaling pathway by targeting SUFU (). Further work will be required to understand if miRNA-324-5p has diagnostic and prognostic value for gastric cancer intervention.
Authors’ contributions
YP, XZ, ZJ, XFa, SM and SL designed the study. YP, XZ, HL, SD, XF, RY, YZ,YY, YC, JW, HF, and WC performed the experiments. YQ collected GC samples, communicated with patients and asked for written informed consent. YW conducted the statistical analysis. HA and DS provided the HFE-145 cell line and contributed to conception of the study. YP, XFa, and ZJ wrote the manuscript. YW, SM, and SL reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and informed consent
This study was approved by the Institutional Animal Care and Use Committee of Shenzhen University, Shenzhen, Guangdong, China.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author upon request.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17–20.
- Ruggieri V, Russi S, Zoppoli P, et al. The role of MicroRNAs in the regulation of gastric cancer stem cells: a meta-analysis of the current status. J Clin Med. 2019;8:639.
- Wang R, Sun Y, Yu W, et al. Downregulation of miRNA-214 in cancer-associated fibroblasts contributes to migration and invasion of gastric cancer cells through targeting FGF9 and inducing EMT. J Exp Clin Cancer Res. 2019;38:20.
- Sun GL, Li Z, Wang WZ, et al. miR-324-3p promotes gastric cancer development by activating Smad4-mediated Wnt/beta-catenin signaling pathway. J Gastroenterol. 2018;53:725–739.
- Crawford M, Batte K, Yu L, et al. MicroRNA 133B targets pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem Biophys Res Commun. 2009;388:483–489.
- Schultz J, Lorenz P, Gross G, et al. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557.
- Wang Y, Lee AT, Ma JZ, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205–13215.
- Dixon-McIver A, East P, Mein CA, et al. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PloS One. 2008;3:e2141.
- Gu C, Zhang M, Sun W, et al. Upregulation of miR-324-5p Inhibits Proliferation and Invasion of Colorectal Cancer Cells by Targeting ELAVL1. Oncol Res. 2019;27:515–524.
- Ferretti E, De Smaele E, Miele E, et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. Embo J. 2008;27:2616–2627.
- Kuo WT, Yu SY, Li SC, et al. MicroRNA-324 in human cancer: miR-324-5p and miR-324-3p have distinct biological functions in human cancer. Anticancer Res. 2016;36:5189–5196.
- Clements WM, Wang J, Sarnaik A, et al. beta-Catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Res. 2002;62:3503–3506.
- Ebert MP, Fei G, Kahmann S, et al. Increased beta-catenin mRNA levels and mutational alterations of the APC and beta-catenin gene are present in intestinal-type gastric cancer. Carcinogenesis. 2002;23:87–91.
- Franco AT, Johnston E, Krishna U, et al. Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res. 2008;68:379–387.
- Nagy TA, Wroblewski LE, Wang D, et al. beta-Catenin and p120 mediate PPARdelta-dependent proliferation induced by Helicobacter pylori in human and rodent epithelia. Gastroenterology. 2011;141:553–564.
- Song X, Xin N, Wang W, et al. Wnt/beta-catenin, an oncogenic pathway targeted by H pylori in gastric carcinogenesis. Oncotarget. 2015;6:35579–35588.
- Peng Y, Zhang X, Ma Q, et al. MiRNA-194 activates the Wnt/beta-catenin signaling pathway in gastric cancer by targeting the negative Wnt regulator, SUFU. Cancer Lett. 2017;385:117–127.
- Adiseshaiah P, Lindner DJ, Kalvakolanu DV, et al. FRA-1 proto-oncogene induces lung epithelial cell invasion and anchorage-independent growth in vitro, but is insufficient to promote tumor growth in vivo. Cancer Res. 2007;67:6204–6211.
- Aran D, Camarda R, Odegaard J, et al. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat Commun. 2017;8:1077.
- Rosenbluh J, Wang X, Hahn WC. Genomic insights into WNT/beta-catenin signaling. Trends Pharmacol Sci. 2014;35:103–109.
- Sanchez-Tillo E, de Barrios O, Siles L, et al. beta-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc Nat Acad Sci USA 2011; 108:19204–19209.
- Xu HS, Zong HL, Shang M, et al. MiR-324-5p inhibits proliferation of glioma by target regulation of GLI1. Eur Rev Med Pharmacol Sci. 2014;18:828–832.
- Gu C, Zhang M, Sun W, et al. Up-regulation of miR-324-5p inhibits proliferation and invasion of colorectal cancer cells by targeting ELAVL1. Oncol Res. 2019;27:515–524.
- Cao L, Xie B, Yang X, et al. MiR-324-5p suppresses hepatocellular carcinoma cell invasion by counteracting ECM degradation through post-transcriptionally downregulating ETS1 and SP1. PloS One. 2015;10:e0133074.
- Namkung J, Kwon W, Choi Y, et al. Molecular subtypes of pancreatic cancer based on miRNA expression profiles have independent prognostic value. J Gastroenterol Hepatol. 2016;31:1160–1167.
- Chang CC, Lin CC, Hsieh WL, et al. MicroRNA expression profiling in PBMCs: a potential diagnostic biomarker of chronic hepatitis C. Dis Markers. 2014;2014:367157.
- Fernandez-Zapico ME. Primers on molecular pathways GLI: more than just Hedgehog? Pancreatology Off J Int Assoc Pancreatology. 2008;8:227–229.
- Meng X, Poon R, Zhang X, et al. Suppressor of fused negatively regulates beta-catenin signaling. J Biol Chem. 2001;276:40113–40119.
- Min TH, Kriebel M, Hou S, et al. The dual regulator Sufu integrates Hedgehog and Wnt signals in the early Xenopus embryo. Dev Biol. 2011;358:262–276.
- Taylor MD, Liu L, Raffel C, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310.
- Dacic S, Sasatomi E, Swalsky PA, et al. Loss of heterozygosity patterns of sclerosing hemangioma of the lung and bronchioloalveolar carcinoma indicate a similar molecular pathogenesis. Arch Pathol Lab Med. 2004;128:880–884.
- Reifenberger J, Wolter M, Knobbe CB, et al. Somatic mutations in the PTCH, SMOH, SUFUH and TP53 genes in sporadic basal cell carcinomas. Br J Dermatol. 2005;152:43–51.
- Shahi MH, Rey JA, Castresana JS. The sonic hedgehog-GLI1 signaling pathway in brain tumor development. Expert Opin Ther Targets. 2012;16:1227–1238.
- Yan R, Peng X, Yuan X, et al. Suppression of growth and migration by blocking the Hedgehog signaling pathway in gastric cancer cells. Cell Oncol. 2013;36:421–435.
- Zhou H, Wang K, Hu Z, et al. TGF-beta1 alters microRNA profile in human gastric cancer cells. Chin J Cancer Res. 2013;25:102–111.
- Peng Y, Zhang X, Feng X, et al. The crosstalk between microRNAs and the Wnt/beta-catenin signaling pathway in cancer. Oncotarget. 2017;8:14089–14106.
- Piovan C, Palmieri D, Di Leva G, et al. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol. 2012;6:458–472.
- Chen J, Shin VY, Siu MT, et al. miR-199a-5p confers tumor-suppressive role in triple-negative breast cancer. BMC Cancer. 2016;16:887.
- Ma L, Young J, Prabhala H, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256.
