ABSTRACT
RNA pumilio RNA binding family member 1 (circPUM1) has been reported to play important roles in the tumorigenesis of several cancers. However, the underlying molecular role of circPUM1 in non-small cell lung cancer (NSCLC) progression remains unknown. The qRT-PCR and western blot were used to evaluate the expression of RNAs and proteins. In vitro cell proliferation assays, flow cytometric and glucose metabolism analyses were performed to test the effects of circPUM1 and its target on NSCLC cell growth and glycolysis. The interaction between microRNA (miR)-590-5p and circPUM1 or methyltransferase like 3 (METTL3) was analyzed by using dual-luciferase reporter, pull-down or RNA immunoprecipitation (RIP) assays. Murine xenograft model was established to conduct in vivo experiments. CircPUM1 was highly expressed in NSCLC tissues and cell lines. CircPUM1 knockdown suppressed cell proliferation, cell cycle and glycolysis in vitro. Moreover, circPUM1 directly bound to miR-590-5p, and miR-590-5p inhibitor reversed the inhibitory effects of circPUM1 knockdown on NSCLC carcinogenesis. Additionally, miR-590-5p suppressed NSCLC progression by directly targeting and regulating METTL3. Importantly, circPUM1 could regulate METTL3 in NSCLC cells through miR-590-5p. In addition, it was also proved circPUM1 silencing impeded tumor growth and glycolysis in the murine xenograft model by regulating miR-590-5p/METTL3 axis. CircPUM1 promoted NSCLC tumor growth and glycolysis through sequestering miR-590-5p and up-regulating METTL3, providing an improved understanding of NSCLC tumorigenesis and a potential therapeutic target for NSCLC therapy.
KEYWORDS:
Introduction
Lung cancer is one of prevalent and lethal malignancy, ranking first among all cancers in cancer-related deaths in the world and China [Citation1,Citation2], and approximately 80 to 85% of lung cancer case are non-small cell lung carcinomas (NSCLC) [Citation3]. Although great improvements in diagnosis and therapeutic strategies, the five-year survival rate for advanced NSCLC remains unsatisfactory with around 15% [Citation4], due to the impact of high rate of metastasis, recurrence and drug resistance [Citation5,Citation6]. Hence, it is necessary to study the pathogenesis and search for effective therapeutic targets for improving NSCLC prognosis.
Circular RNAs (circRNAs) are a class of non-coding RNAs molecules possessing ring structures which have no 5ʹ-3ʹ polarity and polyA tail; besides, circRNAs are highly represented in the eukaryotic transcriptome and often show specie-, tissue- and cell-specific expressed [Citation7,Citation8], which suggest important functions for these molecules. Recently, emerging evidence has exhibited that circRNAs may play a role in multiple disease processes, including cancer development, through the regulation of various cellular crucial biological behaviors associated with carcinogenesis and metabolism [Citation9–11]. Importantly, the significance of circRNAs in the progression of NSCLC has also begun to be discussed. For example, circP4HB was highly expressed in NSCLC, and facilitated NSCLC aggressiveness and epithelial-mesenchymal transition (EMT) through absorbing miR-133a-5p [Citation12]. Circ_0067934 promoted NSCLC tumorigenesis via regulating cell proliferative, migratory, and invasive capacities [Citation13]. Thus, targeting circRNAs may be promising candidates for enhancing the efficacy of NSCLC treatments. Circular RNA pumilio RNA binding family member 1 (circPUM1) is a functional circRNA, a recent study showed circPUM1 was highly expressed in lung adenocarcinoma, and enhanced cell tumorigenesis in vivo and in vitro [Citation14]. Here, we suspected that circPUM1 might also involve in NSCLC.
MicroRNAs (miRNAs) are a kind of single-stranded non-coding RNAs of about 22 nucleotide, which are critical regulators of various biological processes by targeting the 3ʹ-untranslated region (UTR) of the mRNA [Citation15]. Besides that, miRNAs can impact cancer development via innumerous mechanisms [Citation16], and dysregulated miRNAs expression are now known to be involved in the tumorigenesis of NSCLC [Citation17,Citation18]. MiR-590-5p is cancer-related miRNA and has been proven to be related to various cancers. In NSCLC, it was demonstrated be down-regulated, and served as a tumor suppressor to suppress NSCLC cell malignant biological behaviors [Citation19–21]. Methyltransferase like 3 (METTL3) is a RNA methyltransferase responsible for N6-methyladenosine (m6A) modification in coding and non-coding RNAs, thus involving in mRNA biogenesis, translation, and decay control [Citation22,Citation23]. Recent findings indicated that METTL3 can promote the expression of several crucial oncoproteins and its high expression enhanced survival, and metastasis of human lung cancer cells [Citation23,Citation24]. However, the role of circPUM1, miR-590-5p and METTL3 in glycolysis and the interactions among them in NSCLC remain unclear.
In the present study, we attempted to test the expression profile and pathological role of circPUM1 in NSCLC malignant and glycolytic phenotypes in vitro and in vivo, and investigated whether circPUM1 might function as competing endogenous RNA (ceRNA) for miR-590-5p to regulate METTL3 expression in NSCLC, thus involving in the tumorigenesis of NSCLC.
Materials and methods
Clinical specimens
The NSCLC tissues and matched non-cancerous tissues (N = 62) were obtained from The First Affiliated Hospital of Zhengzhou University by resection. All specimens were instantly frozen in liquid nitrogen and stored at −80°C. We have obtained authorization from the Ethics Committee of the The First Affiliated Hospital of Zhengzhou University, and each enrolled individual had signed written informed consent.
Cell culture
Human NSCLC cell lines (H1650 and A549) and non-cancerous lung cell lines (BEAS-2B) were obtained from Beijing Institute for Cancer Research Collection (Beijing, China), and then grown in a 5% CO2 atmosphere at 37°C with RPMI-1640 medium (Invitrogen San Diego, CA, USA) containing fetal bovine serum (FBS, 10%) and 1% penicillin-streptomycin acquired from Sigma (St Louis, MO, USA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Whole-RNA was extracted with the help of Trizol reagent (Invitrogen) from targeted tissues or cells. First strand complementary DNAs (cDNAs) was generated using the PrimeScript RT reagent Kit (Takara Bio Inc, Kusatsu, Japan). Then qRT-PCR analyses used SYBR Select Master Mix (Takara) was performed. The relative expression was assessed by the 2−ΔΔCt method normalizing to U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primer sequences were listed: circPUM1: F 5ʹ-CCAAGCCTGTGGAGGATTTCT-3ʹ, R 5ʹ-CACATCACCCTCCTCCTTCAA-3ʹ; METTL3: F 5ʹ-TTAGCCTTCGGGGTGTCCG-3ʹ, R 5ʹ-CTGCAGGAGGCTTTCTACCC-3ʹ; GADPH: F 5ʹ-CCCACATGGCCTCCAAGGAGTA-3ʹ, R 5ʹ-GTGTACATGGCAACTGTGAGGAGG-3ʹ; U6: F 5ʹ- TTCACGAATTTGCGTGTCAT-3ʹ, R 5ʹ-CGCTCGGCAGCACATATAC-3ʹ. miR-590-5p: F 5ʹ-GAGCTTATTCATAAAAGT-3ʹ, R 5ʹ-TCCACGACACGCACTGGATACGAC-3ʹ; miR-330-5p: F 5ʹ- TCTCTGGGCCTGTGTC-3ʹ, R 5ʹ-CCAGTTTTTTTTTTTTTTTGCCTAAG-3ʹ; miR-767-5p: F 5ʹ-GCCGAGTGCACCATGGTTGTCTG-3ʹ, R 5ʹ-CTCAACTGGTGTCGTGGA-3ʹ.
Cell transfection
The circPUM1-specific siRNA (si-circPUM1-1, si-circPUM1-2) and negative control siRNA (si-NC), pcDNA3.1 vector encoding METTL3 (pcDNA-METTL3) and plasmids containing scrambled sequences (pcDNA), the mimic and inhibitor of miR-590-5p (miR-590-5p or anti-miR-590-5p) and negative controls (miR-NC or anti-NC) were synthesized by Invitrogen (San Diego, CA, USA). Lentiviral plasmids of a short hairpin RNA (shRNA) against circPUM1 (sh-circPUM1) or scrambled control (sh-NC) were procured from GeneCopoeia (Rockville, MD, USA). The transfection was carried out on H1650 and A549 cells using Lipofectamine 2000 (Invitrogen).
Cell proliferation analysis
The cell counting kit-8 (CCK-8) assay was employed to assess cell proliferation. In brief, transfected cells were seeded in 96-well plates. CCK-8 solution (10 μL) was added to each well, and incubated for another 2 h. Finally, the absorbance at 450 nm was tested.
For colony formation assay, transfected cells (1000/well) were plated into a 6-well plate with RPMI 1640 medium and allowed to form colonies for 2 weeks. Afterward, cells were fixed with 10% methanol and stained with 0.1% crystal violet, and the colonies were photographed and counted using a microscope.
Cell cycle assay
After transfection, H1650 and A549 cells were fixed in 75% ice-cold ethanol for 24 h, followed by staining with propidium iodide (PI) (BD Biosciences, San Jose, CA, USA) for 30 min, away from light. Finally, cell cycle was quantified using flow cytometry.
Western blot
The extraction of proteins from tissues and cells was conducted using RIPA lysis solution (Beyotime, Shanghai, China). Subsequently, approximately 30 μg extracted proteins were separated using a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) minigel. Then proteins were shifted to polyvinylidene difluoride (PVDF) membranes and blocked in 5% nonfat milk for 2 h at 37°C. Thereafter, membranes were interacted with primary antibody at 4°C overnight, and then incubated with HRP-conjugated secondary antibody (1:3000, Sangon, Shanghai, China). The primary antibodies obtained from Abcam (Cambridge, MA, USA) were listed below: CyclinD1 (1:10,000, ab134175), c-myc (1:2000, ab32072), glucose transporter 1 (GLUT1) (1:10,000, ab40084), hexokinase-2 (HK2) (1:5000, ab104836), METTL3 (1:1000, ab195352) and β-actin (1:5000, ab8226).
Glycolysis analysis
Transfected H1650 and A549 cells (5 × 103/well) were placed in a 6-well plate, then supernatants of cell culture media were subjected to the levels of glucose and lactate using a Glucose uptake colorimetric assay kit or Lactate Assay Kit II (Sigma) following the instructions of manufacturer. The relative glucose uptake and lactate production were analyzed ground on the standard curve and normalized by the percentage of the control group.
Measurement of extracellular acidification rate (ECAR)
The extracellular acidification rate (ECAR) was measured by using Seahorse XF Glycolysis Stress Test Kit (Seahorse Bioscience, Agilent, California, USA). Following transfection, H1650 and A549 cells were placed in a Seahorse XF 96 cell culture microplate without glucose. After baseline detection, saturating amounts of glucose, oligomycin, and the glucose analog 2-deoxyglucose (2-DG) were sequentially added into per well at indicated time points. Finally, Seahorse XF-96 Wave software was employed to assess the data.
RNA pull-down assay
Biotinylated-circPUM1 probe (circPUM1 probe) and biotinylated-oligonucleotide probe (Oligo probe) were synthesized by GenePharma Company (Shanghai, China). Approximately 1 × 107 cells were lysed by lysis buffer and incubated with 3 μg biotinylated-probes for 2 h at 37°C. Afterward, the lysate was incubated with streptavidin magnetic beads (Thermo Fisher Scientific, Inc., Waltham, MA, USA) to pull down the biotin-coupled RNA complex. Finally, the RNA complexes bound to the beads were eluted and isolated and subjected to qRT-PCR analysis.
RNA immunoprecipitation (RIP) assay
H1650 and A549 cells were homogenized by RIP lysis buffer, then cell lysate was co-immunoprecipitated with Ago2 antibody and negative control IgG antibody conjugated magnetic beads (Millipore, Billerica, MA, USA). Following proteinase K buffer incubation, the immunoprecipitated RNA was isolated and purified RNA was analyzed using qRT-PCR assay.
Dual-luciferase reporter assay
The specific sequences of circPUM1 and METTL3 3ʹ-UTR harboring the complementary site of miR-590-5p were cloned into the psiCHECK-2 vector (Promega, Madison, WI, USA) to generate the corresponding wild-type (WT) or the mutated (MUT) luciferase reporter constructs, named WT-circPUM1, MUT-circPUM1, WT-METTL3 3ʹ-UTR or MUT-METTL3 3ʹ-UTR. Then miR-590-5p mimic or miR-NC with these constructs were introduced into H1650 and A549 cells. After 48 h, relative luciferase activity was evaluated by Dual-Luciferase Reporter Analysis system (Promega)
Xenograft experiments in vivo
A549 cells (2 × 106 cells/mouse) stably transfected with sh-NC or sh-circPUM1 were subcutaneously injected into the right flank of BALB/C nude mice (4–6 weeks old, N = 10) to establish murine xenograft models. This animal experiment was manipulated according to the protocols approved by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University. Tumor volume was detected every 3 days. At day 35, the mice were sacrificed by cervical dislocation after deep anesthesia with 2% isoflurane, and tumors were removed, weighed and collected for subsequent molecule analyses. The study was manipulated according to the protocols approved by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University.
Statistical analysis
Data from thrice-repeated experiments were expressed as mean ± standard deviation (SD). The comparison between two groups was performed with Student’s t-test, and one-way analysis of variance (ANOVA) was used to compare the data between more than two groups. Kaplan-Meier analysis and the log-rank test were utilized to calculate survival curves. Pearson’s correlation coefficient assay was used to analyze the expression correlation. Differences were considered significant at P < 0.05.
Results
The expression profile of circPUM1 in NSCLC
Using qRT-PCR, the expression levels of circPUM1 in clinical tissues (NSCLC tissues and para-carcinoma tissues) were detected. By contrast with normal tissues, circPUM1 expression was significantly elevated in NSCLC tissues (), among them, 75.81% was up-regulated while 24.19% was down-regulated in cancer tissue specimens based on the median level (). Besides that, further analysis suggested that higher circPUM1 expression was more commonly observed in specimens with advanced tumor-node-metastasis (TNM) stages (). Afterward, patients were grouped as follows: a high circPUM1 expression group (N = 30) and a low circPUM1 expression group (N = 32) according to the median level, and kaplan-Meier analysis exhibited patients with high circPUM1 expression showed a shorter overall survival than that with low circPUM1 expression (). Moreover, circPUM1 expression in two NSCLC cell lines (H1650 and A549) was markedly up-regulated relative to expression in BEAS-2B cells, a non-cancerous lung cell line (). In addition, the endogenous of circPUM1 resistant to efficient RNase R digestion was also detected in H1650 and A549 cells (). Meanwhile, subcellular fractionation assay suggested that circPUM1 was preferentially localized at cytoplasm in H1650 and A549 cells (). These results indicated circPUM1 was a stable circRNA and might play a pivotal role in NSCLC progression.
Figure 1. The expression profile of circPUM1 in NSCLC. (a, b) qRT-PCR analysis of circPUM1 expression in NSCLC tissues and para-carcinoma tissues. (c) qRT-PCR analysis of circPUM1 expression in NSCLC tissues according to TNM staging. (d) Kaplan-Meier overall survival curves for 62 patients with NSCLC classified according to relative circPUM1 expression level. (e) qRT-PCR analysis of circPUM1 expression in two NSCLC cell lines (H1650 and A549) and non-cancerous lung cell line BEAS-2B. (f, g) qRT-PCR analysis of circPUM1 expression in H1650 and A549 cells treated with or without RNsae R. (h, i) qRT-PCR indicating the distribution of circPUM1, GAPDH, and U6 in the cytoplasmic and nuclear fractions of H1650 and A549 cells. *P < 0.05
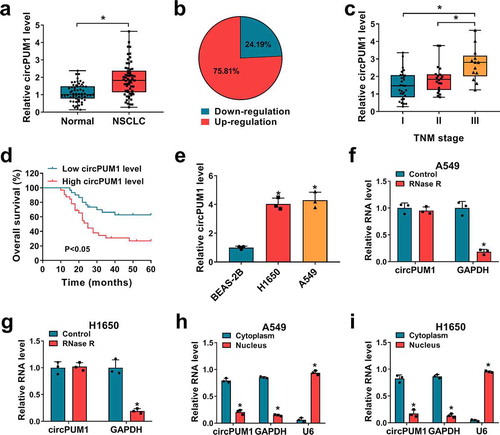
CircPUM1 knockdown suppresses cell growth and glycolysis in NSCLC
To explore the involvement of circPUM1 in NSCLC progression, two specific siRNAs targeting circPUM1 (si-circPUM1-1 and si-circPUM1-2) were designed (). The qRT-PCR analysis showed the expression of circPUM1 was markedly decreased in A549 and H1650 cells after transfection with si-circPUM1-1 or si-circPUM1-2 (). After that, a CCK-8 assay indicated that circPUM1 knockdown significantly suppressed cell proliferation in A549 and H1650 cells (); meanwhile, colony formation assay demonstrated that knockdown of circPUM1 significantly inhibited the colony-forming ability of A549 and H1650 cells (). Furthermore, flow cytometric analysis revealed that cell cycle was arrested when A549 and H1650 cells were transfected with si-circPUM1 ( and ). Thereafter, western blot analysis suggested the expression levels of CyclinD1 and c-myc were markedly reduced in circPUM1-decreased A549 and H1650 cells (). Cumulatively, circPUM1 knockdown suppressed cell growth in NSCLC.
Figure 2. CircPUM1 knockdown suppresses cell growth in NSCLC. (a) Two specific siRNAs targeting circPUM1 were designed. A549 and H1650 cells were transfected with si-circPUM1-1, si-circPUM1-2 or si-NC. (b) qRT-PCR analysis of circPUM1 expression in cells. (c) CCK-8 assay of cell growth rate at different time points of 0, 24, 48, and 72 h. (d) Colony formation assay of cell proliferation. (e, f) Flow cytometry assay of cell cycle. (g, h) Western blot analysis of CyclinD1 and c-myc protein levels in cells. *P < 0.05
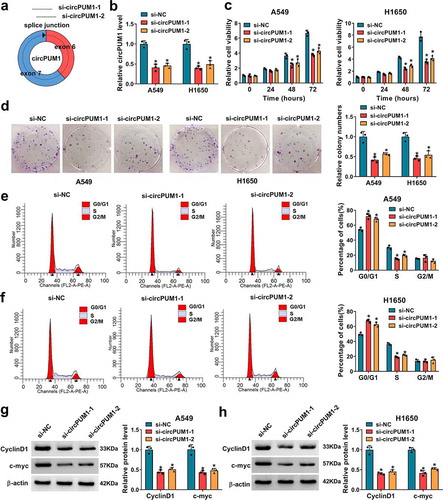
To further investigate the effect of circPUM1 on cell glycolysis in NSCLC, we firstly tested the levels of glucose and lactate in cells. Results showed circPUM1 down-regulation reduced glucose uptake () and lactate production () in A549 and H1650 cells. In addition, a glycolysis stress test suggested that down-regulation of circPUM1 decreased the level of glycolysis, glycolytic capacity and glycolytic reserve in A549 and H1650 cells with the treatment of glucose, oligomycin or 2-DG (), revealing knockdown of circPUM1 exhibited decreased ECAR. Moreover, it was also proved circPUM1 silencing suppressed the expression levels of GLUT1 and HK2 in A549 and H1650 cells ( and e). Taken together, knockdown of circPUM1 dampened glycolysis in NSCLC cells.
Figure 3. CircPUM1 knockdown suppresses cell glycolysis in NSCLC. (a, b) Measurements of glucose uptake and lactate production in A549 and H1650 cells transfected with si-circPUM1 or si-NC using commercial kits. (c) The glycolysis stress test of ECAR in A549 and H1650 cells transfected with si-circPUM1 or si-NC. (d, e) Western blot analysis of GLUT1 and HK2 protein levels in si-circPUM1 or si-NC transfected A549 and H1650 cells. *P < 0.05
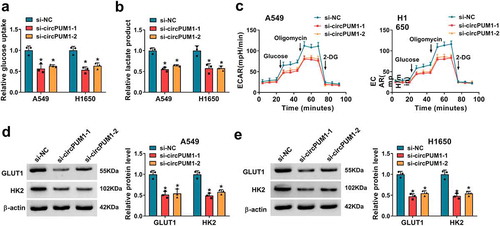
CircPUM1 sequesters miR-590-5p in NSCLC cells
Accumulating evidence has showed that circRNAs can function as miRNA sponges and subsequently abolish the function of the corresponding miRNA [Citation25,Citation26]. We next studied whether circPUM1 regulated the biological behaviors of NSCLC cells through sponging miRNAs. Two databases (circBank and circinteractom) were employed to predict the potential target miRNAs, among which 3 miRNAs were selected from the overlap between the databases (). Subsequently, RNA pull-down assay with a biotinylated circPUM1 probe was conducted to investigate whether circPUM1 could directly bind these candidate miRNAs. Results showed a significantly enhanced fold-change of miR-590-5p for circPUM1 capture in A549 and H1650 cells (). Subsequent RIP assay suggested circPUM1 and miR-590-5p could be enriched by Ago2 antibody relative to IgG antibody in A549 and H1650 cells (). Thus, miR-590-5p was selected and the complementary sites existed between miR-590-5p and circPUM1 were displayed in . Afterward, the dual-luciferase reporter assay was performed and we found the luciferase activity was significantly reduced in A549 and H1650 cells co-transfected with WT-circPUM1 and miR-590-5p mimic () (the transfection efficiency of miR-590-5p was shown in Figure S1a), suggesting the direct interaction between circPUM1 and miR-590-5p. Besides that, it was proved that miR-590-5p expression was up-regulated in circPUM1-decreased A549 and H1650 cells (). Moreover, miR-590-5p was lowly expressed in NSCLC tissues and cell lines (), and a negative correlation between circPUM1 and miR-590-5p expression in NSCLC tissues was observed (). Thus, we confirmed that circPUM1 sequesters miR-590-5p and negatively regulated its expression in NSCLC cells, and miR-590-5p decrease might be associated with NSCLC progression.
Figure 4. CircPUM1 sequesters miR-590-5p in NSCLC cells. (a, b) A schematic drawing showing the overlap of the target miRNAs of circPUM1 predicted by circBank and circinteractom. (c) The relative levels analysis of 3 miRNA candidates in A549 and H1650 cell lysates using qRT-PCR after pull-down assay. (d) RIP assay for circPUM1 and miR-590-5p levels in A549 and H1650 cells transfected with Ago2 or IgG. (e) Schematic illustration showing the binding sites between circPUM1 and miR-590-5p. (f) Luciferase activity detection in A549 and H1650 cells co-transfected with the reporter plasmid and the indicated miRNAs using the dual-luciferase reporter assay. (g) qRT-PCR analysis of miR-590-5p expression in A549 and H1650 cells transfected with si-circPUM1-1 or si-NC. (h, i) qRT-PCR analysis of miR-590-5p expression in NSCLC tissues and para-carcinoma tissues, as well as in NSCLC cell lines (H1650 and A549) and non-cancerous lung cell line BEAS-2B. (j) Pearson’s correlation coefficient assay for the correlation between miR-590-5p and circPUM1 expression in NSCLC tissues. *P < 0.05
Silencing miR-590-5p reverses the inhibitory effects of si-circPUM1 on NSCLC cell growth and glycolysis
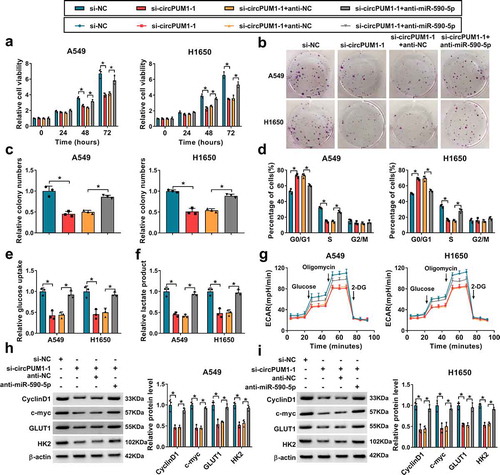
To investigate whether the effects of circPUM1 on NSCLC were mediated by miR-590-5p, first, A549 and H1650 cells were transfected with anti-miR-590-5p or anti-NC, and qRT-PCR analysis showed miR-590-5p expression was significantly decreased in anti-miR-590-5p-transfected cells (Figure S1b). Next, the anti-miR-590-5p was transfected into si-circPUM1-1-expressing A549 and H1650 cells to conduct rescue assay. Results showed the inhibitory effects of si-circPUM1 on A549 and H1650 cell proliferation (–), cell cycle (), glucose uptake (), lactate production (), ECAR () were reversed by the miR-590-5p inhibition. Western blot analysis also indicated the reduction of CyclinD1, c-myc, GLUT1 and HK2 expression induced by circPUM1 knockdown also was attenuated by the introduction of miR-590-5p inhibitor ( and ). Therefore, we demonstrated that miR-590-5p antagonized circPUM1-regulated NSCLC progression in vitro.
Figure 5. Silencing miR-590-5p reverses the inhibitory effects of si-circPUM1 on NSCLC cell growth and glycolysis. The anti-miR-590-5p or anti-NC was transfected into si-circPUM1-1-expressing A549 and H1650 cells. (a-c) CCK-8 assay and colony formation assay of cell proliferation. (d) Flow cytometry assay of cell cycle. (e, f) Analyses of glucose uptake and lactate production in cells using commercial kits. (g) The glycolysis stress test of ECAR in cells. (h, i) Western blot analysis of CyclinD1, c-myc, GLUT1 and HK2 protein levels in cells. *P < 0.05
MiR-590-5p directly targets METTL3 in NSCLC cells
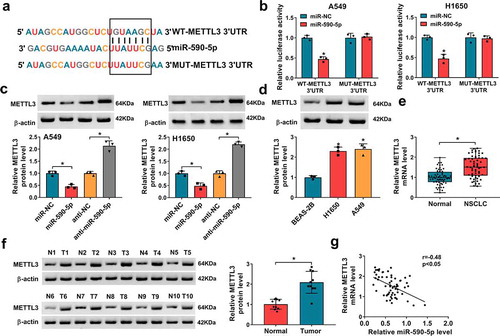
Using the Targetscan and miRanda online databases, it was discovered that miR-590-5p might target METTL3 due to the complementary DNA regions between them (). Further dual-luciferase reporter assay showed miR-590-5p mimic significantly reduced the luciferase activity of the WT-METTL3 3ʹUTR in A549 and H1650 cells, whereas there was no effect on cells transfected with a MUT-METTL3 3ʹ-UTR (). Moreover, transfection with miR-590-5p mimic markedly repressed the expression of METTL3, while miR-590-5p inhibitor markedly enhanced the METTL3 expression in A549 and H1650 cells (). In addition, the expression pattern of METTL3 in NSCLC was detected, METTL3 was increased in NSCLC cell lines () and tissues ( and ), which was negatively correlated with miR-590-5p expression (). Altogether, these data revealed that miR-590-5p directly targeted METTL3 and inversely modulated its expression, and METTL3 increase might be related to NSCLC tumorigenesis.
Figure 6. MiR-590-5p directly targets METTL3 in NSCLC cells. (a) Schematic illustration showing the binding sites between METTL3 and miR-590-5p. (b) Dual-luciferase reporter assay of luciferase activity analysis in A549 and H1650 cells co-transfected with the reporter plasmid and the indicated miRNAs. (c) Western blot analysis of METTL3 in A549 and H1650 cells transfected with miR-NC, miR-590-5p, anti-NC, or anti-miR-590-5p. (d-f) Western blot and qRT-PCR analysis of METTL3 expression in NSCLC tissues and para-carcinoma tissues, as well as in NSCLC cell lines (H1650 and A549) and non-cancerous lung cell line BEAS-2B. (g) Pearson’s correlation coefficient assay for the correlation between miR-590-5p and METTL3 expression in NSCLC tissues. *P < 0.05
MiR-590-5p inhibits cell growth and glycolysis in NSCLC via targeting METTL3
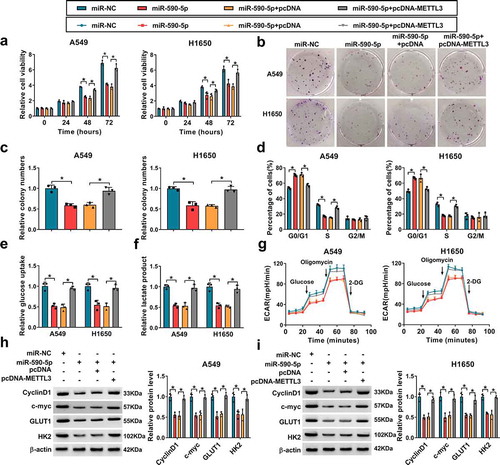
Based on the aforementioned results, miR-590-5p directly targeted METTL3, we hypothesized that METTL3 might function as an important in miR-590-5p-mediated development and progression of NSCLC. qRT-PCR analysis indicated that the expression of METTL3 was remarkably increased after pcDNA-METTL3 (Figure S1c). Then METTL3 was overexpressed in miR-590-5p-transfected A549 and H1650 cells, and rescue assay was performed. It was found miR-590-5p overexpression suppressed A549 and H1650 cell proliferation (–), which was reversed by METTL3 up-regulation. Results of flow cytometry showed the overexpression of METTL3 promoted cell cycle in miR-590-5p-transfected A549 and H1650 cells (). Additionally, miR-590-5p re-expression also suppressed glucose uptake () and lactate production (), as well as reduced ECAR (), while these conditions were abated by METTL3 up-regulation. Moreover, METTL3 overexpression also attenuated miR-590-5p restoration-evoked decrease of CyclinD1, c-myc, GLUT1 and HK2 expression in A549 and H1650 cells ( and ). Collectively, these results verified miR-590-5p inhibited cell growth and glycolysis by regulating METTL3 in NSCLC.
Figure 7. MiR-590-5p inhibits cell growth and glycolysis in NSCLC via targeting METTL3. A549 and H1650 cells were transfected with miR-NC, miR-590-5p, miR-590-5p + pcDNA, or miR-590-5p + pcDNA-METTL3. (a-c) Cell proliferation analysis using CCK-8 and colony formation assays. (d) Flow cytometry assay of cell cycle. (e, f) Glucose uptake and lactate production detection in cells using commercial kits. (g) The glycolysis stress test of ECAR in cells. (h, i) Western blot analysis of CyclinD1, c-myc, GLUT1 and HK2 protein levels in cells. *P < 0.05
CircPUM1 can indirectly regulate METTL3 via miR-590-5p in NSCLC cells
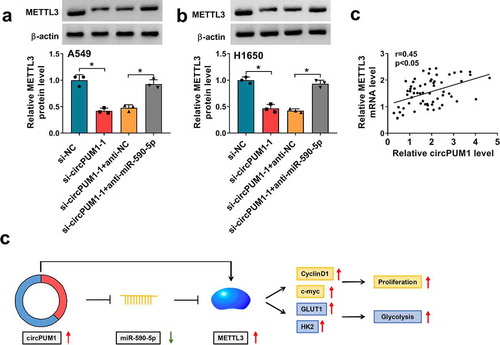
Given the data above, we then explored whether specific crosstalk existed between circPUM1 and METTL3 through competition for miR-590-5p binding. As presented of western blot analysis, knockdown of circPUM1 reduced the expression level of METTL3 in A549 and H1650 cells, while this reduction was rescued by the decrease of miR-590-5p ( and ). Moreover, METTL3 expression level was positively correlated with METTL3 in NSCLC tissues (), further suggesting that circPUM1 could indirectly regulate METTL3 via miR-590-5p. All these results revealed circPUM1 promoted NSCLC cell growth and glycolysis through miR-590-5p/METTL3 axis in vitro ().
Figure 8. CircPUM1 can indirectly regulate METTL3 via miR-590-5p in NSCLC cells. (a, b) Western blot analysis of METTL3 in A549 and H1650 cells transfected with si-NC, si-circPUM1-1, si-circPUM1-1 + anti-NC, or si-circPUM1-1 + anti-miR-590-5p. (c) Pearson’s correlation coefficient assay for the correlation between circPUM1 and METTL3 expression in NSCLC tissues. (d) The schematic diagram of circPUM1/miR-590-5p/METTL3 axis in NSCLC cell growth and glycolysis in vitro. *P < 0.05
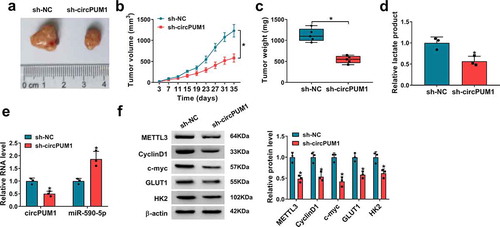
CircPUM1 silencing hinders tumor growth and glycolysis in vivo
Next, we investigated the effects of circPUM1 on tumor growth and glycolysis in vivo. The murine xenograft models were established via injecting A549 cells (2 × 106 cells/mouse) containing sh-NC or sh-circPUM1 into the right flank of BALB/C nude mice. The transfection efficiency of sh-circPUM1 was presented in Figure S1d. Afterward, we found down-regulation of circPUM1 decreased tumor volume and weight than control group (–), and lactate production was lower in sh-circPUM1 group than sh-NC group (). Moreover, qRT-PCR and western blot analyses indicated circPUM1 and METTL3 expression were decreased, while miR-590-5p expression was increased in tumors from sh-circPUM1 groups compared with control groups, and the expression levels of CyclinD1, c-myc, GLUT1 and HK2 were reduced by circPUM1 silencing in xenograft tumors ( and ). Therefore, we validated knockdown of circPUM1 could suppress tumor growth and glycolysis in vivo via miR-590-5p/METTL3 axis.
Figure 9. CircPUM1 silencing hinders tumor growth and glycolysis in vivo. (a-c) CircPUM1 knockdown impeded xenograft growth in nude mice. (d) Lactate production analysis in tumors with the Lactate Assay Kit II. (e, f) qRT-PCR and western blot analyses of circPUM1, miR-590-5p, METTL3, CyclinD1, c-myc, GLUT1 and HK2 expression in excised tumors. *P < 0.05
Discussion
As the most common cause of cancer-related death in the world, there were approximately 1,378,400 deaths due to lung cancer annually, which exceed those of prostate, colorectal and breast cancer combined [Citation1,Citation27]. NSCLC is the mainstay of lung cancer, complete resection, when possible, is still the first choice for NSCLC treatment currently; however, the survival rate of advanced NSCLC remains unsatisfactory [Citation4]. Therefore, investigating new biological targets or molecular mechanisms that enhance the efficacy of NSCLC treatments is necessary.
Previous evidence has reported that aberrant expression of circRNAs is implicated in NSCLC tumorigenesis. For instance, circPTK2 acted as a tumor suppressor to impair cell EMT and metastasis by blocking TGF-β through regulating TIF1γ [Citation28]. CircPVT1 showed high expression in NSCLC, and promoted cell malignant phenotypes via up-regulating E2F2 expression in a miR-125b-dependent manner [Citation29]. In this study, circPUM1 was found to show high expression profile in NSCLC tissues, especially in advanced TNM stages, and highly expressed circPUM1 was associated with poor prognosis of patients with NSCLC. Besides, circPUM1 expression was also higher in NSCLC cells, further in vitro assays exhibited knockdown of circPUM1 impaired cell proliferation, cell cycle and decreased CyclinD1 and c-myc expression, thus suppressing cell growth. Glycolysis is recognized as an important resistance factor for anticancer therapy. Aerobic glycolysis, also known as Warburg effect, is a phenomenon whereby many types of cancers showed a preference of glucose over the oxygen for their energy substrate, thus meeting their metabolic needs for sustained cell proliferation [Citation30,Citation31]. Besides that, increasing evidence has shown that aerobic glycolysis is one of the metabolic hallmark of malignant tumor cells, and foster tumor malignancy [Citation32]. Recently, several studies have revealed that circRNAs, such as circSLC25A16 [Citation33], hsa_circ_0002130 [Citation34] and circ-ACACA [Citation35], intimately promoted glycolysis rate through the modulation of glycolysis enzymes, including GLUT1, HK2 and LDHA, thus supporting cell growth and survival in NSCLC. Targeting cancer cell glycolytic metabolism may be a promising strategy for developing therapeutic interventions. In this work, we found a reduction of glucose uptake and lactate production in circPUM1 down-regulated cells, besides, it was also proved that down-regulation of circPUM1 reduced ECAR and decreased the expression levels of GLUT1 and HK2 in vitro. GLUT1 facilitates the transport of glucose across the plasma membranes of mammalian cells. HK2 is a glycolytic enzyme, hexokinases phosphorylate glucose to produce glucose-6-phosphate (G6P), the first step in most glucose metabolism pathways. ECAR is primarily a measure of lactate production and can be equated to the glycolytic rate. Therefore, circPUM1 silencing might inhibit glycolysis by suppressing GLUT1 and HK2. Importantly, xenograft assays exhibited that circPUM1 silencing hindered NSCLC tumor growth and glycolysis in vivo. CircPUM1 is associated with the progression of NSCLC.
Increasing studies have demonstrated the involvement of circRNAs in competitive regulatory interactions, known as ceRNA networks, whereby circRNAs can function as miRNA sponges, abolishing their inhibitory effects on the expression of target mRNAs [Citation25,Citation26]. For example, circRNA_100367 targeted miR-217 to relieve the suppression of Wnt3 expression to promote the tiumorigenesis and radioresistance in esophageal squamous cell carcinomas [Citation36]. CircRNA cRAPGEF5 was demonstrated to suppress the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway [Citation37]. Thus, we suspected circPUM1-mediated regulatory functions might operate through a ceRNA mechanism. Subsequently, circPUM1 competitively bound with miR-590-5p to up-regulate the expression of its target METTL3. In present study, miR-590-5p expression also was decreased, which was consistent with previous studies; besides that miR-590-5p re-expression suppressed cell growth and glycolysis. Moreover, we also proved silencing of miR-590-5p reversed the inhibitory effects of circPUM1 knockdown on NSCLC tumorigenesis. Previous studies showed that METTL3 expression was increased in NSCLC, and acted as an oncogene to promote cell growth, metastasis and drug resistance [Citation24,Citation38,Citation39]. Importantly, several studies have suggested that contributed to glycolysis and ATP generation in cancer by stabilizing HK2 and GLUT1 expression [Citation40], activating mTOR signal pathway [Citation41], or enhancing the expression of cell metabolism factor PDK4 [Citation42]. In the present study, we verified that miR-590-5p directly targeted METTL3. Furthermore, rescue assay indicated miR-590-5p suppressed NSCLC progression via directly targeting and regulating METTL3.
In summary, it was demonstrated that circPUM1 facilitated the growth and glycolysis in NSCLC cells via targeting miR-590-5p/METTL3 axis, suggesting a potential therapeutic target for NSCLC treatment.
Disclosure
No potential conflict of interest was reported by the author(s).
Supplemental Material
Download Zip (246.5 KB)Acknowledgments
None.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132.
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30.
- Rocco D, Della Gravara L, Battiloro C, et al. The role of combination chemo-immunotherapy in advanced non-small cell lung cancer. Expert Rev Anticancer Ther. 2019;19:561–568.
- Schuchert MJ, Normolle DP, Awais O, et al. Factors influencing recurrence following anatomic lung resection for clinical stage I non-small cell lung cancer. Lung Cancer. 2019;128:145–151.
- Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. 2017;17:637–658.
- Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148.
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338.
- Patop IL, Kadener S. circRNAs in Cancer. Curr Opin Genet Dev. 2018;48:121–127.
- Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol. 2019;12:90.
- Yang Z, Jiang S, Shang J, et al. LncRNA: shedding light on mechanisms and opportunities in fibrosis and aging. Ageing Res Rev. 2019;52:17–31.
- Wang T, Wang X, Du Q, et al. The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun. 2019;513:904–911.
- Wang J, Li H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci. 2018;22:3053–3060.
- Chen J, Xu S, Chen S, et al. CircPUM1 promotes the malignant behavior of lung adenocarcinoma by regulating miR-326. Biochem Biophys Res Commun. 2019;508:844–849.
- Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500.
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269.
- Sui J. MiRNA-30 play an important role in Non-Small Cell Lung Cancer (NSCLC). Clin Lab. 2020;66. DOI:10.7754/Clin.Lab.2019.190712
- Bica-Pop C, Cojocneanu-Petric R, Magdo L, et al. Overview upon miR-21 in lung cancer: focus on NSCLC. Cell Mol Life Sci. 2018;75:3539–3551.
- Xu BB, Gu ZF, Ma M, et al. MicroRNA-590-5p suppresses the proliferation and invasion of non-small cell lung cancer by regulating GAB1. Eur Rev Med Pharmacol Sci. 2018;22:5954–5963.
- Chang Z. Downregulation of SOX2 may be targeted by miR-590-5p and inhibits epithelial-to-mesenchymal transition in non-small-cell lung cancer. Exp Ther Med. 2019;18:1189–1195.
- Khandelwal A, Seam RK, Gupta M, et al. Circulating microRNA-590-5p functions as a liquid biopsy marker in non-small cell lung cancer. Cancer Sci. 2020;111:826–839.
- Bokar JA, Shambaugh ME, Polayes D, et al. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247.
- Lin S, Choe J, Du P, et al. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–345.
- Du M, Zhang Y, Mao Y, et al. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun. 2017;482:582–589.
- Du WW, Zhang C, Yang W, et al. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7:4183–4191.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388.
- Siegel R, Ward E, Brawley O, et al. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236.
- Yu S, Qi Y, Jiang J, et al. APTR is a prognostic marker in cirrhotic patients with portal hypertension during TIPS procedure. Gene. 2018;645:30–33.
- Li X, Zhang Z, Jiang H, et al. Circular RNA circPVT1 promotes proliferation and invasion through sponging miR-125b and activating E2F2 signaling in non-small cell lung cancer. Cell Physiol Biochem. 2018;51:2324–2340.
- Ruiz-Perez MV, Sanchez-Jimenez F, Alonso FJ, et al. Glutamine, glucose and other fuels for cancer. Curr Pharm Des. 2014;20:2557–2579.
- Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482.
- Enzo E, Santinon G, Pocaterra A, et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. Embo J. 2015;34:1349–1370.
- Shangguan H, Feng H, Lv D, et al. Circular RNA circSLC25A16 contributes to the glycolysis of non-small-cell lung cancer through epigenetic modification. Cell Death Dis. 2020;11:437.
- Ma J, Qi G, Li L. A novel serum exosomes-based biomarker hsa_circ_0002130 facilitates osimertinib-resistance in non-small cell lung cancer by sponging miR-498. Onco Targets Ther. 2020;13:5293–5307.
- Wu W, Xi W, Li H, et al. Circular RNA circ‑ACACA regulates proliferation, migration and glycolysis in non‑small‑cell lung carcinoma via miR‑1183 and PI3K/PKB pathway. Int J Mol Med. 2020;45:1814–1824.
- Liu J, Xue N, Guo Y, et al. CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging (Albany NY). 2019;11:12412–12427.
- Chen Q, Liu T, Bao Y, et al. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway. Cancer Lett. 2020;469:68–77.
- Jin D, Guo J, Wu Y, et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12:135.
- Wei W, Huo B, Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag Res. 2019;11:1177–1187.
- Shen C, Xuan B, Yan T, et al. m(6)A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19:72.
- Lin Y, Wei X, Jian Z, et al. METTL3 expression is associated with glycolysis metabolism and sensitivity to glycolytic stress in hepatocellular carcinoma. Cancer Med. 2020;9:2859–2867.
- Li Z, Peng Y, Li J, et al. N(6)-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat Commun. 2020;11:2578.
