ABSTRACT
A central player in meiotic chromosome dynamics is the conserved Polo-like kinase (PLK) family. PLKs are dynamically localized to distinct structures during meiotic prophase and phosphorylate a diverse group of substrates to control homolog pairing, synapsis, and meiotic recombination. In a recent study, we uncovered the mechanisms that control the targeting of a meiosis-specific PLK-2 in C. elegans. In early meiotic prophase, PLK-2 localizes to special chromosome regions known as pairing centers and drives homolog pairing and synapsis. PLK-2 then relocates to the synaptonemal complex (SC) after crossover designation and mediates chromosome remodeling required for homolog separation. What controls this intricate targeting of PLK-2 in space and time? We discuss recent findings and remaining questions for the future.
Meiosis proceeds through stepwise chromosome rearrangement
Sexually reproducing organisms rely on the correct execution of meiosis, a specialized cell division that produces haploid gametes such as sperm and eggs. The reduction in chromosome number during meiosis is achieved by a single round of DNA replication, followed by two consecutive nuclear divisions. Unique to meiosis is the protracted prophase, in which chromosomes pair and synapse with their homologous partners through the formation of a zipper-like protein scaffold called the synaptonemal complex (SC) [Citation1,Citation2]. Concomitant with pairing and synapsis, meiotic recombination initiates with induction of programmed DNA double-strand breaks (DSBs) [Citation3]. A subset of these breaks is eventually processed to form crossovers [Citation4], which physically link each pair of homologs and enable their proper separation during meiosis I. This process also allows for new combinations of alleles, thereby promoting genetic variation that underlies eukaryotic evolution.
Polo-like kinases coordinate meiotic chromosome dynamics
During meiosis, as in mitosis, PLKs regulate many aspects of cell cycle events that are essential for successful chromosome segregation. The sole PLK in budding yeast, Cdc5, is required for crossover formation and SC disassembly, thereby permitting cells to exit pachytene [Citation5,Citation6]. The Polo kinase in fission yeast, Plo1, is excluded from the spindle pole body (SPB) during meiotic prophase [Citation7,Citation8], although the significance of SPB remodeling on telomere-led “horsetail” movement and homolog pairing remains to be determined. In Drosophila and mouse oocytes, prolonged meiotic arrest requires the inhibition of PLK1 [Citation9,Citation10], consistent with its conserved role in prophase exit [Citation11,Citation12]. While the Drosophila Polo is not detected in oocytes until the very late stage of meiotic prophase [Citation10], mammalian PLK1 localizes to the SC in pachytene and promotes its disassembly [Citation13]. During meiosis I, PLKs are recruited to kinetochores through meiosis-specific adaptors (mouse Meikin, budding yeast Spo13, and fission yeast Moa1) and play critical roles in mono-orientation of sister kinetochores and sister chromatid cohesion [Citation5,Citation14–17]. Interestingly, the catalytically active form of Drosophila Polo is sequestered away from chromosomes during female meiosis I by a meiosis-specific protein, Matrimony, and inhibiting the Polo activity at centromeres is essential for protecting cohesion during meiosis I [Citation10,Citation18]. Thus, although the details may vary depending on the organism, PLKs play universally important roles in coordinating the complex behavior of meiotic chromosomes.
PLK-2 is a meiosis-specific paralog of PLK-1 in C. elegans
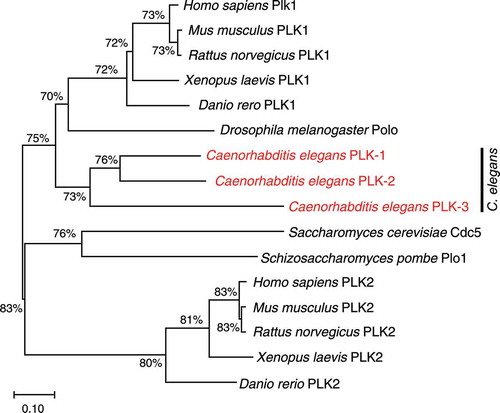
Perhaps the most pervasive role for PLK has been demonstrated during C. elegans meiosis, in which meiosis-specific PLK-2 regulates homolog pairing, synapsis, programmed DSB formation, and chromosome remodeling that defines the pattern of cohesin release during meiotic divisions [Citation19–23]. PLK-2 is one of three PLK orthologs in C. elegans that are closely related to the mammalian PLK1 [Citation24]. In a phylogenetic analysis, PLK-1, PLK-2, and PLK-3 are grouped together with PLK1 orthologs, rather than members of the vertebrate PLK2 subfamily (), suggesting that PLK-2 and PLK-3 arose from gene duplication events in nematodes. While the function of PLK-3 is unknown, PLK-1 and PLK-2 play redundant roles in establishing the cell polarity in embryos [Citation25]. PLK-1 can also localize to pairing centers and partially substitute for PLK-2’s function to promote homolog pairing and synapsis [Citation19,Citation20], corroborating that PLK-2 is a meiosis-specific paralog of PLK-1.
Figure 1. Phylogenetic tree of members of the Polo-like kinase family
Two PLK-2 docking sites are generated by distinct priming kinases
To ensure that PLK-2 coordinates distinct meiotic events at the right space and time, multiple layers of control are in place to direct PLK-2 to specific subcellular locations during meiotic progression. Localization of PLKs is largely governed by their Polo-box domain (PBD), which binds a phosphorylated protein motif (S-[pS/T]-P/X) [Citation26,Citation27]. Thus, “priming” these motifs by phosphorylation provides an initial layer of control for targeting PLKs. In C. elegans, a meiosis-specific Ser/Thr kinase, CHK-2, primes the recruitment of PLK-2 to pairing centers by phosphorylating a family of zinc finger proteins (HIM-8, ZIM-1, ZIM-2, and ZIM-3) [Citation28] that bind specific DNA sequence motifs enriched at pairing centers [Citation29,Citation30]. Recent work has shown that another PBD-docking site is present within an SC component, SYP-1 [Citation31], which is phosphorylated by the cyclin-dependent kinase, CDK-1, just before meiotic onset [Citation32]. Thus, two major PBD-binding sites are generated in early meiotic prophase (), each primed by CDK-1 and CHK-2 that are essential for mitotic and meiotic cell cycle progression, respectively.
Figure 2. Multiple layers of control to regulate dynamic PLK-2 localization during meiotic prophase in C. elegans.
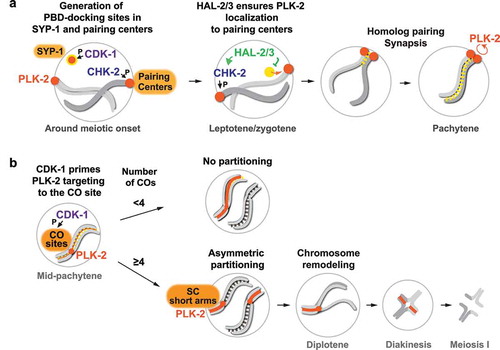
PLK-2 is preferentially targeted to pairing centers by nucleoplasmic regulation
Interestingly, of the two PLK-docking sites, PLK-2 is preferentially targeted to pairing centers in early meiotic prophase where it acts locally to drive homolog pairing and synapsis. The SC is capable of recruiting PLK-2 only when the four pairing center proteins (HIM-8, ZIM-1, ZIM-2, and ZIM-3) are absent at this stage of meiosis [Citation19,Citation32,Citation33]. How do pairing centers win over this competition and recruit PLK-2 at the onset of meiosis? An insight into this problem came from the identification of the nucleoplasmic protein complex consisting of HAL-2 and HAL-3, which functions to promote full activation of CHK-2 and to antagonize PLK-2 activity in the nucleoplasm [Citation33,Citation34]. Mutations in hal-2 or hal-3 result in the displacement of PLK-2 from the pairing center into the nucleoplasm, despite residual CHK-2 activity in the nucleus, and the unleashed PLK-2 activity in the nucleoplasm leads to inappropriate phosphorylation of PLK-2 substrates and defects in axis assembly, homolog pairing, and synapsis [Citation33,Citation34]. Remarkably, these phenotypes in hal-2 mutants can be rescued by mutating the PBD-binding motif on SYP-1 [Citation32], revealing that a key function of the HAL-2/3 complex is to prevent precocious association between PLK-2 and the SC in early prophase (). Thus, PLK-2 is sequestered to the pairing centers through an active mechanism that involves HAL-2/3, and this is critical for proper homolog pairing and synapsis.
This role of HAL-2/3 is analogous to the function of Matrimony in Drosophila female meiosis. Matrimony was initially discovered as a gene that is haplo-insufficient for achiasmate segregation in Drosophila oocytes [Citation35] and was later shown to be a negative regulator of Polo kinase [Citation10]. Matrimony directly binds Polo and limits its activity on chromosomes by sequestering the catalytically active Polo kinase to the microtubule spindle during meiosis I [Citation10,Citation18,Citation36]. Loss of Matrimony leads to precocious loss of sister chromatid cohesion, and this can be rescued by lowering Polo kinase activity [Citation10,Citation18]. Similarly, defects in synapsis and meiotic recombination in hal-3 mutants are partially rescued by the null mutation of plk-2 [Citation33], demonstrating that spatial regulation of PLK activity is essential for a successful outcome of meiosis in both Drosophila and C. elegans. HAL-2 and HAL-3 are not conserved outside of the Caenorhabditis species and share similar characteristics to a rapidly evolving group of meiosis-specific proteins that regulate key cell cycle kinases [Citation37]. However, unlike Matrimony, HAL-2 and HAL-3 do not possess PBD-docking motifs or domains with recognizable functions. Thus, it remains mysterious how the HAL-2/3 complex functions to regulate the interaction between PLK-2 and SYP-1. It is worth noting that, although HAL-2 and HAL-3 are largely found in the nucleoplasm during normal meiosis, they colocalize with SC aggregates known as “polycomplexes” in the absence of chromosome axes [Citation34]. Thus, the HAL-2/3 complex appears to have an affinity to the SC and is likely to act on the soluble pool of SYP proteins before their loading onto chromosomes. Future work will be necessary to determine how HAL-2 and HAL-3 regulate CHK-2 activity and PLK-2’s association with the SC.
Recruitment of PLK-2 to the SC requires crossover formation and a self-reinforcing mechanism
While the PBD-docking motifs within HIM-8/ZIMs are dephosphorylated as CHK-2 activity is weakened after chromosomes pair and synapse [Citation28], SYP-1 phosphorylation by CDK-1 persists along the length of chromosomes throughout pachytene [Citation31,Citation32]. However, PLK-2 is unable to relocate to the SC in late pachytene, unless crossover designation occurs [Citation32]. Thus, the priming phosphorylation of SYP-1 by CDK-1 is not sufficient for the spatiotemporal control to recruit PLK-2 to the SC. What prevents the stable association between PLK-2 and the SC prior to crossover formation is currently unknown. In our analysis, a kinase-dead mutant of PLK-2 precociously spreads to the SC in early pachytene [Citation32], indicating that PLK-2 activity is necessary for maintaining its tight association with pairing centers (). It is possible that the rapid, dynein-driven motion of pairing centers, sustained by the concentrated PLK-2 activity at the pairing center [Citation19,Citation20,Citation38–40], serves as a thermodynamic barrier that must be overcome before PLK-2 can access its binding sites on SYP-1. As the inactivation of CHK-2 and cessation of pairing center movement are coupled to SC assembly and crossover formation [Citation28,Citation41–43], this feedback mechanism may help license the PLK-2 recruitment to the SC upon crossover formation.
Then, how does the crossover event transmit signals across the length of chromosomes to recruit PLK-2 to the SC? Recent cytological analysis of recombination sites in C. elegans has revealed that PLK-2 colocalizes with pro-crossover proteins within the SC “bubble” [Citation44]. Consistent with this finding, our evidence suggests that PLK-2 is first recruited to crossover sites in a CDK-1-dependent manner before spreading along the SC () [Citation32]. It remains to be determined which proteins at the crossover site are phosphorylated by CDK-1 to generate docking sites for PLK-2. In budding yeast, Cdc5 is recruited to the crossover site through a direct binding to Exo1 and is important for MutLɣ-mediated crossover resolution [Citation45]. Thus, localization of PLK to crossover-designated recombination intermediates might be a conserved feature of crossover regulation, and potential functions for PLK-2 at the crossover site will be an important avenue for future research.
Earlier evidence has shown that crossover-designated sites transmit a signal to enrich for PLK-2 and stabilize the SC in cis [Citation21–23]. This change in the dynamic state of the SC has been shown to depend on PLK-2 [Citation21–23], which retains SYP-1 phosphorylation on chromosomes harboring crossovers using its own kinase activity [Citation32]. How PLK-2 reinforces its own docking sites and spreads from the crossover site to the SC is currently unclear. Although a self-priming and binding mechanism for PLK can promote its own localization [Citation27], PLK-2 was unable to phosphorylate the PBD-docking motif on SYP-1 in our in vitro kinase assays [Citation32]. Thus, we favor a scenario where PLK-2 activity indirectly supports the retention of phosphorylated SYP-1 by affecting the SC turnover and/or locally modulating CDK-1 activity.
PLK-2 localization is responsive to both nucleus-wide and chromosome-autonomous signaling
In holocentric C. elegans, which lacks defined centromeres [Citation46], a single crossover event partitions the chromosome into two distinct domains (long and short arms) and defines where cohesion will be released during meiosis I () [Citation47–53]. However, it remains poorly understood how the presence of crossovers is signaled to partition meiotic chromosomes. One of the earliest events that differentiate the long versus short arm is the recruitment of PLK-2 to the SC short arm, which mediates axis remodeling and asymmetric SC disassembly [Citation19,Citation21,Citation31]. Intriguingly, this asymmetric appearance of PLK-2 and SYP-1 phosphorylation depends on the number of crossover-designated sites within each nucleus [Citation32], suggesting that the symmetry breaking of bivalent chromosomes is controlled by nucleus-wide signaling. In nuclei with low numbers of crossovers (<4), no partitioning occurs, and PLK-2 localizes along the entire length of chromosomes that have designated crossovers, while it is lost from chromosomes without crossovers. However, in nuclei with higher number of crossovers (≥4), PLK-2 and SYP-1 phosphorylation exhibit the asymmetric localization on the SC short arm (). These findings have been further confirmed by analysis of the phosphorylation of a HORMA domain protein, HIM-3, whose asymmetric enrichment to the SC short arm also depends on the PBD-docking motif on SYP-1 and the number of crossovers in each nucleus [Citation54].
How the interplay between nucleus-wide and chromosome-autonomous signaling partitions the chromosome relative to the crossover site awaits further study. However, these results are consistent with a speculative model, in which crossover-designated sites nucleate and compete for limiting factors that stabilize the SC in a chromosome-autonomous manner. Sub-threshold levels of crossovers may recruit high levels of such factors, which spill over along the entire chromosome length. This may deplete the soluble pools of SYP proteins below the critical concentration required to maintain the SC on chromosomes without crossovers, leading to their desynapsis [Citation21,Citation23]. A more intense competition is expected as the number of crossovers rises, which will decrease the amount of SC-stabilizing factors recruited to each chromosome. Then, a self-reinforcing mechanism to amplify their effects may yield a faster equilibrium and help establish the short arm relative to the crossover site. The idea of inter-chromosomal competition during crossover formation was initially formulated by Mather in 1936 [Citation55], although it has been largely disfavored due to conflicting evidence. It is possible that this specialized mechanism has been adapted in holocentric nematodes to break the symmetry across the length of holocentric chromosomes and define their segregation pattern in meiosis I.
Concluding remarks
Recent studies have demonstrated how subcellular localization of PLK in meiotic prophase is controlled by elaborate mechanisms that involve not only priming kinases but also other meiotic regulators, a self-reinforcing feedback, and a nucleus-wide crossover-counting mechanism. However, many important questions remain to be addressed. How is the enzymatic activity of PLK-2 regulated during meiotic progression? Although PLK-2-dependent phosphorylation is detected only after the onset of the meiotic program [Citation19,Citation20,Citation56], the mechanism by which PLK-2 is activated remains elusive. Moreover, it is largely unknown how PLK-2 plays such multifaceted functions in orchestrating meiotic chromosome dynamics. Future identification of PLK substrates will illuminate how this conserved protein kinase family ensures accurate chromosome segregation during meiosis.
The authors declare no competing interests.
Acknowledgments
We thank members of the Yumi Kim laboratory for critical reading of this manuscript. We acknowledge support from the National Institutes of Health to YK (R35GM124895).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20(1):525–558.
- Bhalla N, Dernburg AF. Prelude to a division. Annu Rev Cell Dev Biol. 2008;24(1):397–424.
- de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47(1):563–599.
- Meiotic Recombination HN. The Essence of Heredity. Cold Spring Harb Perspect Biol. 2015;7:a016618.
- Clyne RK, Katis VL, Jessop L, et al. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol. 2003;5(5):480–485.
- Sourirajan A, Lichten M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 2008;22(19):2627–2632.
- Krapp A, Del Rosario EC, Simanis V. The role of Schizosaccharomyces pombe dma1 in spore formation during meiosis. J Cell Sci. 2010;123(19):3284–3293.
- Ohta M, Sato M, Yamamoto M. Spindle pole body components are reorganized during fission yeast meiosis. Mol Biol Cell. 2012;23(10):1799–1811.
- Shen W, Ahmad F, Hockman S, et al. Female infertility in PDE3A -/- mice. Cell Cycle. 2010;9(23):4720–4734.
- Xiang Y, Takeo S, Florens L, et al. The inhibition of polo kinase by matrimony maintains G2 arrest in the meiotic cell cycle. PLoS Biol. 2007;5(12):e323. .
- Zitouni S, Nabais C, Jana SC, et al. Polo-like kinases: structural variations lead to multiple functions. Nat Rev Mol Cell Biol. 2014;15(7):433–452.
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10(4):265–275.
- Jordan PW, Karppinen J, Handel MA. Polo-like kinase is required for synaptonemal complex disassembly and phosphorylation in mouse spermatocytes. J Cell Sci. 2012;125:5061–5072.
- Attner MA, Miller MP, Ee L-S, et al. Polo kinase Cdc5 is a central regulator of meiosis I. Proc Natl Acad Sci U S A. 2013;110(35):14278–14283.
- Galander S, Barton RE, Borek WE, et al. Reductional Meiosis I Chromosome Segregation Is Established by Coordination of Key Meiotic Kinases. Dev Cell. 2019;49(4):526–541. e5. .
- Miyazaki S, Kim J, Yamagishi Y, et al. Meikin-associated polo-like kinase specifies Bub1 distribution in meiosis I. Genes Cells. 2017;22(6):552–567.
- Kim J, Ishiguro K-I, Nambu A, et al. Meikin is a conserved regulator of meiosis-I-specific kinetochore function. Nature. 2015;517(7535):466–471. .
- Bonner AM, Hughes SE, Hawley RS. Regulation of Polo Kinase by Matrimony Is Required for Cohesin Maintenance during Drosophila melanogaster Female Meiosis. Curr Biol. 2020;30(4):715–722. e3. .
- Harper NC, Rillo R, Jover-Gil S, et al. Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev Cell. 2011;21(5):934–947.
- Labella S, Woglar A, Jantsch V, et al. Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev Cell. 2011;21(5):948–958.
- Pattabiraman D, Roelens B, Woglar A, et al. Meiotic recombination modulates the structure and dynamics of the synaptonemal complex during C. elegans meiosis. PLoS Genet. 2017;13(3):e1006670.
- Nadarajan S, Lambert TJ, Altendorfer E, et al. Polo-like kinase-dependent phosphorylation of the synaptonemal complex protein SYP-4 regulates double-strand break formation through a negative feedback loop. Elife. 2017;6:e23437.
- Machovina TS, Mainpal R, Daryabeigi A, et al. A Surveillance System Ensures Crossover Formation in C. elegans. Curr Biol. 2016;26(21):2873–2884.
- Chase D, Golden A, Heidecker G, et al. Caenorhabditis elegans Contains a Third Polo-Like Kinase Gene. DNA Seq. 2000;11(3–4):327–334.
- Nishi Y, Rogers E, Robertson SM, et al. Polo kinases regulate C. elegans embryonic polarity via binding to DYRK2-primed MEX-5 and MEX-6. Development. 2008;135(4):687–697.
- Elia AEH, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299(5610):1228–1231.
- Park J-E, Soung N-K, Johmura Y, et al. Polo-box domain: a versatile mediator of polo-like kinase function. Cell Mol Life Sci. 2010;67(12):1957–1970.
- Kim Y, Kostow N, Dernburg AF. The Chromosome Axis Mediates Feedback Control of CHK-2 to Ensure Crossover Formation in C. elegans. Dev Cell. 2015;35(2):247–261.
- Phillips CM, Wong C, Bhalla N, et al. 8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 2005;123(6):1051–1063.
- Phillips CM, Dernburg AF. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev Cell. 2006;11(6):817–829.
- Sato-Carlton A, Nakamura-Tabuchi C, Chartrand SK, et al. Phosphorylation of the synaptonemal complex protein SYP-1 promotes meiotic chromosome segregation. J Cell Biol. 2018;217(2):555–570.
- Brandt JN, Hussey KA, Kim Y. Spatial and temporal control of targeting Polo-like kinase during meiotic prophase. J Cell Biol. 2020;219(11):e202006094.
- Roelens B, Barroso C, Montoya A, et al. Spatial Regulation of Polo-Like Kinase Activity During Caenorhabditis elegans Meiosis by the Nucleoplasmic HAL-2/HAL-3 Complex. Genetics. 2019;213(1):79–96.
- Zhang W, Miley N, Zastrow MS, et al. 2 promotes homologous pairing during Caenorhabditis elegans meiosis by antagonizing inhibitory effects of synaptonemal complex precursors. PLoS Genet. 2012;8(8):e1002880.
- Harris D, Orme C, Kramer J, et al. A Deficiency Screen of the Major Autosomes Identifies a Gene (matrimony) That Is Haplo-insufficient for Achiasmate Segregation in Drosophila Oocytes. Genetics. 2003;165(2):637–652.
- Bonner AM, Hughes SE, Chisholm JA, et al. Binding of Drosophila Polo kinase to its regulator Matrimony is noncanonical and involves two separate functional domains. Proc Natl Acad Sci U S A. 2013;110(13):E1222–31. .
- Galander S, Marston AL. Meiosis I Kinase Regulators: conserved Orchestrators of Reductional Chromosome Segregation. Bioessays. 2020;42(10):e2000018.
- Wynne DJ, Rog O, Carlton PM, et al. Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J Cell Biol. 2012;196(1):47–64.
- Baudrimont A, Penkner A, Woglar A, et al. Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet. 2010;6(11):e1001219. .
- Sato A, Isaac B, Phillips CM, et al. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139(5):907–919.
- Rosu S, Zawadzki KA, Stamper EL, et al. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet. 2013;9(8):e1003674.
- Stamper EL, Rodenbusch SE, Rosu S, et al. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet. 2013;9(8):e1003679.
- Woglar A, Daryabeigi A, Adamo A, et al. Matefin/ SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLoS Genet. 2013;9(3):e1003335.
- Woglar A, Villeneuve AM. Dynamic Architecture of DNA Repair Complexes and the Synaptonemal Complex at Sites of Meiotic Recombination. Cell. 2018;173(7):1678–1691. e16. .
- Sanchez A, Adam C, Rauh F, et al. Exo1 recruits Cdc5 polo kinase to MutLγ to ensure efficient meiotic crossover formation. Proc Natl Acad Sci U S A. 2020;117(48):30577–30588. .
- Melters DP, Paliulis LV, Korf IF, et al. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012;20(5):579–593.
- Martinez-Perez E, Schvarzstein M, Barroso C, et al. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 2008;22(20):2886–2901.
- Altendorfer E, Láscarez-Lagunas LI, Nadarajan S, et al. Crossover Position Drives Chromosome Remodeling for Accurate Meiotic Chromosome Segregation. Curr Biol. 2020;30(7):1329–1338. e7. .
- Nabeshima K, Villeneuve AM, Colaiácovo MP. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J Cell Biol. 2005;168(5):683–689.
- Rogers E, Bishop JD, Waddle JA, et al. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J Cell Biol. 2002;157(2):219–229.
- Kaitna S, Pasierbek P, Jantsch M, et al. The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous Chromosomes during meiosis. Curr Biol. 2002;12(10):798–812.
- de Carvalho CE, Zaaijer S, Smolikov S, et al. 1 antagonizes the Aurora B kinase in C. elegans. Genes Dev. 2008;22(20):2869–2885.
- Ferrandiz N, Barroso C, Telecan O, et al. Spatiotemporal regulation of Aurora B recruitment ensures release of cohesion during C. elegans oocyte meiosis. Nat Commun. 2018;9(1):834. .
- Sato-Carlton A, Nakamura-Tabuchi C, Li X, et al. Phosphoregulation of HORMA domain protein HIM-3 promotes asymmetric synaptonemal complex disassembly in meiotic prophase in Caenorhabditis elegans. PLoS Genet. 2020;16(11):e1008968.
- Mather K Competition between bivalents during chiasma formation. Proceedings of the Royal Society of London Series B. 1936; 120:208–227.
- Penkner AM, Fridkin A, Gloggnitzer J, et al. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 2009;139(5):920–933. .
- Stecher G, Tamura K, Kumar S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol Biol Evol. 2020;37(4):1237–1239.
