ABSTRACT
Circular RNAs (circRNAs) have recently been described as key regulators in the progression of non-small cell lung cancer (NSCLC), and this study aimed to investigate the functional role of circMAT2B in NSCLC. CircMAT2B expression in NSCLC tissues and cell lines was investigated using RT-qPCR analysis. A series of functional experiments, including MTT assay, colony formation assay, wound healing assay and transwell assay, were carried out to investigate the effects of circMAT2B knockdown/overexpression on the malignant traits of NSCLC cells. Western blot analysis was performed to detect the expression of EMT-related proteins. Dual-luciferase reporter assay and RIP assay were further carried out to analyze the interaction between circMAT2B and miR-431 in NSCLC. We observed that circMAT2B was overexpressed in NSCLC tissues and cell lines, and high expression of circMAT2B was closely associated with large tumor size, advanced TNM stage and poor prognosis of NSCLC patients. Further functional experiments showed that circMAT2B knockdown markedly inhibited the proliferation, migration, invasion and EMT of NSCLC cells, whereas circMAT2B overexpression led to the opposing results. Mechanistically, circMAT2B could directly interact with miR-431, and subsequently decrease miR-431 expression in NSCLC. The effects of circMAT2B overexpression in NSCLC cells were abrogated by miR-431 restoration. Our findings revealed the novel oncogenic roles of circMAT2B in NSCLC by sponging miR-431.
Background
Lung cancer is a leading cause of cancer-associated mortality worldwide. Among all lung cancer cases, approximately 85% of patients are diagnosed with non-small cell lung cancer (NSCLC) [Citation1]. NSCLC includes adeno- and squamous cell carcinoma. Although great achievements have been made in the diagnosis and therapeutic methods, the long-term prognosis of NSCLC patients remains largely dismal, particularly in the advanced stages [Citation2]. Therefore, it is imperative to elucidate the molecular mechanisms of NSCLC and develop novel anti-NSCLC agents.
Circular RNAs (circRNAs) are a naturally occurring type of RNAs that, unlike linear RNAs, are formed by a covalently closed loop with neither 5ʹ-3ʹ polarities nor polyadenylated tails [Citation3]. circRNAs are stable and resistant to RNase R, and often exhibit tissue/developmental-stage-specific expression. circRNAs were first regarded as products of splicing errors [Citation4], but recently, emerging evidence shows that they play important roles in the carcinogenesis and the malignant behaviors of human cancers [Citation5]. Recent studies have demonstrated that a novel circRNA, circMAT2B, can promote the progression of hepatocellular carcinoma and colorectal cancer [Citation6,Citation7]. In this study, we aimed to investigate the functional role of circMAT2B in NSCLC.
Materials and methods
Ethics approval and consent to participate
The study was authorized by the ethical committee of the the First Affiliated Hospital, West China Hospital, Sichuan University (Ethical approval number: 2021 KT No. 16 (2021KT16)). The study was performance in accordance with the Declaration of Helsinki and all methods were performed in accordance with the relevant guidelines and regulations. Informed consent was obtained from all participants prior to the study.
Patients and tissue samples
NSCLC tissues and the matched adjacent normal tissues were collected from 106 patients who underwent operation at West China Hospital (Chengdu, China). None of these patients underwent chemotherapy or radiotherapy prior to surgery. After collection, all tissue samples were immediately snap-frozen in liquid nitrogen and stored at −80°C.
Cell culture and transfection
Four NSCLC cell lines (A549, H1299, H1975 and SPCA1) and a normal human bronchial epithelial cell line (16HBE), bought from the Type Culture Collection of Chinese Academy of Sciences (Shanghai, China), were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with fetal bovine serum (FBS; HyClone, Logan City, UT, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin in a humidified incubator at 37°C and 5% CO2.
Specific knockdown of circMAT2B was achieved using a small interfering (si)RNA designed and chemically synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China) to target the back-splice junction. To construct circMAT2B overexpression plasmid, circMAT2B cDNA sequence was synthesized and cloned into a pLVX-cir vector (Shanghai Genomeditech Co., Ltd., Shanghai, China). An empty plasmid served as the negative control. miR-431 mimics and mimics negative control (NC) were obtained from Shanghai GenePharma Co., Ltd (Shanghai, China). Cell transfection was carried out using Lipofectamine 2000 reagent (Invitrogen).
cDNA synthesis
For the cDNA synthesis protocol, we added 5 μL of total RNA to a mixture including 1 μL of 10 mM dNTP mix at neutral pH, 1 μL of 0.25 μg/μL random hexamers, and 6 μL of DEPC-treated water. The PCR mixtures were incubated at 65°C for 5 min and chilled on ice. After adding a mixture of 4 μL of First-strand Buffer (5×), 2 μL of 0.1 M dithiothreitol (DTT), and 1 μL of MMLV reverse transcriptase (at room temperature), cDNA synthesis was done at 25°C for 10 min, 37°C for 50 min and 70°C for 15 min. The cDNA was stored at −70°C until used. In addition, by using PrimeScript RT Master Mix kits (Takara, Japan), cDNA was synthesized according to the manufacturer’s recommendation. In short, 5 uL of total RNA was added to a mixture containing 5 uL of 5× Master mix and 11 uL of DEPC-treated water. cDNA synthesis was performed at 37°C for 15 min, and 85°C for 5 s [Citation8].
Quantitative real-time RT-PCR (qRT-PCR) analysis
Total RNA was isolated using TRIzol reagent (Invitrogen) from cultured cells or tissues, and reverse transcribed to cDNA using the PrimeScript RT reagent Kit (TaKaRa, Dalian, China) according to the manufacturer’s protocol. Real-time PCR amplification was then performed using the SYBR Premix Ex Taq II kit (TaKaRa) on the MyIQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) as described by the manufacturer. Briefly, the total qPCR volume was 20 µl, which contained 2 µl of cDNA template (100 ng), 10 µl of 2X SYBR Premix Ex Taq II, 0.8 µl of primers (10 µM each), and 6.4 µl of ddH2O [Citation9]. The PCR conditions were as follows: 95°C for 5 s; 55°C for 30 s; and 72°C for 30 s for 45 cycles. All RT-PCR reactions were performed in triplicate. The gene primers were purchased from TaKaRa. The primer sequences were for MAT2B-F:5′-TGCATCCAGCACA AACATGTCCT-3ʹ, MAT2B-R:5′-ATGCGTGCAAGAACCCAGCA-3ʹ and miR-431-F: GATG AGTTTTCCCAGGA-3ʹ miR-431-R: GAATCAGTCTGCGTATCCA-3ʹ. The target gene expression level was calculated using the 2-ΔΔCt method [Citation10] as a convenient way to analyze the relative changes in gene expression from real-time quantitative PCR experiments, which was normalized to GAPDH or U6.
MTT assay
Cell proliferation was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyl-2 H-tetrazoliumbromide (MTT) assay. Cells were grown in 96-well plates at a density of 5,000 cells/well. 20 µl MTT solution (5 mg/l; Sigma-Aldrich, St. Louis, MO, USA) was added to each well at 24, 48, 72 and 96 h. After incubation for additional 4 h, the medium was removed, and 150 μl DMSO (Sigma-Aldrich) was added to each well. The absorbance of each well was then read at 570 nm on an ELx808 microplate reader (BioTek Instruments, Inc., Winooski, VT, USA).
Colony formation assay
Cells (n = 500) were plated into the 6-well plate and cultured using complete medium for two weeks. The medium was changed every 3 days. The cell colonies were immobilized with 4% paraformaldehyde and stained with 0.1% crystal violet. Visible colonies were then counted.
Wound healing assay
3 × 105 cells were seeded in a 6-well plate. Once the cultures reached 85% confluency, the artificial wounds was implemented by a sterile 200 μl plastic pipette tips. After rinsing with PBS, the cells were allowed to migrate, and photographs were taken at 0 h and 48 h.
Transwell assay
5 × 104 cells were seeded in the upper chamber of transwell inserts (8 μm pore size; BD Biosciences, San Jose, CA, USA) with serum-free medium. The lower chamber was filled with medium containing 10% FBS as a chemoattractant. After incubation for 24 h, the cells remaining in the top chamber were removed by cotton swabs, while the cells that went through the membrane were fixed by 4% paraformaldehyde and stained with 0.1% crystal violet. The chamber after staining was observed under a microscope.
Western blot analysis
Total protein was extracted using RIPA protein extraction reagent (Beyotime, Shanghai, China). Protein lysates were separated by gel electrophoresis, and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Bradford, MA, USA). Next, the membranes were blocked in 5% nonfat milk for 1.5 h, and then incubated with primary anti-GAPDH antibody (1:1000, Invitrogen, Carlsbad, CA, USA), and anti-E-cadherin antibody (610181A, BD Biosciences, San Jose, CA, USA) at 4°C overnight and then incubated with anti-rabbit antibodies (1:5,000; Santa Cruz Biotechnology, Inc.) were used as the secondary antibodies at room temperature for 2 h. Then, the membranes were incubated with secondary antibodies for 2 h. The protein bands were visualized using an enhanced chemiluminescence kit (Santa Cruz Biotechnology, Inc., Dallas, TX, USA). GAPDH was used as a protein-loading control.
Dual-luciferase reporter assay
The sequence of circMAT2B or ZEB1 mRNA containing wild-type (wt) or mutant (mut) miR-431 binding sites was amplified by PCR and inserted into the pmiR-RB-REPORT™ luciferase reporter vector (Guangzhou RiboBio Co., Ltd.). Cells were seeded in a 24-well plate and co-transfected with the corresponding plasmids and miR-431 mimics or mimics NC using Lipofectamine 2000 reagent. After 48 h, the luciferase activities were analyzed with the Dual Luciferase Reporter Assay System (Promega).
RNA immunoprecipitation (RIP) assay
RIP assay was performed by using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore). Then the cell lysates were incubated with magnetic beads which are conjugated with human anti-Argonaute2 (AGO2) antibody (Millipore, Billerica, MA, USA) or negative control IgG at 4 °C overnight. Finally, the RNA in beads complexes was isolated and subjected to RT-qPCR analysis.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 6.0 software (GraphPad Software Inc., San Diego, CA, USA) and SPSS 18.0 software (SPSS Inc., Chicago, IL, USA). The significant difference was calculated by Student’s t test and one-way analysis of variance (ANOVA) between groups. Kolmogorov-Smirnov test was used to determine the normality of the distribution of data in each group. Survival curves were generated using the Kaplan-Meier method and log-rank test. P values of less than 0.05 were considered as statistically significance.
Results
circMAT2B is upregulated in NSCLC
Through RT-qPCR analysis, we found that, compared with adjacent normal tissues, circMAT2B was significantly upregulated in NSCLC tissues ()). Besides, as shown in ), compared with normal 16HBE cells, NSCLC cell lines (A549, H1299, H1975 and SPCA1) displayed higher expression of circMAT2B. We then evaluated the clinical value of circMAT2B in NSCLC. These 106 NSCLC patients were allocated into two groups based on the mean expression of circMAT2B, and as shown in , high expression of circMAT2B was significantly correlated with large tumor size (P= 0.032) and advanced TNM stage (P= 0.004) of NSCLC patients. Kaplan-Meier survival analysis showed that high expression of circMAT2B was closely associated with poor overall survival of NSCLC patients (P= 0.026; )).
Table 1. Relationship between circMAT2B expression and clinicopathological factors of NSCLC patients
Figure 1. circMAT2B is upregulated in NSCLC. (a) The expression of circMAT2B in NSCLC tissues and adjacent normal tissues, detected by RT-qPCR analysis. (b) The expression of circMAT2B in NSCLC cell lines and 16HBE cells. (c) The relationship between circMAT2B expression and overall survival of NSCLC patients. *P< 0.05 vs. normal tissues or 16HBE cells
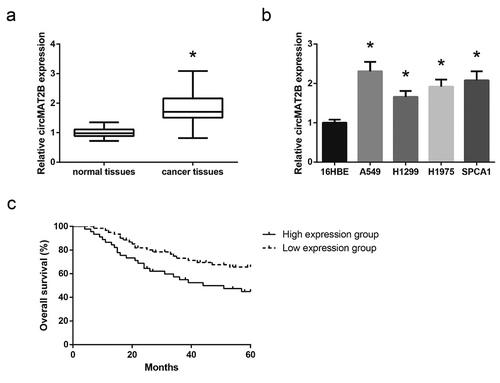
circMAT2B knockdown inhibits NSCLC cell proliferation and invasion
A549 cells were then transfected with the siRNA that specifically targeted the junction site of circMAT2B, and 48 h post-transfection, circMAT2B was knocked-down by over 60% ()). By MTT assay, we observed a significant decrease in the proliferation of A549 cells following circMAT2B knockdown ()). circMAT2B knockdown also greatly decreased the numbers of visible colonies ()). In addition, as indicated by wound healing assay and transwell assay, circMAT2B knockdown also led to the impairment of migration and invasion in A549 cells ()).
Figure 2. circMAT2B knockdown inhibits NSCLC cell proliferation and invasion. (a) The expression of circMAT2B in A549 cells after transfection. (b) The proliferation of A549 cells after transfection, detected by MTT assay. (c) The colony-forming capacity of A549 cells after transfection, detected by colony formation assay. (d) The migration of A549 cells after transfection, detected by wound healing assay. (e) The migration and invasion of A549 cells after transfection, detected by transwell assay. *P< 0.05 vs. si-NC-transfected cells
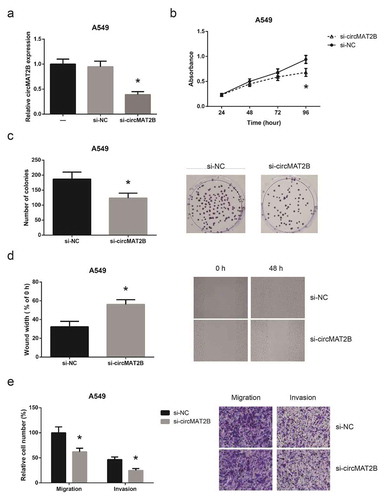
circMAT2B overexpression promotes NSCLC cell proliferation and invasion
We then overexpressed circMAT2B in H1299 cells ()), and we observed that overexpression of circMAT2B markedly promoted the proliferation of H1299 cells ()). The colony-forming capacity of H1299 was also significantly enhanced by circMAT2B overexpression ()). We then noticed that the migration and invasion of H1299 cells were notably promoted when circMAT2B was overexpressed ()).
Figure 3. circMAT2B overexpression promotes NSCLC cell proliferation and invasion. (a) The expression of circMAT2B in H1299 cells after transfection. (b) The proliferation of H1299 cells after transfection. (c) The colony-forming capacity of H1299 cells after transfection. (d) The migration of H1299 cells after transfection. (e) The migration and invasion of H1299 cells after transfection. *P< 0.05 vs. empty vector-transfected cells
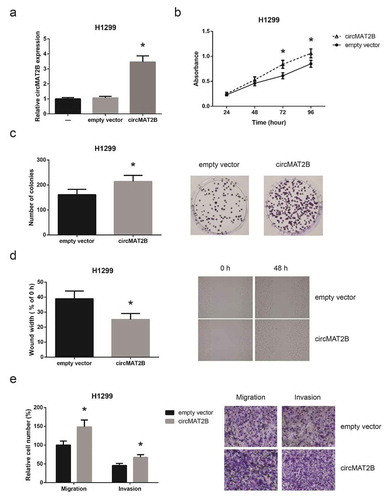
circMAT2B directly binds to miR-431 and decreases miR-431 expression in NSCLC
We further found that circMAT2B was preferentially localized in the cytoplasm of A549 and H1299 cells ()), providing a prerequisite for its function as a ceRNA. According to the Starbase online software (http://starbase.sysu.edu.cn/index.php), circMAT2B possessed a potential complementary sequence to miR-431 seed region ()). We further confirmed that miR-431 was much lower in NSCLC tissues than in normal adjacent tissues ()). The expression of circMAT2B and miR-431 showed a significant inverse correlation in NSCLC tissues ()). Moreover, as shown in ), circMAT2B knockdown increased miR-431 expression in A549 cells; in contrast, miR-431 expression was decreased in H1299 cells with circMAT2B overexpression. We then confirmed the binding interaction by dual-luciferase reporter assay, and the results demonstrated that co-transfection of miR-431 mimics reduced the luciferase reporter activity of circMAT2B-wt by approximately 50% in A549 and H1299 cells, while it had no evident effect on that of circMAT2B-mut ()). RIP assay further showed that circMAT2B and miR-431 were largely enriched by anti-AGO2 in comparison to anti-IgG ()).
Figure 4. circMAT2B directly binds to miR-431 and decreases miR-431 expression in NSCLC. (a) The subcellular localization of circMAT2B in A549 and H1299 cells. (b) The putative miR-431 binding site in circMAT2B sequence. (c) The expression of miR-431 in NSCLC tissues and adjacent normal tissues. (d) The negative correlation between the expression of circMAT2B and miR-431 in NSCLC tissues. (e) The expression of miR-431 in A549 and H1299 cells after transfection. (f) The luciferase reporter activity of circMAT2B-wt or circMAT2B-mut in A549 and H1299 cells after co-transfection. (g) The amount of circMAT2B and miR-431 pulled down by AGO2 or IgG in A549 and H1299 cells. *P< 0.05 vs. normal tissues or mimics NC-transfected cells; #P< 0.05 vs. si-NC or empty vector-transfected cells; @P< 0.05 vs. IgG group
miR-431 restoration blocks circMAT2B function in NSCLC cells
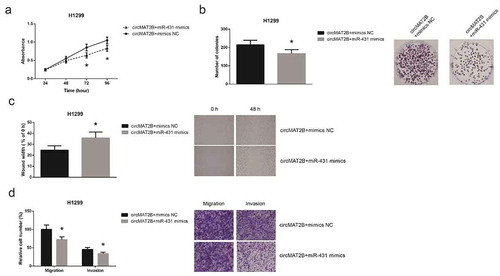
To explore whether circMAT2B exerts its oncogenic role in NSCLC partly by interacting with miR-431, rescue experiments were then performed. As shown in ), the enhanced proliferation and colony-forming capacity of H1299 cells were effectively abrogated by co-transfection with miR-431 mimics. Moreover, miR-431 restoration also remarkably diminished the effects of circMAT2B on the migration and invasion of H1299 cells ()).
Figure 5. miR-431 restoration blocks circMAT2B function in NSCLC cells. (a) The proliferation of H1299 cells after co-transfection. (b) The colony-forming capacity of H1299 cells after co-transfection. (c) The migration of H1299 cells after co-transfection. (d) The migration and invasion of H1299 cells after co-transfection. *P< 0.05 vs. circMAT2B overexpression plasmid+mimics NC-transfected cells
circMAT2B promotes EMT in NSCLC cells by sponging miR-431
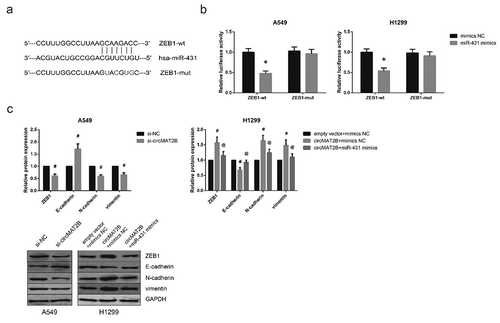
Through the online database TargetScan (http://www.targetscan.org/vert_71/), ZEB1 was predicted as a potential target gene of miR-431 ()), and dual-luciferase reporter assay further showed that co-transfection with miR-431 mimics remarkably decreased the luciferase activity of ZEB1-wt but not ZEB1-mut in A549 and H1299 cells ()). Moreover, through western blot analysis, we observed that circMAT2B knockdown markedly increased E-cadherin expression, while decreased the expression levels of ZEB1, N-cadherin and vimentin in A549 cells ()). In addition, the enhanced EMT of H1299 cells with circMAT2B overexpression was notably diminished by miR-431 restoration.
Figure 6. circMAT2B promotes EMT in NSCLC cells by sponging miR-431. (a) The putative miR-431 binding site in ZEB1 mRNA sequence. (b) The luciferase reporter activity of ZEB1-wt or ZEB1-mut in A549 and H1299 cells after co-transfection. (c) The expression of EMT-related proteins in A549 and H1299 cells after transfection, detected by western blot analysis. *P< 0.05 vs. mimics NC-transfected cells; #P< 0.05 vs. si-NC or empty vector+mimics NC-transfected cells; @P< 0.05 vs. circMAT2B overexpression plasmid+mimics NC-transfected cells
Discussion
NSCLC remains a major health problem. To date, many circRNAs have been reported to serve as oncogenic stimuli or tumor suppressors in the progression of NSCLC. For example, Zhang et al. reported that circular RNA ZFR exhibited a carcinogenic role in NSCLC [Citation11], while Wei et al. showed the role of circPTPRA in suppressing NSCLC progression [Citation12]. circRNAs have potential as biomarkers for the diagnosis, prognosis, and treatment of NSCLC [Citation13].
Our investigations identified a novel circRNA, circMAT2B, in NSCLC progression. circMAT2B was previously identified as an oncogene in some human cancers, and herein, we also showed that circMAT2B was highly expressed and associated with adverse clinicopathological factors and poor prognosis of patients in NSCLC. Functionally, circMAT2B knockdown suppressed the malignant traits of NSCLC cells, while circMAT2B overexpression led to the opposing results. Metastasis is a major cause of cancer-related deaths, and EMT is a central mechanism contributing to cancer metastasis [Citation14,Citation15]. This process is accompanied by the loss of epithelial markers and a gain in mesenchymal markers. This study also verified that circMAT2B is closely related to EMT in NSCLC.
Several lines of studies have demonstrated that circRNAs can function as miRNA “sponges” to regulate target mRNAs [Citation16]. Xu et al. previously identified miR-431 as a tumor suppressor in lung cancer [Citation17], and through bioinformatics prediction and further experimental verification, this study confirmed that miR-431 could be sponged and negatively regulated by circMAT2B in NSCLC. We further found that restoration of miR-431 reversed the oncogenic effects of circMAT2B overexpression in NSCLC. ZEB1 is a transcription factor in EMT, and its increased expression is associated with metastasis in lung cancer [Citation18]. ZEB1 was previously identified as a target of miR-431 in hepatocellular carcinoma [Citation19], and herein, this binding relation was also confirmed in NSCLC.
In summary, we firstly demonstrated that circMAT2B functions as a tumor promoter in NSCLC partly by sponging miR-431. Our findings provided a novel molecular mechanism underlying the progression of NSCLC. circMAT2B could serve as a promising therapeutic target for NSCLC patients.
Authors’ contributions
HW,LF,DC,YZ,YX, and BF conceived and designed the experiments, performed the analysis;, contributed to references collecting and contributed to writing. All authors read and approved the final manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure statement
The authors declare that they have no competing interests.
References
- Inamura K. Lung cancer: understanding its molecular pathology and the 2015 WHO classification. Front Oncol. 2017;7:193.
- Gridelli C, Rossi A, Carbone DP, et al. Non-small-cell lung cancer. Nat Rev Dis Primers. 2015;1:15009.
- Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148.
- Cocquerelle C, Mascrez B, Hetuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160.
- Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14(5):514–521.
- Li Q, Pan X, Zhu D, et al. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70(4):1298–1316.
- Zhao JP, Chen LL. Circular RNA MAT2B induces colorectal cancer proliferation via sponging miR-610, resulting in an increased E2F1 expression. Cancer Manag Res. 2020;12:7107–7116.
- Yu K, Park S, Chang Y, et al. Evaluation of commercial complementary DNA synthesis kits for detecting human papillomavirus. Korean J Clin Lab Sci. 2019;51:309–315.
- Zhang C, Gao GR, Lv CG, et al. Protease-activated receptor-2 induces expression of vascular endothelial growth factor and cyclooxygenase-2 via the mitogen-activated protein kinase pathway in gastric cancer cells. Oncol Rep. 2012 Nov;28(5):1917–1923.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- Zhang H, Wang X, Hu B, et al. RNA ZFR accelerates non-small cell lung cancer progression by acting as a miR-101-3p sponge to enhance CUL4B expression. Artif Cells Nanomed Biotechnol. 2019;47(1):3410–3416.
- Wei S, Zheng Y, Jiang Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193.
- Li C, Zhang L, Meng G, et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2019;145(12):2875–2889.
- Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer. 2006;6(6):449–458.
- Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu Rev Pathol. 2018;13:395–412.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
- Xu CM, Chen LX, Gao F, et al. MiR-431 suppresses proliferation and metastasis of lung cancer via down-regulating DDX5. Eur Rev Med Pharmacol Sci. 2019;23(2):699–707.
- Larsen JE, Nathan V, Osborne JK, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest. 2016;126(9):3219–3235.
- Sun K, Zeng T, Huang D, et al. MicroRNA-431 inhibits migration and invasion of hepatocellular carcinoma cells by targeting the ZEB1-mediated epithelial-mensenchymal transition. FEBS Open Bio. 2015;5:900–907.
