ABSTRACT
Bladder cancer (BLCA) is a common malignant urothelial cancer in the world. Although circular RNAs (circRNAs) involve in regulating BLCA progression, the role of a novel circular RNA circSETD3 in regulating BLCA pathogenesis has not been studied. The expression of circSETD3, miR-641, PTEN mRNA in BLCA tissues and cell lines were measured using RT-qPCR. The gain-of-function experiments were performed in vitro and in vivo to detect the effects of circSETD3 on cell proliferation, migration, EMT, and stemness maintenance. Besides, rescue experiments were performed to demonstrate the regulatory mechanism of circSETD3/miR-641/PTEN in BLCA cell malignant phenotypes in vitro. CircSETD3 was remarkably downregulated in the cancerous clinical tissues and cell lines, in contrast with their normal counterparts, and circSETD3 tended to be deficient in BLCA patients with larger tumor size, advanced clinical stages, positive lymph metastasis and worse prognosis. In addition, circular isoforms of circSETD3 were more resistant to RNase R+ and actinomycetes D treatment compared to their linear isoforms, and circSETD3 mainly distributed in the cytoplasm of the BLCA cells. Further gain-of-function experiments showed that circSETD3 acted as a tumor suppressor to suppress BLCA cell proliferation, migration, EMT and stemness, and the underlying mechanisms had also been elucidated. Mechanistically, circSETD3 sponged miR-641 to upregulate PTEN, resulting in the blockage of BLCA progression. Our findings indicated that circSETD3 acted as a vital tumor suppressor in BLCA via regulating the miR-641/PTEN axis.
Introduction
Bladder cancer (BLCA) is the tenth common malignant urothelial cancer in the world, which brings huge health burden for males. It has estimated that 549, 000 new cases and 200,000 death in 2018, and about four times in men than in women [Citation1]. The main risk factors of BLCA include exposure to environmental chemical substances, water pollution, and smoking [Citation1–3]. According to the difference in prognosis, management, and therapeutic objectives, BLCA has been classed into categories, including non-muscle-invasive tumors, muscle-invasive tumors, and metastatic tumors [Citation4]. Despite the optimal therapeutic strategies have been chose in corresponding categories, the outcome still exhibits very poor [Citation5–8]. Therefore, it is urgent to explore novel biomarkers for BLCA diagnosis and therapy.
In recent years, numerous evidences have demonstrated a subpopulation cancer stem cells (CSCs) in BLCA act as accelerators to promote tumorigenesis and development of BLCA [Citation9,Citation10]. CSCs exhibit the self-renew and stemness maintain to promote BLCA cell active, metastasis, and therapeutic resistance [Citation11,Citation12]. ALDH1, Nestin, OCT4, Nanog, CD44, and CD144 have been reported to act as the stemness markers in multiple tumors [Citation13]. Therefore, the dysregulation of the stemness markers can act as indicators of the stemness phenotype change. Moreover, epithelial-to-mesenchymal transition (EMT) not only provides an important process to generate and maintain stemness, but also contributes in tumor invasion, heterogeneity, and chemoresistance in tumor [Citation14]. Thus, to investigate the abnormal of stemness maintenance and EMT process is significance for understanding the mechanism of tumor progression and improving the prognosis of BLCA patients.
Circular RNAs (circRNAs) are a group of non-coding RNAs with stable circular structure without 5ʹ-3ʹ polarity and ployA tail [Citation15]. circRNAs commonly generate from exons of eukaryotic cells, and mostly distribute in cytoplasm fractionation [Citation16]. Commonly, circRNAs exert the function with gene-regulatory in cancer through acting as efficient miRNAs spongers [Citation17,Citation18]. For example, Song et al. have demonstrated that has_circ_101996 promotes cervical cancer cell proliferation and invasion by sponging miR-8075 to increase TPX2 expression [Citation19]. Besides, Ding et al. have reported has_circ_0001955 accelerates Hepatocellular carcinoma (HCC) cell proliferation, migration, and invasion by acting as a sponger of miR-145-5p [Citation20]. Previous research has indicated circSETD3 acts as a tumor suppressor in HCC via sponging miR-421 to inhibit cell growth in vitro and in vivo [Citation21]. However, circSETD3 has been proved plays as an oncogene in nasopharyngeal carcinoma and non-small cell lung cancer by promoting cell migration, invasion and chemical resistance [Citation22,Citation23]. Whereas, the function of circSETD3 in tumorigenesis and progression of BLCA remains unknown. Hence, we wanted to explore the function and mechanism of circSETD3 in BLCA.
Taken together, in this study, we explore the tumor inhibitory function of circSETD3 in BLCA through regulating miR-641/PTEN axis to repress cell growth, migration, invasion, EMT process and stemness maintenance. Our finding provided the potential diagnostic and therapeutic molecular marker for BLCA.
Materials and methods
Clinical specimen
A total of 45 paired BLCA tissues and adjacent non-tumor tissues were separated from BLCA patients who underwent surgical resection at the Hospital of Chengdu University of Traditional Chinese Medicine between November 2015 and June 2020. And their clinicopathological characteristics were collected and analyzed in this study. All patients that enrolled in this study have written informed consent. Moreover, this study was approved by the Ethics Committee of the Hospital of Chengdu University of Traditional Chinese Medicine. The samples were obtained and immediately snap-frozen in liquid nitrogen and then stored at −80°C for subsequent experiment.
Cell culture
Human bladder epithelial cells HCV29 and BLCA cell lines T24, J87, SW780, UMUC3 were obtained from the Chinese Academy of Sciences (Shanghai, China), 293 T cells were purchased from ATCC (Manassas, VA, USA). All cells were maintained in DMEM medium (Gibco, NY, USA) with 10% fetal bovine serum (FBS) (Gibco, NY, USA) supplemented with 1% penicillin/streptomycin (). Then, all cells were incubated at 37°C with 5% CO2.
Cell transfection
PcDNA-CircSETD3 and its corresponding control pcDNA3.1, PTEN-specific small interfering RNA (si-PTEN) and its negative control (si-NC), miR-641 mimic and its corresponding control NC mimic were designed and synthesized from Genepharma (Shanghai, China). T24 and UMUC3 cells were transfected with pcDNA3.1 vector, siRNA, or mimic using Lipofectamine 2000 (Invitrogen, CA, USA).
RNA extraction and real-time quantitative reverse transcription PCR (qRT-PCR)
Trizol reagent (Life Technologies, CA, USA) was used to isolate total RNA from cells and tissues. And the cDNA was generated from RNA using Prime-Script RT reagent Kit (TaKaRa, Dalian, China) following the manufacturer’s instructions. Then, cDNA products were amplified using SYBR Green qPCR Master Mix (TaKaRa, Dalian, China) according to the manufacture’s protocol. Primer sequences in this study were shown as: CircSETD3: F, 5ʹ-TGA AGA AGA TGA AGT TCG GTA T-3ʹ; R, 5ʹ-GTG CCA GAT TTC TGA GTT TT-3ʹ. SETD3: F, 5ʹ-GAC AGA CTC TAC GCC ATG-3ʹ; R, 5ʹ-GTC TCC CAG CAA GTG TTC-3ʹ. MiR-641: F, 5ʹ-GGG GAA AGA CAT AGG ATA GAG T-3ʹ; R, 5ʹ-CAG TGC GTG TCG TGG AG-3ʹ. PTEN: F, 5ʹ-CTG CAG AAA GAC TTG AAG GCG-3ʹ; R, 5ʹ-TGC TTT GAA TCC AAA AAC CTT ACT-3ʹ. ALDH1: F, 5ʹ-CCC GTG GCG TAC TAT GGA TG-3ʹ; R, 5ʹ-CAG TGC AGG CCC TAT CTT CC-3ʹ. OCT4: F, 5ʹ-GGC CAC ACG TAG GTT CTT GA-3ʹ; R, 5ʹ-ATA CCT TCC CAA ATA GAA CCC C-3ʹ.CD133: F, 5ʹ-CTG TGC GGG AAC TCC TTT TC-3ʹ; R, 5ʹ-AGC GAG TAC TCA GGT TGC AC-3ʹ. CD44: F, 5ʹ-GCC CAA TGC CTT TGA TGG AC-3ʹ; R, 5ʹ-CCC ATG TGA GTG TCT GGT AGC-3ʹ. GAPDH: F, 5ʹ-ACT CCT CCA CCT TTG ACG C-3ʹ; R, 5ʹ-GCT GTA GCC AAA TTC GTT GTC-3ʹ. U6: F, 5ʹ-CTC GCT TCG GCA GCA CA-3ʹ; R, 5ʹ-AAC GCT TCA CGA ATT TGC G-3ʹ. U6 was used as a control for miRNA, and GAPDH was used as control for other genes. Relative expression was quantified by the 2−∆∆Ct methods.
Ribonuclease (RNase) R and actinomycin D digestion
The transcription inhibitory experiments were performed using RNase R and actinomycin D. Briefly, the RNA was incubated with 3 U/µg RNase R (Sigma-Aldrich, MO, USA) for 30 min at room temperature. Similarly, RNA was incubated with 2 mg/mL actinomycin D (R & D Systems, Shanghai, China) for 6, 12, 24 h. Then, the expression of circSETD3 and SETD3 was examined using qRT-PCR. Moreover, the RNA of nucleus and cytoplasm was extracted using the PARISTM Kit (Thermo, MA, USA). The expression of cricSETD3 was detected using qRT-PCR. GAPDH was used as the control of cytoplasm, and the U6 was used as the control of cell nuclei, respectively.
Cell counting kit-8 (CCK-8) assay
Cell viability of T24 and UMUC3 cells was determined using CCK-8 kit (Beyotime, Shanghai, China) following manufacturer’s protocol. Briefly, 5000 cells were plated onto 96-well plates. And the 10 µL CCK-8 solution was added into each well at 0 h, 24 h, 48 h, 72 h, and incubated for 2 h. Then the absorbance at 450 nm was examined.
Colony formation assay
5000 cells were plated on 6-well plates and incubated in at 37°C with 5% CO2 for two weeks, and the medium was replaced by fresh medium every three days. Then, the clones were fixed with methanol and stained 0.1% crystal violet (Beyotime, Shanghai, China) for 20 min. And the colony number was counted under a microscope (Leica, Wetzlar, Germany).
Migration assay
Cell migration was performed using transwell chamber. Briefly, 2 × 105 cells were plated into upper chamber. And 500 µl medium supplemented with 20% FBS was added into lower chamber, then cells were incubated at 37°C with 5% CO2 for 48 h. The cells on the front of chamber were removed by sterile swabs, and the migrated and invaded cells on the opposite were fixed with methanol and stained 0.1% crystal violet (Beyotime, Shanghai, China) for 20 min. Then, the migrated and invaded cells were counted and imaged using an inverted microscope.
Microsphere assay
1000 cells were plated onto non-adhesive 6-well plates and cultured in serum-free DMEM medium supplemented with 2% B27 (Invitrogen, MA, USA), 20 ng/mL EGF (Invitrogen, MA, USA), 20 ng/mL bFGF (Invitrogen, MA, USA), and 5 µg/mL insulin (Invitrogen, MA, USA) for 10 days. Then, the spheres were counted and imaged in a microscope (Leica, Wetzlar, Germany).
Western blotting assay
Protein of tissues and cells was isolated using RIPA lysis buffer (Beyotime, Shanghai, China) according to the manufacturer’s protocol. protein was separated on 10% SDS-PAGE gels and transferred to polyvinylidene fluoride (PVDF) membrane. And then the membrane was blocked with 5% skimmed milk and incubated with primary antibodies overnight at 4°C. Then the membrane was incubated with a horseradish peroxidase secondary antibody (Abcam, MA, USA) at room temperature for 1 h. Finally, proteins were visualized with ECL chemiluminescence detection kit (Bio-Rad, CA, USA). All antibodies were purchased from Abcam (MA, USA), including PTEN (ab267787), E-cadherin (ab76055), N-cadherin (ab76011), vimentin (ab92547), GAPDH (ab8245).
RNA immunoprecipitation (RIP) assay
Magna RIPTM RNA-Binding Protein Immunoprecipitation kit (Millipore, Billerica, MA, USA) was used to accomplish RIP assay. Briefly, cell lysates were incubated with Argonaute-2 (Ago2) antibody or Immunoglobulin G (IgG) antibody and then added Sepharose beads (BioRad, CA, USA) into the mixtures. The expression of circSETD3 and miR-641 was examined using qRT-PCR.
Luciferase reporter assay
The wild or mutant sequences of circSETD3 or PTEN targeted miR-641 were cloned into the pGl3 luciferase reporter vector. Then the vectors were co-transfected with miR-641 mimic or NC mimic into 293 T cells using Lipofectamine 2000 (Invitrogen, CA, USA) and incubated for 48 h. Then, the luciferase activity was detected using a Dual-Glo luciferase assay system (Promega, WI, USA) according to the manufacturer’s protocol.
Animal model
Four to six-weeks-old male C57BL/6 mice were randomly divided into two groups with six mice each group. Briefly, circSETD3-inhibited T24 cells were subcutaneously transplanted into the back of mice at density of 1 × 106 cells. And the tumor growth was observed and calculated every 7 days. At day 28, the mice were sacrificed and then the tumors were isolated from mice for experiments. Animal experiments in this study was approved by the Ethics Committee of the Hospital of Chengdu University of Traditional Chinese Medicine.
Statistical analyses
The statistical analyses in this study were accomplished by GraphPad Prism version 8.0 (GraphPad Software). Student’s t-test and one-way ANOVA were used to compare the differences between two or multiple groups. Pearson correlation analysis was use to investigate the correlations among circSETD3, miR-641, and PTEN. Kaplan-Meier plot was used to detect overall survival of BLCA patients. Values were presented as means ± standard (SD). P < 0.05 was considered as statistic significant.
Results
Downregulation of circ-SETD3 in bladder cancer tissues and cell lines
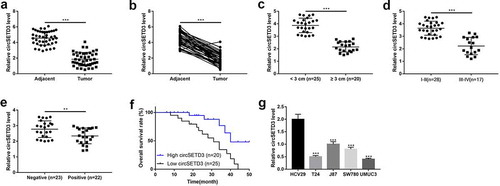
First of all, we explored the expression of circSETD3 in 45 paired BLCA tissues and the corresponding adjacent normal tissues. We found that the expression of circSETD3 significantly decreased in BLCA tissues compared to adjacent non-tumor tissues ()). Moreover, we analyzed the association between circSETD3 expression levels and the pathological characteristics of BLCA patients, and the results indicated that BLCA patients with larger tumor size ()), higher grade of TNM stage ()) and positive lymph node metastasis ()) tended to have low-expressed circSETD3. Consistently, the survival curve analysis showed that BLCA patients with high expression of cicSETD3 exhibited optimistic survival time than BLCA patients with low expression of circSETD3 ()). The above clinical results were indirectly supported by the cellular results, which showed that, compared to the normal HCV29 cells, circSETD3 was strongly downregulated in BLCA cell lines (T24, J87, SW780, UMUC3). These finding suggested that aberrant expression of circSETD3 was closely associated with BLCA development.
Figure 1. Downregulation of circ-SETD3 in bladder cancer tissues and cell lines
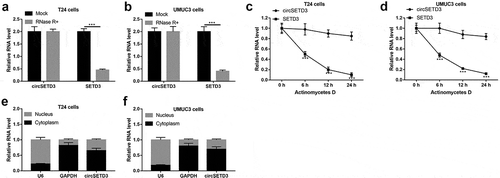
Stability and structure of circSETD3
We further investigated the stability of the circular structure of circSETD3 and its distribution in the BLCA cells. RNase R+ and actinomycetes D was used to analyze the stability of the circular structure, and the results showed that the linear isoform of SETD3 was obviously digested by RNase R+, which had little impacts on the circular isoform of circSETD3 ()). Consistently, circSETD3 was more stable than linear SETD3 underwent actinomycetes D treatment (), suggesting that circular isoform of SETD3 (circSETD3) was much more resistant to both RNase R+ and actinomycetes D treatment. Besides, the cyto-distribution of circSED3 was analyzed, and we noticed that circSED3 mainly located in the cytoplasm but not in the nucleus of the BLCA cells ()). Taken together, our results indicated that circSETD3 had stable circular structure and the majority of circSETD3 distributed in the cytoplasm of BLCA cells.
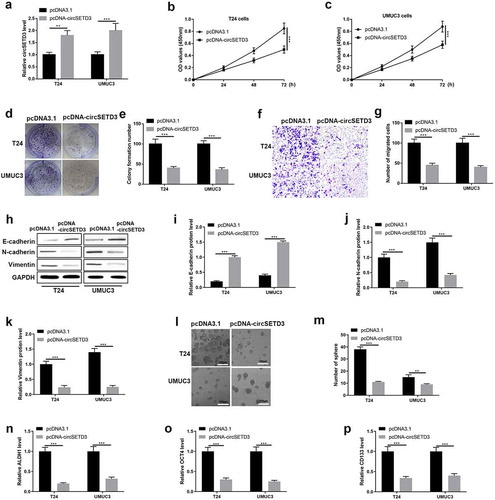
Overexpression of circSETD3 suppresses bladder cancer cell growth, migration, and stemness in vitro
The gain-of-function experiment was employed to determine the function of circSETD3 in BLCA cells. The transfected efficiency was performed using qRT-PCR, and we found upregulation of circSETD3 were observed in T24 cells and UMUC3 cells with transfected with pcDNA-circSETD3 ()). Cell viability was tested using CCK-8 assay, and cell viability was inhibited by overexpression of circSETD3 ()). Besides, cell colony formation ability was detected using colony formation assay, the results were shown in ), which indicated that circSETD3 restrained colonies formation abilities in BLCA cells. Also, transwell assay results showed that the number of migrated cells was reduced by overexpression of circSETD3 (). Besides, the protein level of epithelial cell marker E-cadherin was upregulated by overexpression of circSETD3, however, the protein levels of mesenchymal cell markers N-cadherin and vimentin were downregulated by overexpression of circSETD3 (). Furthermore, microsphere assay showed that the number of sphere formation was reduced by overexpression of circSETD3 ()), and the mRNA levels of stemness associated biomarkers (ALDH1, OCT4, CD133, and CD44) were also decreased by upregulating circSETD3 in both T24 and UMUC3 cells ()).
Figure 3. Overexpression of circSETD3 suppresses bladder cancer cell growth, migration, stemness in vitro.
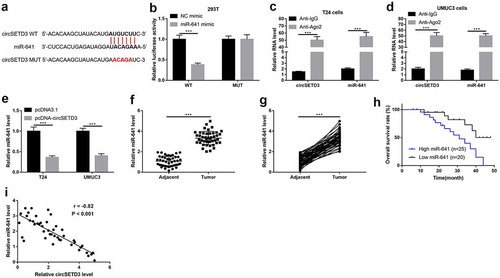
CircSETD3 functions as RNA sponge for miR-641
We next investigated whether circSETD3 acted as a tumor suppressor via sponging miRNAs, and miR-641 was a speculated candidate for circSETD3 by using the StarBase (http://starbase.sysu.edu.cn/). As shown in ), 3ʹ-UTR of cirsSETD3 targeted with miR-641, and the relationship between circSETD3 and miR-641 was verified using the luciferase reporter assay and RIP assay. As shown in ), the luciferase activity was suppressed by wild type circSETD3-pGl3 luciferase reporter vector was co-transfected with miR-641 mimic, but the luciferase activity showed no obvious change by mutant type circSETD3-pGl3 luciferase reporter vector was co-transfected with NC mimic or miR-641 mimic. Moreover, circSTED3 and miR-641 were obviously enriched by Ago2 antibody in both T24 cells and UMCM3 cells (). Furthermore, qRT-PCR results exhibited upregulation of circSETD3 significantly inhibited miR-641 expression both in T24 cells and UMUC3 cells ()). Also, we examined the expression of miR-641 in BLCA tissues and adjacent non-tumor tissues, and found that miR-641 was strongly increased in tumor tissues compared to adjacent non-tumor tissues (). Besides, overall survival curve showed that BLCA patients with high miR-641 expression exhibited shorter survival time ()). The Spearman correlation analysis indicated that circSETD3 expression negatively associated with miR-641 expression in BLCA tissues ()), suggesting that endogenous interactions between circSETD3 and miR-641 were existed in BLCA.
Figure 4. CircSETD3 acts as a sponger by binding miR-641
Upregulated miR-641 reverses the regulating effects of circSETD3 overexpression on BLCA cells
Then, the rescue experiment was used to test the regulating effects of the circSETD3/miR-641 axis on BLCA cells. As shown in ), the expression of miR-641 was significantly upregulated both in T24 cells and UMUC3 cells by transfecting the cells with miR-641 mimic. After that, the cell viability was determined using CCK-8 assay, and the results showed that cell viability was strongly reduced by circSETD3 upregulation both in T24 cells and UMUC3 cells, but the inhibitory effects of circSETD3 upregulation on cell viability was reversed by miR-641 mimic ()). Besides, as shown in )), the BLCA cell colony formation abilities were inhibited by circSETD3 upregulation, which were also restored by upregulating miR-641. Similarly, circSETD3 also restrained cell migration by sponging miR-641 in the BLCA cells ()). Furthermore, by performing RT-qPCR, we noticed that circSETD3 overexpression downregulated the mRNA levels of E-cadherin, while upregulated N-cadherin and vimentin mRNA to hamper EMT process, and the inhibiting effects of circSETD3 on EMT were abrogated by upregulating miR-641 (–j) In addition, the mRNA levels of stemness markers (ALDH1, OCT4, CD133, CD44) was inhibited by circSETD3 upregulation, which were reversed by miR-641 mimic in the BLCA cells ()). Taken together, circSETD3 played a tumor suppressor in BLCA cells via targeting miR-641 to modulate cell growth, migration, invasion, EMT process, and stemness.
Figure 5. Upregulated miR-641 mitigates circSETD3 effects in bladder cancer cells
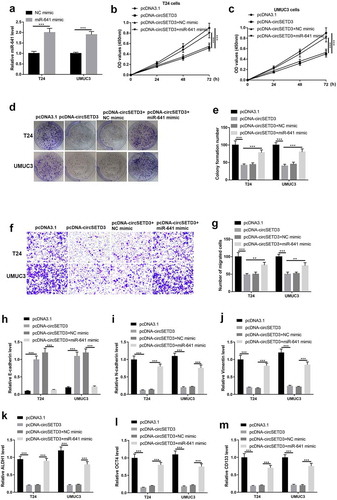
MiR-641 directly targets with PTEN
PTEN was predicated as a downstream target for miR-641 by using StarBase (http://starbase.sysu.edu.cn/). As shown in ), miR-641 potentially bound to the 3ʹUTR of PTEN mRNA, the following luciferase activity reporter assay indicated that the luciferase activity was reduced by wild type PTEN-pGl3 luciferase reporter vector was co-transfected with miR-641 mimic, however, the luciferase activity no obvious change by mutant type PTEN-pGl3 luciferase reporter vector was co-transfected with NC mimic or miR-641 mimic ()). And the mRNA and protein expression of PTEN were seriously inhibited by miR-641 mimic ()). In addition, we found the mRNA levels of PTEN were significantly decreased in BLCA tissues compared to adjacent non-tumor tissues ()). Furthermore, the BLCA patients with high PTEN expression showed optimistic survival time than patients with low PTEN expression ()). And the Spearman correlation analyses showed negative correlation between miR-641 expression and PTEN expression ()). Collectively, miR-641 degraded PTEN by targeting its 3ʹUTR.
Figure 6. MiR-641 directly targets with PTEN
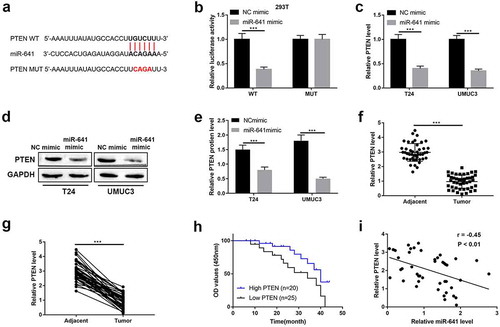
CircSETD3 suppresses BLCA cell growth, migration, stemness through regulating the miR-641/PTEN axis
We further verified the mechanisms that circSETD3 acted as tumor suppressor via regulating miR-641/PTEN axis. Firstly, we found the mRNA and protein levels of PTEN were promoted by upregulating circSETD3 both in T24 cells and UMUC3 cells (–c)). Besides, the Spearman correlation analyses showed positive correlation between circSETD3 expression and PTEN expression in BLCA tissues ()). Moreover, PTEN downregulation significantly inhibited PTEN and upregulated miR-641 expression both in T24 and UMUC3 cells (). Then, the rescue experiment was employed to corroborate the effects of circSETD3 on BLCA cells via regulating miR-641/PTEN axis. As shown in ), cell viability was seriously repressed by circSETD3 upregulation, but the inhibitory effect of circSETD3 upregulation was reversed by PTEN downregulation both in T24 and UMUC3 cells. Moreover, the number of colony formation was reduced by circSETD3 upregulation, but the inhibitory effect of circSETD3 upregulation was reversed by PTEN downregulation both in T24 and UMUC3 cells (). Furthermore, the number of migrated cells was reduced by circSETD3 upregulation, which were also reversed by silencing PTEN (). The mRNA level of epithelial cell marker E-cadherin was decreased, and the mRNA levels of mesenchymal cell markers N-cadherin and vimentin were increased by circSETD3 upregulation, but the inhibitory effects of circSETD3 upregulation on EMT process were reversed by PTEN downregulation both in T24 and UMUC3 cells (). The mRNA levels of stemness markers (ALDH1, OCT4, CD133, CD44) were also inhibited by circSETD3 upregulation, but the inhibitory effect of circSETD3 upregulation was reversed by PTEN downregulation both in T24 and UMUC3 cells (). Overall, our data indicated that circSETD3 acted as tumor suppressor in BLCA cells by sponging miR-641 to accelerate PTEN expression.
Figure 7. CircSETD3 suppresses bladder cancer cell growth, migration, stemness through regulating miR-641/PTEN axis
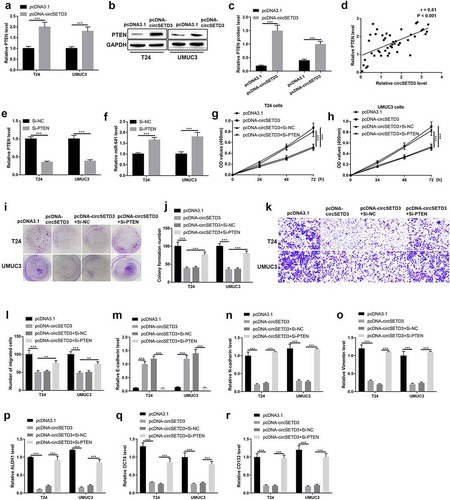
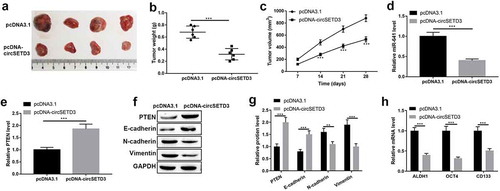
Overexpression of circSETD3 inhibits tumor growth by modulating miR-641/PTEN axis in vivo
Importantly, the tumor inhibitory effect of circSETD3 was determined by in vivo xenograft tumor mice model. The mice were injected with T24 cells with circSETD3 upregulation or empty vector. As shown in –c), tumor growth was suppressed by circSETD3 upregulation. And the expression of miR-641 was inhibited, but the mRNA and protein expression of PTEN were increased by circSETD3 upregulation in vivo (–g)). Moreover, the protein level of epithelial marker E-cadherin was increased, and the protein levels of mesenchymal cell markers N-cadherin and vimentin were reduced by circSETD3 upregulation in vivo ()). In addition, the expressions of ALDH1, OCT4, CD133, CD44 mRNA were downregulated by circSETD3 upregulation in vivo ()). These results suggested that overexpression of circSETD3 could inhibit tumorigenesis in vivo.
Figure 8. Overexpression of circSETD3 inhibits tumor growth by modulating miR-641/PTEN axis in vivo.
Discussion
BLCA is one of the most prevalent malignant tumors which results in 150,000 global deaths per year [Citation24]. Despite improvement of the treatments, the outcome among patients remains unexpected [Citation25]. Therefore, seeking the effective diagnosed and therapeutic biomarker for BLCA patients has important significance. In recent years, a population of stem-like cells which is CSCs have been demonstrated to be associated with high recurrence rate of BLCA patients [Citation26]. Thus, to investigate the stemness properties of BLCA it is necessary for improvement the poor prognosis of BLCA patients.
In the present study, we found circSETD3 significantly downregulated in BLCA tissues and cell lines, and it negatively associated with larger tumor size, advanced clinical stage, and lymphatic metastasis. Besides, high expression of circSETD3 associated with optimistic survival time for BLCA patients. For investigation of the mechanism of circSETD3, we found upregulated circSETD3 seriously inhibited cell growth, migration, EMT process and stemness maintenance through sponging miR-641 to accelerate PTEN expression. Our finding suggested circSETD3 might act as a tumor suppressor through mainly regulating the stemness phenotype and EMT process.
CircRNAs are an important member of non-coding RNAs generate from the pre-mRNA splicing [Citation27]. For a long time circRNAs have been considered as junk-products, but increasing evidences have demonstrated the crucial role of circRNAs in multiple tumors [Citation28]. CircRNAs always act as spongers of miRNAs through directly binding with miRNAs to regulate tumorigenesis and progression [Citation29]. A large number of circRNAs have been reported function as oncogenes or tumor suppressor in BLCA, such as, has_circ_0001361 function as an oncogene through promoting migration and invasion via regulating miR-491-5p/MMP9 axis in BLCA [Citation30]. And Lu et al. suggest circSLC8A1 acts as a tumor suppressor in BLCA by suppressing cell proliferation, migration and invasion in vitro and in vivo [Citation31]. In the past few years, circRNAs sponge miRNAs to regulate cell phenotype is the mostly important regulatory pattern [Citation32]. In our study, circSETD3 function as a tumor suppressor through sponging miR-641 in BLCA. MiR-641 has been reported as a tumor suppressor in human lung cancer [Citation33], and has been reported as an oncogene in cervical cancer [Citation34]. And we firstly demonstrated miR-641 acted as an oncogene in BLCA. PTEN is well-known tumor suppressor in multiple tumors [Citation35]. It has been reported PTEN inhibits tumor progression through inducing cell apoptosis, and inhibiting cell proliferation, migration, invasion, and EMT process in BLCA [Citation29,Citation31,Citation36]. It also has been reported that downregulated PTEN represses cancer stem-like cell properties in breast cancer [Citation37,Citation38]. And in this study, we firstly reported PTEN act as tumor suppressor in BLCA inhibited stemness phenotype and EMT process.
It had been reported that stemness and EMT are the facilitators to promote tumorigenesis, metastasis, and aggressive in multiple tumors [Citation39,Citation40]. Recent research has indicated that EMT is a complex process in distinct cellular states exhibits different functional characteristics, including proliferation, propagation, plasticity, invasion and metastasis [Citation41]. In addition, novel finding suggests that EMT may contribute to immune escape during immune attacks [Citation42]. Not only cell undergoing EMT promotes tumor development, metastasis and drug resistance, but also CSCs accelerate the aggressive behaviors of tumor cells. For example, high expression of cancer stem cell markers OCT4 and NANOG associate with poor prognosis in renal cell carcinoma [Citation43]. And overexpression of cancer stemness maker CD133 correlates with poor prognosis of invasive breast cancer patients [Citation44], and refers to invasive gastric carcinoma [Citation45]. In our finding, we the CSCs markers ALDH1, OCT4 and CD133 were significantly inhibited by overexpression of circSETRD3.
Conclusion
Overall, our study revealed that circSETD3 acted as a tumor suppressor in BLCA through functioning as sponger for miR-641 to upregulate PTEN to inhibit cell growth, migration, invasion, EMT process and stemness maintenance in BLCA. Our findings provide a novel diagnosis and treatment molecular marker, and highlighted a novel mechanism for development and progression of BLCA.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I,et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Antoni S, Ferlay J, Soerjomataram I, et al. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71:96–108.
- Cumberbatch MGK, Jubber I, Black PC, et al. Epidemiology of bladder cancer: a systematic review and contemporary update of risk factors in 2018. Eur Urol. 2018;74:784–795.
- Clark PE, Agarwal N, Biagioli MC, et al. Bladder cancer. J Natl Compr Canc Netw. 2013;11:446–475.
- Alifrangis C, McGovern U, Freeman A, et al. Molecular and histopathology directed therapy for advanced bladder cancer. Nat Rev Urol. 2019;16:465–483.
- Ghandour R, Singla N, Lotan Y. Treatment options and outcomes in nonmetastatic muscle invasive bladder cancer. Trends Cancer. 2019;5:426–439.
- Alfred Witjes J, Lebret T, Compérat EM, et al. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur Urol. 2017;71:462–475.
- Berdik C. Unlocking bladder cancer. Nature. 2017;551:S34–s35.
- Zhang R, Xia J, Wang Y, et al. Co-expression of stem cell and epithelial mesenchymal transition markers in circulating tumor cells of bladder cancer patients. Onco Targets Ther. 2020;13:10739–10748.
- Zhan Y, Chen Z, He S, et al. Long non-coding RNA SOX2OT promotes the stemness phenotype of bladder cancer cells by modulating SOX2. Mol Cancer. 2020;19:25.
- Wang F, Ma X, Mao G, et al. STAT3 enhances radiation-induced tumor migration, invasion and stem-like properties of bladder cancer. Mol Med Rep. 2021;23:1791–3004.
- Abd-El-Raouf R, Ouf SA, Gabr MM, et al. Escherichia coli foster bladder cancer cell line progression via epithelial mesenchymal transition, stemness and metabolic reprogramming. Sci Rep. 2020;10:18024.
- Kaipio K, Chen P, Roering P, et al. ALDH1A1-related stemness in high-grade serous ovarian cancer is a negative prognostic indicator but potentially targetable by EGFR/mTOR-PI3K/aurora kinase inhibitors. J Pathol. 2020;250:159–169.
- Pradella D, Naro C, Sette C, et al. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer. 2017;16:8.
- Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7.
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388.
- Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565.
- Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127.
- Song T, Xu A, Zhang Z, et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. J Cell Physiol. 2019;234:14296–14305.
- Ding B, Fan W, Lou W. hsa_circ_0001955 enhances in vitro proliferation, migration, and invasion of HCC cells through miR-145-5p/NRAS axis. Mol Ther Nucleic Acids. 2020;22:445–455.
- Xu L, Feng X, Hao X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38:98.
- Tang L, Xiong W, Zhang L, et al. circSETD3 regulates MAPRE1 through miR-615-5p and miR-1538 sponges to promote migration and invasion in nasopharyngeal carcinoma. Oncogene. 2021;40(2):307–321.
- Huang Y, Dai Y, Wen C, et al. circSETD3 contributes to acquired resistance to gefitinib in non-small-cell lung cancer by targeting the miR-520h/ABCG2 pathway. Mol Ther Nucleic Acids. 2020;21:885–899.
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917.
- Garg M. Urothelial cancer stem cells and epithelial plasticity: current concepts and therapeutic implications in bladder cancer. Cancer Metastasis Rev. 2015;34:691–701.
- Aghaalikhani N, Rashtchizadeh N, Shadpour P, et al. Cancer stem cells as a therapeutic target in bladder cancer. J Cell Physiol. 2019;234:3197–3206.
- Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. Embo J. 2019;38:e100836.
- Arnaiz E, Sole C, Manterola L, et al. CircRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol. 2019;58:90–99.
- Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17:19.
- Liu F, Zhang H, Xie F, et al. Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene. 2020;39:1696–1709.
- Lu Q, Liu T, Feng H, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18:111.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691.
- Kong Q, Shu N, Li J, et al. miR-641 functions as a tumor suppressor by targeting MDM2 in human lung cancer. Oncol Res. 2018;26:735–741.
- Zhu Y, Liu B, Zhang P, et al. LncRNA TUSC8 inhibits the invasion and migration of cervical cancer cells via miR-641/PTEN axis. Cell Biol Int. 2019;43:781–788.
- Ding ZS, He YH, Deng YS, et al. MicroRNA-34a inhibits bladder cancer cell migration and invasion, and upregulates PTEN expression. Oncol Lett. 2019;18:5549–5554.
- Shen D, Xu J, Cao X, et al. Long noncoding RNA MAGI2-AS3 inhibits bladder cancer progression through MAGI2/PTEN/epithelial-mesenchymal transition (EMT) axis. Cancer Biomark. 2021;30(2):155–165.
- Li B, Lu Y, Yu L, et al. miR-221/222 promote cancer stem-like cell properties and tumor growth of breast cancer via targeting PTEN and sustained Akt/NF-κB/COX-2 activation. Chem Biol Interact. 2017;277:33–42.
- Baker A, Wyatt D, Bocchetta M, et al. Notch-1- PTEN-ERK1/2 signaling axis promotes HER2+ breast cancer cell proliferation and stem cell survival. Oncogene. 2018;37:4489–4504.
- Zhang H, Cui X, Cao A, et al. ITGA3 interacts with VASP to regulate stemness and epithelial-mesenchymal transition of breast cancer cells. Gene. 2020;734:144396.
- Sun L, Cao J, Chen K, et al. Betulinic acid inhibits stemness and EMT of pancreatic cancer cells via activation of AMPK signaling. Int J Oncol. 2019;54:98–110.
- Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226.
- Terry S, Savagner P, Ortiz-Cuaran S, et al. New insights into the role of EMT in tumor immune escape. Mol Oncol. 2017;11:824–846.
- Rasti A, Mehrazma M, Madjd Z, et al. Co-expression of cancer stem cell markers OCT4 and NANOG predicts poor prognosis in renal cell carcinomas. Sci Rep. 2018;8:11739.
- Joseph C, Arshad M, Kurozomi S, et al. Overexpression of the cancer stem cell marker CD133 confers a poor prognosis in invasive breast cancer. Breast Cancer Res Treat. 2019;174:387–399.
- Attia S, Atwan N, Arafa M, et al. Expression of CD133 as a cancer stem cell marker in invasive gastric carcinoma. Pathologica. 2019;111:18–23.