ABSTRACT
Non-small cell lung cancer (NSCLC) is the highest incidence and mortality of malignant tumors worldwide and has become a global public health problem. Long non-coding RNAs (LncRNAs) are expected to participate in the progression of NSCLC. This study aims to explore the effects and underlying mechanisms of LncRNA HOXC-AS2 on NSCLC cell proliferation, apoptosis, and migration. The Cell Counting Kit-8 (CCK-8) and clone formation assay were used to measure the A549 and HCC827 cell proliferation. The cell apoptosis and migration was respectively analyzed by flow cytometry and transwell assay. RNA immunoprecipitation (RIP) was used to detect the interaction between HOXC-AS2 and HOXC13. The expression of β-catenin, α-SMA, MMP-1, MMP-2 expression, E-cadherin, and Ki-67 expression were determined by Western blot or immunohistochemistry (IHC) assay. We found that HOXC-AS2 was significantly up-regulated in NSCLC tissues. Knockdown of HOXC-AS2 expression resulted in significant decreases in NSCLC cell proliferation, migration, and epithelial-mesenchymal transition (EMT) process marker proteins, simultaneously activated A549 and HCC827 cell apoptosis. RIP assay suggested that HOXC13 was a functional target for HOXC-AS2. And HOXC-AS2 and HOXC13 could positively regulate each other. Compared with the normal tissues, the mRNA level of HOXC13 was increased in NSCLC tissues. HOXC13 silencing counteracted increases of A549 and HCC827 cell proliferation and migration, as well as a decrease of cell apoptosis induced by HOXC-AS2 overexpression. Moreover, HOXC-AS2 silencing reduced tumor growth rate and Ki-67 expression in vivo. Taken together, HOXC-AS2 knockdown inhibited NSCLC cell proliferation and migration, as well as stimulated NSCLC cell apoptosis through regulation of HOXC13 expression.
KEYWORDS:
1 Introduction
Lung cancer is the malignant tumor with the highest morbidity and mortality in the world, which seriously threatens human life and health. Notably, non-small cell lung cancer (NSCLC) accounts for 80% to 85% of all lung cancers [Citation1]. The 5-year survival rate of lung cancer patients is still less than 20% due to its high rates of invasione and migration [Citation2]. The study has shown that the 5-year survival rate of lung cancer patients with metastasis is only 5%, while the 5-year survival rate of lung cancer patients without metastasis can reach 57% [Citation2]. Therefore, it is urgent to develop early diagnose and effective intervene preventing tumors from metastasize in NSCLC.
Long non-coding RNA (LncRNAs) are a class of RNA molecules that are longer than 200 nt but do not have the ability to encode proteins [Citation3,Citation4]. The regulation of LncRNA on cell proliferation, apoptosis, differentiation and metabolism predicts its special status in tumorigenesis and development. Related studies showed many LncRNAs were abnormally expressed during occurrence and development of NSCLC injury, such as XIST, H19, PCAT6, and GIAT4RA [Citation5–8]. Our previous high-throughput sequencing and bioinformatics analysis suggested that LncRNA HOXC cluster antisense RNA 2 (HOXC-AS2, chr12, 53,993,810–53,995,899) was highly expressed in NSCLC tissues. So far, HOXC-AS2 was reported to upregulate in glioma cells and tissues. And HOXC-AS2/miR-876-5p/ZEB1 axis were involved in regulation of EMT and glioblastoma mulitiforme (GBM) in glioma [Citation9]. In addition, emerging literature discovered that HOXC-AS2 have excellent diagnostic value for gastric adenocarcinoma (GAC) by analyzing the Cancer Genome Atlas (TCGA) datasets [Citation10]. However, the biological effect and underlying mechanism of HOXC-AS2 in NSCLC malignant progression remain to be investigated.
In this study, we studied the effects of HOXC-AS2 on NSCLC cell proliferation, migration, and apoptosis, and then explored the molecular mechanism of HOXC-AS2 in NSCLC. In addition, we examined its effect on the regulation of HOXC13 gene, which is abnormally expressed in multiple cancers, and plays an important regulatory role in tumor proliferation and migration [Citation11–13].
2 Materials and methods
2.1 Human NSCLC tissues and cell lines
The 20 NSCLC tissues and 10 paracancerous tissues were both collected from the fresh tumor tissues of NSCLC patients who underwent surgical resection in sichuan tumor hospital from January 2018 to December 2018. NSCLC tissues and adjacent tissues were collected from different NSCLC patients. None of the patients received chemotherapy or radiotherapy before surgery. This study was reviewed by the ethics committee of Sichuan cancer hospital. All patients and their families provided written informed consent.
The NSCLC cell lines HCC827, which was sensitive to icotinib and contained an EGFR exon 19 deletion (DelE746-A750) and A549 were purchased from Purcello (Wuhan, China). The cells were cultured in highglucose Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA) and 100 μg/mL penicillin double antibody (Hyclone, Los Angeles, USA) in a humidified incubator with 5% CO2 and 37°C.
2.2 Mice
16 female nude mice (four-week-old) were purchased from Dashuo Animal Experiment Co., Ltd. (Chengdu, Sichuan) and raised in the West China Animal Experiment Center of Sichuan University. The feeding environment was 24 ± 1°C, relative humidity 55 ± 5%, and light/darkness for 12 h circulation. The mice are allowed to eat and drink freely. This study was approved by the Sichuan University Ethics Committee.
2.3 Construction of nude mice implanted tumor model
Ten nude mice were randomly divided into two groups (n = 5/group), namely shRNA-NC group and shRNA-HOXC-AS2 group. The PBS suspended HOXC-AS2 stable low-expression HCC827 cells and the negative control cells (4 × 106 cells/100 μL) were subcutaneously injected into nude mice (total injection of 2 × 106 cells). Then, the nude mice were fed normally and observed the inoculation site for leaks. Observation indicators: weight, diet, mental, and tumor morphology were daily recorded. The diameter of the tumor was measured with a vernier caliper. Four weeks after the successful establishment of the model, pentobarbital sodium (50 mg/kg) was injected intraperitoneally to anesthetize the nude mice. The nude mice were sacrificed, the tumor tissue was removed, and some tumors were kept in −80°C for subsequent RT-qPCR analysis. Part of the tumor tissues were fixed with 10% paraformaldehyde for subsequent immunohistochemistry (IHC) experiments. Tumor volume calculation formula: V = 1/2 × L × D2 (L represents the largest diameter measured, D represents the smallest diameter measured).
2.4 Real-time fluorescence quantitative polymerase chain reaction (RT-qPCR)
Total RNA were isolated using TRIzol® reagent (Thermo Fisher, Massachusetts, USA). SYBR Pemix Ex Taq kit (Bao Biological Engineering, Dalian, China) was used following the guidelines. The reverse transcriptional reaction condition was as follows: 95°C for 10 min; 40 cycles of 95°C for 5 s, 60°C for 30 s, and 70°C for 60 s. The sequences of specific miRNA RT primers (Invitrogen, Carlsbad, CA, USA) were as follows: HOXC-AS2 forward, 5ʹ-CAA CTG CAT GTG GCC TGT AG-3ʹ and reverse, 5ʹ-GCA GGC CTT AGC TGG ATT TG-3ʹ, HOXC13 forward, 5ʹ-GGA GTT CGC CTT CTA CC-3ʹand reverse, 5ʹ-GAC TGT CCC AGC CAT TG-3ʹ, β-actin forward, 5ʹ- CAT GTA CGT TGC TAT CCA GGC-3ʹ and reverse, 5ʹ-CTC CTT AAT GTC ACG CCA CGA T-3ʹ. The relative gene expression level was determined using the 2−ΔΔCt method on ABI software, Foster City, CA. β-actin mRNA level as an internal control. The data were analyzed using Bio-Rad CFX Manager software.
2.5 Western blot analysis
Total protein from A549 and HCC827 cells was separated by 10% SDS polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes (Millipore, Boston, MA, USA). The membranes were blocked with 5% nonfat milk and incubated with corresponding protein antibodies or a rabbit anti-β-actin monoclonal antibody. Then, the membranes were subsequently incubated with an HRP Goat anti-Rabbit IgG (1:20,000; Boster, Wuhan, China; BA1054). The net optical density was analyzed with the gel Image processing system (Image-pro Plus 6.0). Primary antibodies used were as follows: E-cadherin (Abcam, Cambridge, UK; cat. no. ab1416, 1:2000), β-catenin (Abcam, Cambridge, UK; cat. no. ab32572, 1:2000), α-SMA (Abcam, Cambridge, UK; cat. no. ab5831, 1:2000), MMP-1 (Abcam, Cambridge, UK; cat. no. ab52631, 1:2000), MMP-2 (Abcam, Cambridge, UK; cat. no. ab97779, 1:2000), and β-actin (Abcam, Cambridge, UK; cat. no. ab5694, 1:2000). The luminescence reaction is displayed using Pierce ECL luminescence kit (Affinity Biosciences, Cincinnati, Ohio, USA) and β-actin was used as an internal control.
2.6 Cell vitality assay
The cell vitality of A549 and HCC827 cells was measured using the Cell Counting Kit 8 (CCK-8, Thermo Fisher Scientific) according to the manufacturer’s instructions. To be brief, the log phase cells were collected and wash with PBS. Then, the cells were digested with trypsin (7 × 104/mL) and were plated in 96-well plates (100 μL/well). The seeded cells were incubated at 37°C and 5% CO2 for 48 hours. Then, the supernatant was removed. The CCK-8 solution was diluted using serum-free fresh medium at a ratio of 10:1. 100 μL CCK-8 solutions were added to each well and further incubated for 1 h at 37°C. Finally, the cell vitality was measured using enzyme-linked immune monitor at 450 nm.
2.7 Clone formation
A549 and HCC827 cells were seeded in 6-well plates (1000 cells/well) and placed in a 5% CO2 and 37°C incubator. The cells were observed every 12 h, and stopped the culture when there are visible clones. Finally, the cells were fixed with formaldehyde and stained with crystal violet. The numbers of clone-forming cells counted using an optical inverted microscope.
2.8 Apoptosis assay
The apoptosis of A549 and HCC827 cells was assessed by Annexin V FITC/PI staining flow cytometry according to the manufacturer’s instructions. The HT22 cells were washed with PBS (Invitrogen, Carlsbad, CA, USA) and adjusted the cell concentration to 7 × 104 cells/mL. The cells were then resuspended with 500 μL Binding Buffer, 5 μL Annexin-V-FITC, and 5 μL PI. The samples were protected from light in 4°C for 15 min and then analyzed by flow cytometry with excitation at 488 nm and emission measured at 560 nm.
2.9 Transwell assay
The cell density was adjusted to 1 × 105/mL using serum-free MEM medium. Place the Transwell chamber was placed in a 24-well plates. 200 μL/well cells were add in the upper chamber of Transwell, and 600 μL DMEM medium containing 10% serum was added in the lower chamber of Transwell. Then, the cells were placed in incubator (5% CO2 and 37°C) for 48 h. Finally, the cells were fixed with 4% paraformaldehyde for 20 min, and stained with 0.1% crystal violet for 30 min. Fluorescence inverted microscope (Thermo Fisher, Massachusetts, USA) was used to observe the number of cells in 5 random visions.
2.10 RNA immunoprecipitation (RIP)
A549 and HCC827 cells were lysed in RIP lysis buffer. Magnetic beads were pre-incubated with antibodies for 30 min at room temperature and the cell lysates was immunoprecipitated with beads for 6 h at 4°C. Then, RNA was purified using TRIzol® reagent (Thermo Fisher, Massachusetts, USA) and detected by RT-qPCR analysis.
2.11 Immunohistochemistry (IHC)
Paraffin sections of nude mouse tumor tissue were dewaxed with xylene and dehydrated with gradient alcohol. Antigen repair was performed with 0.01 mol/L citrate buffer solution. 3% hydrogen peroxide was used to block endogenous peroxidase. After cooling naturally at room temperature, the paraffin sections were washed with 0.1 mol/L PBS and blocked with 5% BSA. Then, the paraffin sections were incubated overnight with rabbit anti-Ki-67 (1:100). Finally, the paraffin sections were subsequently incubated with HRP-labeled goat anti-rabbit secondary antibody. The proteins were detected at room temperature using a DAB immunohistochemistry color development kit (Boshide Biological, Wuhan, China). ImagePro Plus image analysis software was used to analyze the optical density value (OD) of the slice.
2.12 Statistical analysis
The data were represented as means ± standard deviation (SD) and each experiment was performed in triplicate in this study. A one-way ANOVA with Tukey post hoc test of means and Student’s unpaired t-test were used to assess statistical significance, which was done with using SPSS 22.0 software (SPSS, Inc., Chicago, IL, USA). P-value <0.05 was considered statistically significant. P-value <0.01 means the difference is extremely significant.
3 Results
3.1 HOXC-AS2 is upregulated in NSCLC tissues
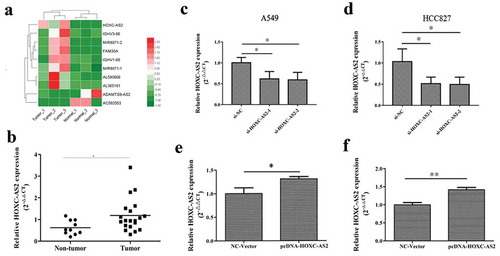
As shown in ), microarray analysis was used to identify top 10 dysregulated lncRNAs in NSCLC tissues. Among them, HOXC-AS2 was significantly upregulated in NSCLC tissues, predicting its expression and biological function in NSCLC tumorigenesis. Then, To investigate the expression level of HOXC-AS2, we determined the levels of HOXC-AS2 in tumors from 20 patients with NSCLC and 10 samples of normal tissues. As shown in ), increased HOXC-AS2 mRNA level was detected in NSCLC tissues compared with normal tissues. Thus, we designed efficient siRNA and overexpression vector to, respectively, promote and inhibit the expression of HOXC-AS2. And they were used to uncover the effect of HOXC-AS2 on NSCLC progression (-f)).
Figure 1. HOXC-AS2 is upregulated in NSCLC tissues. (a) Heat map of the dysregulated expression of the top 10 lncRNAs. (AB) The expression of HOXC-AS2 in human NSCLC tissues (n = 20) and normal tissues (n = 10) was compared by RT-qPCR analysis. (B, C, D, and EC, D, E, and F) HOXC-AS2 mRNA expression in A549 and HCC827 cell lines were detected by RT-qPCR analysis. *P < 0.05 (vs normal tissues/si-NC/NC-Vector), **P < 0.01 (vs NC-Vector). Data were represented as means ± standard deviation (SD). Each experiment was performed in triplicate
3.2 HOXC-AS2 regulates NSCLC cell proliferation, apoptosis, and migration
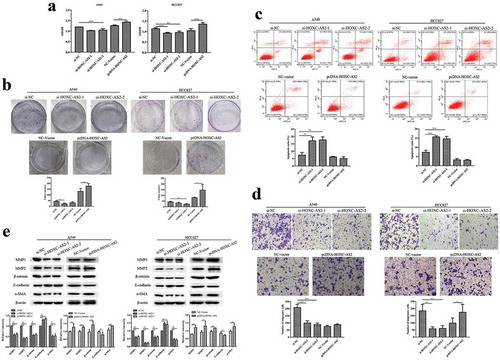
HOXC-AS2 siRNA (si-HOXC-AS2) and pcDNA-HOXC-AS2 were transfected in A549 and HCC827 cell lines. CCK-8 and clone formation assay found that silencing of HOXC-AS2 in A549 and HCC827 cells decreased the ability of cell proliferation (,b)). And overexpression of HOXC-AS2 obviously promoted the cell proliferation (,b)). Moreover, compared with control group, A549 and HCC827 cell apoptosis were increased in si-HOXC-AS2 transfected A549 and HCC827 cells ()). Transwell assay showed that HOXC-AS2 knockdown also weakened A549 cell migration ()). And HOXC-AS2 overexpression strongly stimulated HCC827 cell migration ()). Next, the expressions of EMT marker proteins E-cadherin, β-catenin, α-SMA, MMP-1, and MMP-2 expression were detected using Western blot analysis. The results demonstrated that the silencing of HOXC-AS2 reduced β-catenin, α-SMA, MMP-1, and MMP-2 expression, as well as enhanced E-cadherin expression, which were reversed in pcDNA-HOXC-AS2 transfected A549 and HCC827 cells ()).
Figure 2. HOXC-AS2 regulates NSCLC cell proliferation, apoptosis, and migration. (a and b) The proliferation of A549 and HCC827 cells were evaluated by CCK-8 and clone formation assay. (c) The apoptosis of A549 and HCC827 cells was assessed by Annexin V FITC/PI staining flow cytometry. (d) The migration ability of SW480 and HT29 cells were evaluated by transwell assay. (e) The β-catenin, α-SMA, MMP-1, MMP-2, and E-cadherin expression in A549 and HCC827 cells was determined by Western blot analysis. β-actin is a loading control. *P < 0.05 (vs si-NC/NC-Vector), **P < 0.01 (vs si-NC/NC-Vector), ***P < 0.001 (vs si-NC/NC-Vector). Data were represented as means ± standard deviation (SD). Each experiment was performed in triplicate
3.3 HOXC13 is a target gene of HOXC-AS2
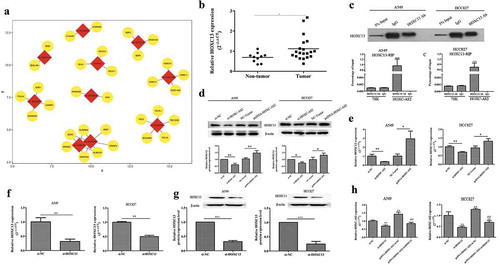
Next, we looked for the possible target gene for HOXC-AS2. Our previous high-throughput sequencing and bioinformatics analysis found that HOXC13 was a positive neighboring gene of HOXC-AS2. Our previous bioinformatics analysis found that HOXC13 was a possible target gene for HOXC-AS2 ()). Furthermore, we found that there was a correlation between HOXC13 and HOXC-AS2 through calculating the Pearson correlation coefficient. Of note, compared with normal tissues, the mRNA level of HOXC13 was significantly increased in NSCLC tissues ()). To further research how HOXC-AS2 regulates the expression of HOXC13 protein, RNA immunoprecipitation (RIP) experiments was performed. The data proved that HOXC13 protein strongly enriched HOXC-AS2 in A549 and HCC827 cells, confirming the target relationship between HOXC-AS2 and HOXC13 ()). And RT-qPCR and Western blot analysis displayed that HOXC-AS2 overexpression obviously activated HOXC13 expression both in protein and RNA levels (,e)). And HOXC13 expression was decreased in si-HOXC-AS2 transfected A549 and HCC827 cells (,e)). More importantly, effective HOXC13 silent cell line was constructed to explore whether HOXC13 can also regulate HOXC-AS2 (,g)). RT-qPCR analysis revealed that HOXC-AS2 expression was significantly reduced in HOXC13-siRNA transfected A549 and HCC827 cells ()). Meanwhile, HOXC13 knockdown suppressed pcDNA-HOXC-AS2-stimulated HOXC-AS2 expression ()).
Figure 3. HOXC13 is a target gene of HOXC-AS2. (a) The predicted target genes of the dysregulated 10 lncRNAs. The dysregulated lncRNAs were as the center marked with red and the predicted target genes were marked with yellow. (AB) The expression of HOXC13 in human NSCLC tissues (n = 20) and normal tissues (n = 10) was compared by RT-qPCR analysis. (BC) RNA immunoprecipitation (RIP) assay was performed to recover the interaction of HOXC13 and HOXC-AS2. ***P < 0.001 (vs IgG). (C, D, and ED-G) The expression of HOXC13 in A549 and HCC827 cells was tested by Western blot and RT-qPCR analysis. β-actin is a loading control. (FH) The expression of HOXC-AS2 in A549 and HCC827 cells were tested via RT-qPCR analysis. *P < 0.05 (vs normal tissues/si-NC/NC-Vector), **P < 0.01 (vs si-NC/NC-Vector), ##P < 0.01(vs pcDNA-HOXC-AS2+ si-NC). Data were represented as means ± standard deviation (SD). Each experiment was performed in triplicate
3.4 HOXC-AS2 regulates NSCLC cell proliferation, apoptosis, and migration via activation of HOXC13
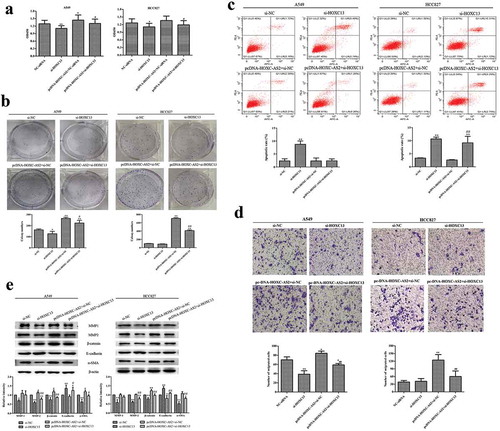
CCK-8 and clone formation results showed that HOXC13-siRNA could significantly inhibit A549 and HCC827 cell proliferation (,b)). Of note, HOXC13 silencing reversed the promotion of HOXC-AS2 overexpression on cell proliferation (,b)). Flow cytometry assay revealed that HOXC13 silencing significantly stimulated the apoptosis of A549 and HCC827 cells. Co-transfection of pc-DNA-HOXC-AS2 and HOXC13-siRNA reversed the inhibition of pc-DNA-HOXC-AS2 on HCC827 cell apoptosis In A549 cells, increased apoptosis when cells transfected with si-HOXC13 was reversed when pcDNA-HOXC-AS2 was co-transfected, while this reversal was not observed in HCC827 cells ()). These results maybe because A549 cells are more sensitive to the regulation of PCDNA-HOxC-AS than HCC827 cells. It also suggests that HOXC13 knockdown may regulate the expression of other apoptosis-related genes and proteins in HCC827 cells. In addition, transwell chamber experiments showed HOXC13 knockdown significantly suppressed the migration of A549 cells. At the same time, HOXC13 knockdown offset HOXC-AS2 overexpression-induced increases of cell migration in A549 and HCC827 cells ()). Concurrently, our results also displayed that HOXC13 silencing overexpression reduced β-catenin, α-SMA, MMP-1, and MMP-2 expression as well as strengthened E-cadherin expression in A549 and HCC827 cells, which was obviously counteracted by pcDNA-HOXC-AS2 cotransfection ()). These results indicated that HOXC-AS2 promoted NSCLC cell proliferation and migration, as well as inhibited cell apoptosis by positively regulating HOXC13 expression.
Figure 4. HOXC-AS2 regulates NSCLC cell proliferation, apoptosis, and migration via activation of HOXC13. (a and b) The proliferation of A549 and HCC827 cells were evaluated by CCK-8 and clone formation assay. (c) The apoptosis of A549 and HCC827 cells was assessed by Annexin V FITC/PI staining flow cytometry. (d) The migration ability of SW480 and HT29 cells were evaluated by transwell assay. (e) The β-catenin, α-SMA, MMP-1, MMP-2, and E-cadherin expression in A549 and HCC827 cells was determined by Western blot analysis. β-actin is a loading control. *P < 0.05 (vs si-NC), **P < 0.01 (vs si-NC), #P < 0.05(vs pcDNA-HOXC-AS2+ si-NC), ##P < 0.01(vs pcDNA-HOXC-AS2+ si-NC). Data were represented as means ± standard deviation (SD). Each experiment was performed in triplicate
3.5 HOXC-AS2 silencing suppresses NSCLC development in vivo
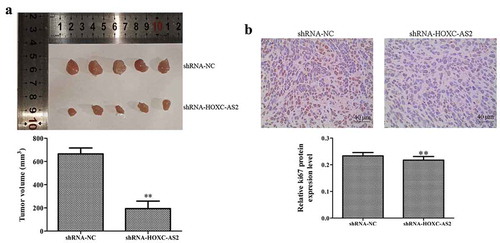
We subcutaneously injected HCC827 cells with shRNA-HOXC-AS2 or negative regulator vector into the nude mice. After 30 days, the mice were killed, and the tumors were examined ()). The data indicated that injection of HCC827 cells loaded with shRNA-HOXC-AS2 led to a significant reduction in tumor growth rate compared with the control groups ()). Moreover, IHC assay showed that the expression of Ki-67 protein in tumor tissues of shRNA-HOXC-AS2 group was obviously lower than that of shRNA-NC group ()).
Figure 5. HOXC-AS2 silencing suppresses NSCLC development in vivo. (a) Compared with the shRNA-NC group, shRNA-HOXC-AS2 inhibited the tumor volume (bar = 10 mm). (b) The expression of Ki-67 was measured using immunohistochemistry (IHC) assay. *P < 0.05 (vs shRNA-NC), **P < 0.01 (vs shRNA-NC). Data were represented as means ± standard deviation (SD). Each experiment was performed in triplicate
4 Discussion
Over the past decade, whole-genome and transcriptome sequencing technologies were used to find differential gene expression and differential splicing of mRNAs [Citation14]. Gene polymorphism analysis associated to disease development may provide useful tools for prognosis, risk rating, and treatment of clinical patients [Citation15,Citation16]. Through high-throughput RNA sequencing (RNA-seq), the authors proved that significant expression differences and splicing events were occurred in exposure to A2E-exposed retinal pigment epithelium cells which involve in specific cellular activities including oxidative stress [Citation17,Citation18]. In particular, RNA-seq is a reliable technique to reveal the expression profiles and functions of tumor genes and moleculars. Previous proved that top 10 dysregulated circRNAs, including circDDX17, were potentially participated in colorectal cancer development and mainly associated with the KEGG pathways [Citation19]. Consequently, our studies screened the differentially expressed top 10 lncRNAs between NSCLC and normal mucosa tissues by RNA-seq. HOXC-AS2, an LncRNA located on chromosome 12q13.13, was significantly up-regulated in NSCLC tissues compared with adjacent tissues. And the results were highly consistent with the RT-qPCR data.
According to previous studies, LncRNAs play a pivotal role in the progression of human NSCLC progression. Recent report proved that LncRNA PCAT6 was up-regulated in human NSCLC specimens. Moreover, PCAT6 promoted the aggressiveness of NSCLC by binding to EZH2 and suppressing LATS2 [Citation7]. Compared with normal tissues, the expression of LncRNA NAPRAL in NSCLC tissues was significantly decreased, and NAPRAL promoted NCI-H929 and A549 cell proliferation via activating P53 transcription [Citation20]. Our previous high-throughput sequencing analysis suggested that HOXC-AS2, an LncRNA located on chromosome 12q13.13, was significantly up-regulated in NSCLC tissues compared with adjacent tissues. More importantly, HOXC-AS2 silencing was found to restrained invasive ability and reduced expression of epithelial-mesenchymal transition (EMT) marker proteins (N-cadharin and Vimentin) in glioma [Citation9]. In pediatric acute myeloid leukemia, HOXC-AS2 was reported to be a translocation partner of ets translocation variant gene 6 (ETV6), which participates in various cancers-related translocations [Citation21,Citation22]. ETV6-HOXC-AS2 cooperation may result in a loss of function of ETV6 or HOXC-AS2, and ultimately plays a key role in gastric cancer [Citation10]. Our studies firstly investigated the role and regulatory mechanism of HOXC-AS2 in NSCLC. The finding suggested that HOXC-AS2 was upregulated in NSCLC tissues. In addition, the depletion of HOXC-AS2 weakened NSCLC cell proliferation and migration, as well as promoted NSCLC cell apoptosis. Meanwhile, we firstly studied the effect of HOXC-AS2 on EMT processes in NSCLC cells. EMT refers to the loss of polarity and connection between epithelial cells triggering the increase the ability migration and invasion [Citation23]. Tumor cells induced the expression of metalloproteinase, which is conducive to the metastasis of tumor cells [Citation24]. The accumulation of β-catenin in the nucleus inhibits E-cadherin expression, thereby promoting the occurrence of EMT [Citation25]. α-smooth muscle actin (α-SMA) is well known to secrete soluble chemokines and participate in the reconstruction and synthesis of tumor extracellular matrix (ECM) [Citation26]. In our results, EMT marker proteins β-catenin, α-SMA, MMP-1, and MMP-2 expression were deduced, as well as E-cadherin expression was enhanced due to HOXC-AS2 knockdown. Similarly, overexpression of HOXC-AS2 showed the negative results.
Homeobox 13 (HOX13) was a member of HOX genes and accumulated evidence have indicated that HOX genes played important roles in many cancer progressions, including breast cancer, colon cancer, breast cancer, and NSCLC [Citation27–29]. Previous studies showed that HOXC13 is significantly elevated in human liposarcomas [Citation30], squamous cell carcinoma [Citation31], lung adenocarcinomas [Citation29], and prostate cancer [Citation32]. We confirmed that HOXC13 was significantly increased in NSCLC tissues. Moreover, HOXC13 silencing suppressed cell proliferation and migration, as well as stimulated the apoptosis of NSCLC cells. Mechanistically, silencing HOXC13 inhibited the proliferation of lung adenocarcinoma cells and induced G1 arrest by down-regulating CCND1 and CCNE1 expression [Citation29]. Simultaneously, HOXC13 was a target gene of miR-503. And HOXC13 promoted esophageal squamous cell carcinoma (ESCC) proliferation and inhibited apoptosis via repressing transcription of CASP3 [Citation33]. Our previous bioinformatics analysis found HOXC13 was a nearby target of HOXC-AS2. In the present study, HOXC13 was a target gene of HOXC-AS2 and co-expressed with HOXC-AS2 as well. Moreover, HOXC13 knockdown obviously offset HOXC-AS2-induced NSCLC proliferation, migration, and apoptosis.
Ki67 plays an extremely important role in the regulation of cell proliferation. Ki67 expresses in the active phase of the cell cycle (G1, S, G2, and M), while its expression is absolutely suppressed in the G0 phase. Various studies confirmed that Ki67 protein expression was related to the prognosis of various malignant tumor patients [Citation34–36]. The expression of Ki67 in breast cancer tissue was significantly correlated with tumor size and lymph node metastasis, suggesting that Ki67 was an indicator of poor prognosis in patients with triple-negative breast cancer (TNBC) [Citation37]. Therefore, we also detected the expression of Ki-67 in the NSCLC nude mice by IHC. The results showed that compared with the negative control group, the Ki-67 expression in the tumor tissue of the HOXC-AS2 silent group was significantly suppressed. And our study further confirmed that HOXC-AS2 silencing dramatically inhibited tumor growth in vivo.
Collectively, our present study highlights the importance of correlation between HOXC-AS2 and HOXC13 both in vitro and in vivo. The HOXC-AS2/HOXC13 mutual regulation has potential to be a therapeutic target for NSCLC treatment.
Acknowledgments
This study was funded by the National Science and Technology Major Project (grant number 2017ZX09304023).
Disclosure statement
The authors declare that they have no conflict of interest.
Additional information
Funding
References
- Rotenberg Z, Weinberger I, Sagie A, et al. Total lactate dehydrogenase and its isoenzymes in serum of patients with non-small-cell lung cancer. Clin Chem. 1988;34:668–70.
- Siegel RL, Miller KD. Cancer Statistics, 2020. Multicenter Study. 2020;70:7–30.
- Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62.
- Donato L, Scimone C, Alibrandi S, et al. Transcriptome Analyses of lncRNAs in A2E-Stressed Retinal Epithelial Cells Unveil Advanced Links between Metabolic Impairments Related to Oxidative Stress and Retinitis Pigmentosa. Antioxidants (Basel). 2020;9:318.
- Sun W, Zu Y, Fu X, et al. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol Rep. 2017;38:3347–3354.
- Lei Y, Guo W, Chen B, et al. Tumor‑released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non‑small cell lung cancer. Oncol Rep. 2018;40:3438–3446.
- Shi X, Liu Z, Liu Z, et al. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018;37:177–187.
- Yang R, Liu N, Chen L, et al. GIAT4RA functions as a tumor suppressor in non-small cell lung cancer by counteracting Uchl3-mediated deubiquitination of LSH. Oncogene. 2019;38:7133–7145.
- Dong N, Guo J, Han S, et al. Positive feedback loop of lncRNA HOXC-AS2/miR-876-5p/ZEB1 to regulate EMT in glioma. Onco Targets Ther. 2019;12:7601–7609.
- Fu T, Ji X, Bu Z, et al. Identification of key long non-coding RNAs in gastric adenocarcinoma. Cancer Biomark. 2020;27:541–553.
- Awgulewitsch A. Overexpression of Hoxc13 in differentiating keratinocytes results in downregulation of a novel hair keratin gene cluster and alopecia. Development. 2001;128:1547–1558.
- Liu M, Zhao S, Lin Q, et al. YAP regulates the expression of Hoxa1 and Hoxc13 in mouse and human oral and skin epithelial tissues. Mol Cell Biol. 2015;35:1449–1461.
- Luo J, Wang Z, Huang J, et al. HOXC13 promotes proliferation of esophageal squamous cell carcinoma via repressing transcription of CASP3. Cancer Sci. 2018;109:317–329.
- Stark R, Grzelak M. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20:631–656.
- Rinaldi C, Bramanti P, Scimone C, et al. Relevance of CCM gene polymorphisms for clinical management of sporadic cerebral cavernous malformations. J Neurol Sci. 2017;380:31–37.
- Scimone C, Donato L, Katsarou Z, et al. Two Novel KRIT1 and CCM2 Mutations in Patients Affected by Cerebral Cavernous Malformations: new Information on CCM2 Penetrance. Front Neurol. 2018;9:953.
- Donato L, D’Angelo R, Alibrandi S, et al. Effects of A2E-Induced Oxidative Stress on Retinal Epithelial Cells: new Insights on Differential Gene Response and Retinal Dystrophies. Antioxidants (Basel). 2020;9:307.
- Donato L, Scimone C. Discovery of GLO1 New Related Genes and Pathways by RNA-Seq on A2E-Stressed Retinal Epithelial Cells Could Improve Knowledge on Retinitis Pigmentosa. Antioxidants (Basel).2020;9:416.
- Li XN, Wang ZJ, Ye CX, et al. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37:325.
- Su P, Wang F, Qi B, et al. P53 Regulation-Association Long Non-Coding RNA (LncRNA PRAL) Inhibits Cell Proliferation by Regulation of P53 in Human Lung Cancer. Med Sci Monit. 2017;23:1751–1758.
- Wada R, Arai H, Kure S, et al. “Wild type” GIST: clinicopathological features and clinical practice. Pathol Int. 2016;66:431–437.
- Rooij JD, Beuling E, Fornerod M, et al. ETV6 Aberrations Are a Recurrent Event in Pediatric Acute Myeloid Leukemia with Poor Clinical Outcome. Blood. 2014;124(21): 1012-1012.
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96.
- Xu Y, Zhang J, Liu X, et al. MMP-2-responsive gelatin nanoparticles for synergistic tumor therapy. Pharm Dev Technol. 2019;24:1–26.
- Kang C. Downregulated microRNA-200a promotes EMT and tumor growth through the wnt/β-catenin pathway by targeting the E-cadherin repressors ZEB1/ZEB2 in gastric adenocarcinoma. Oncol Rep. 2013;29(4):1579–1587.
- Zhang J, Jiang T, Liang X, et al. lncRNA MALAT1 mediated high glucose-induced HK-2 cell epithelial-to-mesenchymal transition and injury. J Physiol Biochem. 2019;75:443–452.
- Li C, Cui J, Zou L, et al. Bioinformatics analysis of the expression of HOXC13 and its role in the prognosis of breast cancer. Oncol Lett. 2020;19:899–907.
- Kasiri S, Ansari KI, Hussain I, et al. Antisense oligonucleotide mediated knockdown of HOXC13 affects cell growth and induces apoptosis in tumor cells and over expression of HOXC13 induces 3D-colony formation. RSC Adv. 2013;3:3260–3269.
- Yao Y, Luo J, Sun Q, et al. HOXC13 promotes proliferation of lung adenocarcinoma via modulation of CCND1 and CCNE1. Am J Cancer Res. 2017;7:1820–1834.
- Chiara AD. Hyperexpression of HOXC13, located in the 12q13 chromosomal region, in well-differentiated and dedifferentiated human liposarcomas. Oncol Rep. 2013;30:2579–86.
- Marcinkiewicz KM, Gudas LJ. Altered Histone Mark Deposition and DNA Methylation at Homeobox Genes in Human Oral Squamous Cell Carcinoma. J Cell Physiol. 2014;229:1405–1416.
- Komisarof J, McCall M, Newman L, et al. A four gene signature predictive of recurrent prostate cancer. Oncotarget. 2017;8:3430–3440.
- Luo J, Wang Z, Huang J, et al. HOXC13 promotes proliferation of esophageal squamous cell carcinoma via repressing transcription of CASP3. Cancer Sci. 2018;109:317–329.
- Yang C, Zhang J, Ding M, et al. Ki67 targeted strategies for cancer therapy. Clin Transl Oncol. 2018;20:570–575.
- Li LT, Jiang G, Chen Q, et al. Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol Med Rep. 2015;11:1566–1572.
- Yerushalmi R, Woods R, Ravdin PM, et al. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 2010;11:174–183.
- Li H, Han X, Liu Y, et al. Ki67 as a predictor of poor prognosis in patients with triplenegative breast cancer. Oncol Lett. 2015;9:149–152.
