ABSTRACT
Long non-coding RNAs (lncRNAs) are crucial drivers in the progression of human diseases such as myocardial infarction (MI). However, the impact of lncRNA MCM3AP antisense RNA 1 (MCM3AP-AS1) on MI remains unknown. This research was determined to explore the effect of MCM3AP-AS1 modulating microRNA-24-3p (miR-24-3p) and eukaryotic translation initiation factor 4 gamma 2 (EIF4G2) on MI. The rat MI models were constructed and, respectively, treated with altered MCM3AP-AS1, miR-24-3p or/and EIF4G2. Subsequently, the cardiac function, myocardial pathological injury, malondialdehyde content and superoxide dismutase activity were determined. The vascular endothelial cells (VECs) were isolated and treated severally, and then proliferation and migration of VECs were measured. MCM3AP-AS1, miR-24-3p, EIF4G2 and vascular endothelial growth factor (VEGF) expressions in myocardial tissues and VECs were assessed. MCM3AP-AS1 and EIF4G2 were upregulated while miR-24-3p and VEGF were downregulated in MI rat myocardial tissues. MCM3AP-AS1 silencing or miR-24-3p elevation improved cardiac function and myocardial pathological injury, suppressed malondialdehyde content, and also enhanced VEGF expression and superoxide dismutase activity in MI rats. In VECs, downregulated MCM3AP-AS1 or upregulated miR-24-3p accelerated cell proliferation and migration. These effects of miR-24-3p upregulation were reversed by overexpressed EIF4G2. Our study summarizes that reduced MCM3AP-AS1 elevates miR-24-3p to promote proliferation and migration of MI rat VECs by inhibiting EIF4G2.
Introduction
Cardiovascular disease is the prevalent cause of death worldwide and causes almost one-third of all deaths [Citation1]. Myocardial infarction (MI) results from the ruptured atherosclerotic plaque resulting in thrombus formation in the lumen of a coronary vessel, which blocks the blood flow to distal myocardium [Citation2]. Infarction contributes to cardiomyocyte apoptosis and necrosis of the tissue in the infarcted area, attracting inflammatory cells that phagocytose dead cells and debris [Citation3]. Although the 30-day mortality of MI has decreased in recent decades, it remains remarkably at 7.8% [Citation4]. Although the broad use of primary percutaneous coronary intervention and improved periprocedural care could improve the survival of MI patients [Citation5], there is still a need for exploring novel biomarkers for MI treatment.
Long noncoding RNAs (lncRNAs), non-protein coding RNAs with >200 nucleotides in length, participate in multiple biological processes [Citation6]. Micro-chromosome maintenance protein 3 (MCM3) refers to an essential modulator in DNA replication. MCM3AP is acetylated MCM3 with the combination of chromatin, and MCM3AP-AS1 is a lncRNA antisense to human MCM3AP gene [Citation7]. MCM3AP-AS1 usually serves as an oncogene during cancer development, such as gastric cancer [Citation8] and prostate cancer [Citation9]. Nevertheless, the impact of MCM3AP-AS1 on the progression of MI remains unexplored. LncRNAs were reported to function as competing endogenous RNAs (ceRNAs) of microRNAs (miRNAs) to play a post-transcriptional regulatory role in gene expression [Citation10]. MiRNAs are noncoding RNAs with a length of 22 nt that repress gene expression via binding the 3ʹ-untranslated region (3ʹUTR) of the target mRNA [Citation11]. MiR-24-3p, one of the miRNAs, has been verified to have cardioprotective impacts on myocardial ischemia/reperfusion (I/R) injury [Citation12]. Moreover, it has also been identified as a biomarker of early detection of acute kidney injury after acute MI [Citation13]. miR-24-3p has a binding relationship with MCM3AP-AS1 through bioinformatics website analysis, there is no related report on MCM3AP-AS1 in myocardial infarction, so MCM3AP-AS1 was chosen as the study subject from innovative considerations. Abnormal expression of eukaryotic initiation factor 4 F (EIF4F) component, such as eukaryotic initiation factor 4 gamma (EIF4G), played a critical role in human diseases [Citation14]. For example, EIF4G2 has been reported to participate in biological processes of glioblastoma cells [Citation15] and osteosarcoma cells [Citation16], and overexpression of EIF4G2 promotes the occurrence of inflammation [Citation17]. Bioinformatics analysis of our study found that EIF4G2 was the target gene of miR-24-3p. For this regard, we aimed to explore the role of MCM3AP-AS1 sponging miR-24-3p during MI development, and we speculated that MCM3AP-AS1 may serve as a ceRNA of miR-24-3p to regulate proliferation and migration of vascular endothelial cells (VECs) via targeting EIF4G2.
Materials and methods
Ethics statement
Animal experiments were strictly in accordance with the Guide to the Management and Use of Laboratory Animals issued by the National Institutes of Health. The protocol of animal experiments was approved by the Institutional Animal Care and Use Committee of The Affiliated Hospital of Hangzhou Normal University.
Experimental animals
Specific pathogen-free (SPF) healthy Sprague Dawley rats weighing 200–220 g (male and female in half) purchased from Hangzhou Ziyuan Laboratory Animal Science and Technology Co., Ltd. (Hangzhou, China) were fed in an SPF environment with temperature at 23°C ± 2°C and free access to food and water.
Establishment of MI rat models
Anesthetized by pentobarbital sodium, the rats were fixed on an operation table in a supine position and connected with an experimental small animal ventilator (Techman Software Co., Ltd., Sichuan, China) through tracheal intubation. Rats’ limbs were connected with a physiological recorder to record the electrocardiogram (ECG) during the surgery. The skin of the 3rd to 4th intercostal of the rat's left chest was cut open and the ribs were separated to expose the heart, and then the left anterior descending (LAD) branch of coronary artery was ligated. ECG was observed and an elevation of ST segment as well as ligation area and heart apex turning white indicated a successful modeling. The incision was sutured and rats without LAD ligation were taken as the controls [Citation18].
Animal grouping
The MI rats (n = 10/group) were, respectively, intramyocardially injected with adenoviruses (150 μL, titer at 1.0 × 1010 PFU/mL), including small interfering RNA (si)-negative control (NC), si-MCM3AP-AS1, agomir NC, miR-24-3p agomir and miR-24-3p agomir + overexpressed (OE)-EIF4G2. Three days after the injection, rats were performed with modeling surgery [Citation19]. The sham group (n = 10) was taken as control (rats were performed with thoracotomy but not ligation). The corresponding adenoviruses were gained from GenePharma Co., Ltd. (Shanghai, China). The rats were grouped as following: MI, si-NC, si-MCM3AP-AS1, agomir NC, miR-24-3p agomir and miR-24-3p agomir + OE-EIF4G2 groups.
Hemodynamic examination
Rats were connected with a ventilator with their limbs connected with ECG monitoring electrodes. A median incision was made on rats’ neck to expose the common carotid artery for passive isolation. Rats were given intravenous injection of heparin and a self-made catheter was used to connect a pressure sensor with the rat's left ventricle through the right carotid artery. The left ventricle systolic pressure (LVSP), left ventricular end diastolic pressure (LVEDP) and the maximum left ventricular pressure rising and dropping rates (± dp/dtmax) were determined.
Collection of myocardial tissue and blood samples
Two hours after modeling, rats were anesthetized by pentobarbital sodium and the hearts were harvested. Myocardial tissues from five rats were fixed with 4% paraformaldehyde for 8 h, normally dehydrated, permeabilized, embedded and sectioned (5-μm thick) for the hematoxylin-eosin (HE) staining; myocardial tissues from another five rats were preserved in liquid nitrogen and transferred into an ultra-low temperature refrigerator for molecular biological detection.
Measurement of malondialdehyde (MDA) content and superoxide dismutase (SOD) activity
Ischemic myocardial tissues were homogenated with lysis buffer (20 mg tissue was added with 150 μL lysis buffer) and normal saline, and the homogenate was centrifuged at 15,000 r/min and 4°C for 5 min. The supernatant was collected for determination of MDA content (thiobarbituric acid method) and SOD activity (kit method). The kits were obtained from JianCheng Bioengineering Institute (Nanjing, China).
HE staining
The myocardial tissue sections were normally dehydrated and permeabilized. The HE staining was conducted as previously described [Citation20] and the results were observed and photographed through a microscope.
Isolation and grouping of vascular endothelial cells (VECs)
Myocardial tissues from MI rats were treated with enzyme digestion solution for 15 min. After natural deposition for 1 min, the supernatant was removed and the centrifugation samples were conducted with primary cell culture. VECs were seeded onto 6-well plates when reached 80–90% cell confluence. The transfection was performed based on protocols of Lipofectamine 2000 reagent (Invitrogen, CA, USA). Serum-free opti minimum Eagle’s medium (MEM, 250 mL, GIBCO, NY, USA) was used to dilute 100 pmol si-NC, si-MCM3AP-AS1, mimic NC, miR-24-3p mimic and miR-24-3p mimic + OE-EIF4G2. The Lipofectamine 2000 reagent (5 mL) was also diluted by 250 mL serum-free opti MEM and the above two were mixed up. The medium was changed after 6 h; and after 48 h of culture, the cells were collected for subsequent experiments. The cells were finally separated into six groups: the blank, si-NC, si-MCM3AP-AS1, mimic NC, miR-24-3p mimic and miR-24-3p mimic + OE-EIF4G2 groups.
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay
VECs were seeded and added with MTT solution (5 mg/mL) for 4-h culture. Next, each well was added with 100 μL dimethyl sulfoxide for 10-min shaking. A microplate reader was used for measurement of optical density (OD) value at 490 nm. This assay was performed based on MTT kit instructions (Beyotime Institute of Biotechnology, Shanghai, China).
Transwell assay
The cell concentration was adjusted to 1 × 106 cells/mL by serum-free medium, and 100 μL cell suspension was seeded into Transwell apical chambers. The basolateral chambers were added with 500 μL cull culture solution for 48-h culture. The non-migrative cells were removed and the chambers were fixed with 40 mL/L paraformaldehyde for 15 min, stained with hematoxylin for 5 min and observed under an inverted microscope. The number of transmembrane cells in three optical fields from each sample was counted.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNAs from tissues and cells were extracted by Trizol kits and reversely transcribed into cDNAs. Subsequently, PCR amplification were performed, and the data were analyzed using 2−ΔΔCt method. The primers () were designed by TaKaRa.
Table 1. Primer sequence for RT-qPCR
Western blot analysis
Total proteins were extracted using radio-immunoprecipitation assay cell lysis buffer (Beyotime). Afterward, the proteins were performed with 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis for 2 h, transferred onto membranes, and blocked with 5% skim milk powder for 2 h, and then incubated with primary antibodies EIF4G2, vascular endothelial growth factor (VEGF), and glyceraldehyde phosphate dehydrogenase (GAPDH) (all 1:1000 and from Santa Cruz Biotechnology, Inc, CA, USA). Relative secondary antibody was added for reaction and enhanced chemiluminescent method was applied for development. The gray values of protein band was assessed with GAPDH as the loading control, and then the relative protein expression was calculated.
Dual luciferase reporter gene assay
Binding sites of MCM3AP-AS1 and miR-24-3p were predicted at StarBase (http://starbase.sysu.edu.cn/). The fragments of lncRNA MCM3AP-AS1 containing the predicted binding sites of miR-24-3p were amplified by PCR and cloned into Hind III –Mlu I restriction sites of the pMIR reporter vector (Promega, Madison, WI, USA). Meanwhile, the QuikChange Multi Site-Directed Mutagenesis kit (Stratagene, Lajolla, CA, USA) was employed to construct a mutant of lncRNA MCM3AP-AS1. MiR-24-3p mimic and its NC (GenePharma) were severally co-transfected with MCM3AP-AS1-wile type (WT) or MCM3AP-AS1-mutant type (MUT) vector into the VECs. Cells were transfected for 48 h, and the luciferase activity was determined using the luciferase detection kits (Promega, WI, USA) and an ultra-micro ultraviolet photometer (Bio-Rad Laboratories, CA, USA).
Binding region of miR-24-3p and EIF4G2 mRNA 3ʹUTR was also predicted at StarBase (http://starbase.sysu.edu.cn/). WT fragments of the 3′UTR of EIF4G2 with putative binding sites of miR-24-3p were cloned into pMIR reporter vector. EIF4G2-3′UTR-MUT reporter containing mutant miR-24-3p binding sites was used and generated using QuikChange Multi Site-Directed Mutagenesis kit.P Cultured VECs were, respectively, co-transfected with miR-24-3p mimic and EIF4G2-3ʹUTR-WT, miR-24-3p mimic and EIF4G2-3ʹUTR-MUT, mimic NC and EIF4G2-3ʹUTR-WT, and mimic NC and EIF4G2-3ʹUTR-MUT for 48 h. The luciferase activity was determined using the luciferase detection kits (Promega).
RNA pull-down assay
Three diverse biotin-labeled miRNA sequences WT miR-24-3p (Bio-miR-24-3p-WT), MUT miR-24-3p (Bio-miR-24-3p-MUT, sequence mutation that complementary to MCM3AP-AS1), and a random miRNA that did not complementary to MCM3AP-AS1 (Bio-NC) were designed and synthesized by GenePharma. The miRNAs were transfected into VECs for 48 h when the cell confluence reached 80–90%. Subsequently, cells were lysed using lysis buffer and the lysate was co-cultured with magnetic beads coated with M-280 streptavidin (Sigma-Aldrich Chemical Company, MO, USA) at 4°C for 3 h. The beads were rinsed and protein-nucleotide complex absorbed by the beads were eluted. Trizol was used to extract the total RNA, and MCM3AP-AS1 expression was assessed using RT-qPCR.
Statistical analysis
All data analyses were conducted using SPSS 21.0 software (IBM Corp. Armonk, NY, USA). The data were expressed as mean ± standard deviation. The unpaired t-test was performed for comparisons between two groups, one-way analysis of variance (ANOVA) was used for comparisons among multiple groups and Tukey’s post hoc test was used for pairwise comparisons after one-way ANOVA. P value < 0.05 was indicative of a statistically significant difference.
Results
Silenced MCM3AP-AS1 or elevated miR-24-3p improves cardiac function of MI rats
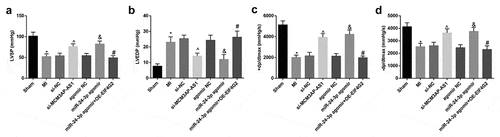
Results of hemodynamic determination () reflected that MI rats had increased LVEDP and decreased LVSP and ± dp/dtmax; injection with si-MCM3AP-AS1 or miR-24-3p agomir decreased LVEDP while increased LVSP and ± dp/dtmax in MI rats; effects of miR-24-3p agomir on these indices were reversed by overexpressed EIF4G2. These results indicated that silenced MCM3AP-AS1 or elevated miR-24-3p improved cardiac function of MI rats and overexpression of EIF4G2 reversed the role of elevated miR-24-3p in cardiac function of MI rats.
Figure 1. Silenced MCM3AP-AS1 or elevated miR–24–3p improves cardiac function of MI rats. A, LVSP of rats in each group; B, LVEDP of rats in each group; C, + dp/dtmax of rats in each group; D, – dp/dtmax of rats in each group; * P < 0.05 vs the sham group, ^ P < 0.05 vs the si-NC group, & P < 0.05 vs the agomir NC group, # P < 0.05 vs the miR–24–3p agomir group; n= 10; the data were expressed as mean ± standard deviation, one-way ANOVA was used for comparisons among multiple groups and Tukey’s post hoc test was used for pairwise comparisons after one-way ANOVA.
Downregulated MCM3AP-AS1 or enhanced miR-24-3p attenuates myocardial injury in MI rats
HE staining showed that () the myocardial strains were clear and not fractured in the sham group, and there were no inflamma-tory cell infiltration and necrosis in myocardial interstitium; rats in the MI, si-NC, agomir NC and miR-24-3p agomir + OE-EIF4G2 groups showed necrotic and fractured fibers, broadened myocardial interstitium and inflammatory cell infiltration in the myocardial tissues; rats in the si-MCM3AP-AS1 and miR-4-3p agomir groups showed partial necrosis and fracture in myocardial fibers, and the inflammatory cells in myocardial interstitium were decreased.
Figure 2. Downregulated MCM3AP-AS1 or enhanced miR–24–3p attenuates myocardial injury in MI rats. A, representative images of HE staining (× 200); B, SOD activity in rat myocardial tissues; C, MDA content in rat myocardial tissues; * P < 0.05 vs the sham group, ^ P < 0.05 vs the si-NC group, & P < 0.05 vs the agomir NC group, # P < 0.05 vs the miR–24–3p agomir group; n = 5; the data were expressed as mean ± standard deviation, one-way ANOVA was used for comparisons among multiple groups and Tukey’s post hoc test was used for pairwise comparisons after one-way ANOVA.
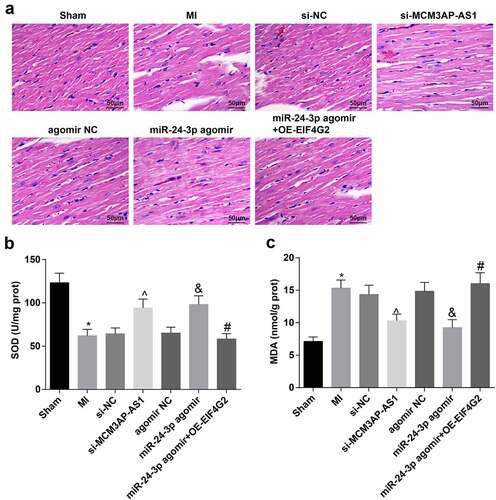
SOD activity and MDA content in rat myocardial tissues were assessed and we found that () MI rats had increased MDA content and decreased SOD activity; after treatment with si-MCM3AP-AS1 or miR-24-3p agomir, MDA content was suppressed and SOD activity was promoted; treatment with OE-EIF4G2 could mitigate the effect of miR-24-3p agomir on MDA content and SOD activity. The above data reflected that downregulated MCM3AP-AS1 or enhanced miR-24-3p attenuated myocardial injury in MI rats, and the impact of miR-24-3p was reversed by overexpressed EIF4G2.
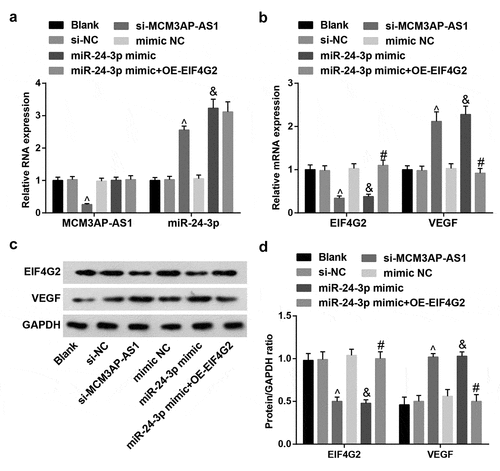
MCM3AP-AS1 and EIF4G2 are upregulated while miR-24-3p and VEGF are downregulated in MI rat myocardial tissues
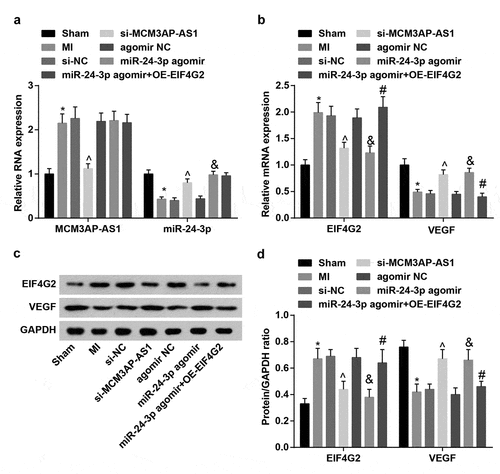
Expression of MCM3AP-AS1, miR-24-3p, EIF4G2 and VEGF in rat myocardial tissues was determined and we observed that () MI rats had higher MCM3AP-AS1 and EIF4G2 expression and lower miR-24-3p and VEGF expression; in response to si-MCM3AP-AS1 injection, MCM3AP-AS1 and EIF4G2 expression was inhibited while miR-24-3p and VEGF expression was elevated in MI rats; it was determined that miR-24-3p agomir treatment downregulated EIF4G2 but upregulated miR-24-3p and VEGF expression; OE-EIF4G2 reversed the impact of miR-24-3p agomir on EIF4G2 and VEGF expression.
Figure 3. MCM3AP-AS1 and EIF4G2 are upregulated while miR–24–3p and VEGF are downregulated in MI rat myocardial tissues. A, expression of MCM3AP-AS1 and miR–24–3p in rat myocardial tissues; B, expression of EIF4G2 and VEGF in rat myocardial tissues; C, protein bands of EIF4G2 and VEGF; D, protein expression of EIF4G2 and VEGF in rat myocardial tissues; * P < 0.05 vs the sham group, ^ P < 0.05 vs the si-NC group, & P < 0.05 vs the agomir NC group, # P < 0.05 vs the miR–24–3p agomir group; n = 5; the data were expressed as mean ± standard deviation, one-way ANOVA was used for comparisons among multiple groups and Tukey’s post hoc test was used for pairwise comparisons after one-way ANOVA.
MCM3AP-AS1 silencing or miR-24-3p elevation promotes VEC proliferation and migration
The morphology and growth characteristics of the cells were observed with an inverted microscope. The primary cultured rat VECs grew in a sub-fusion state on the 3rd d; on the 7th day, when the cells covered the bottom wall of the culture flask, the cells showed a spindle shape and grown like a “paving stone” (Supplementary Figure 1). According to the results of MTT assay (), transfection with si-MCM3AP-AS1 or miR-24-3p mimic in VECs resulted in increased OD value; further transfection with OE-EIF4G2 reversed the role of miR-24-3p mimic in VEC proliferation.
Figure 4. MCM3AP-AS1 silencing or miR–24–3p elevation promotes VEC proliferation and migration. A, VEC proliferation was detected by MTT assay; B, VEC migration was detected by transwell assay; C, comparison of migrative VECs among the groups; ^ P < 0.05 vs the si-NC group, & P < 0.05 vs the mimic NC group, # P < 0.05 vs the miR–24–3p mimic group; N= 3; the data were expressed as mean ± standard deviation, one-way ANOVA was used for comparisons among multiple groups and Tukey’s post hoc test was used for pairwise comparisons after one-way ANOVA.
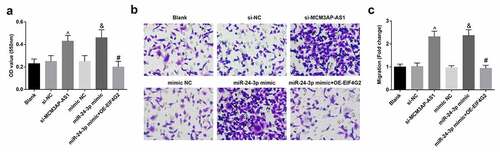
Transwell assay was applied to detect VEC migration and it was found that () for VECs transfected with si-MCM3AP-AS1 or miR-24-3p mimic, number of migrative cells was increased; miR-24-3p mimic-induced promotion of VEC migration was abolished after transfection with OE-EIF4G2. The above outcomes indicated that reduced MCM3AP-AS1 or amplified miR-24-3p accelerated VEC migration, while amplified miR-24-3p-induced promotion on VEC migration was reversed by overexpression of EIF4G2.
These results reflected that MCM3AP-AS1 silencing or miR-24-3p elevation promoted VEC proliferation and migration, and the effect of upregulated miR-24-3p was reversed by overexpressed EIF4G2.
Expression of MCM3AP-AS1, miR-24-3p, EIF4G2 and VEGF in VECs
MCM3AP-AS1, miR-24-3p, EIF4G2 and VEGF expression in VECs from each group was assessed. The results () suggested that treatment of si-MCM3AP-AS1 downregulated MCM3AP-AS1 and EIF4G2 while upregulated miR-24-3p and VEGF; miR-24-3p mimic downregulated EIF4G2 but upregulated miR-24-3p and VEGF; OE-EIF4G2 impaired miR-24-3p mimic-mediated effect on EIF4G2 and VEGF expression.
Figure 5. Expression of MCM3AP-AS1, miR–24–3p, EIF4G2 and VEGF in VECs. A, expression of MCM3AP-AS1 and miR–24–3p in VECs of each group; B, expression of EIF4G2 and VEGF in VECs of each group; C, protein bands of EIF4G2 and VEGF; D, protein expression of EIF4G2 and VEGF in VECs of each group; ^ P < 0.05 vs the si-NC group, & P < 0.05 vs the mimic NC group, # P < 0.05 vs the miR–24–3p mimic group; N = 3; the data were expressed as mean ± standard deviation, one-way ANOVA was used for comparisons among multiple groups and Tukey’s post hoc test was used for pairwise comparisons after one-way ANOVA.
Interaction among MCM3AP-AS1, miR-24-3p and EIF4G2
A particular binding region between MCM3AP-AS1 and miR-24-3p sequence was predicted by a bioinformatic software (). Results of dual luciferase reporter gene assay reflected that () miR-24-3p mimic could diminish the luciferase activity of MCM3AP-AS1-WT, indicating a binding relation between MCM3AP-AS1 and miR-24-3p.
Figure 6. Interaction among MCM3AP-AS1, miR–24–3p and EIF4G2. A, binding sites of MCM3AP-AS1 and miR–24–3p were predicted at http://starbase.sysu.edu.cn/; B, binding relation between MCM3AP-AS1 and miR–24–3p was confirmed by dual luciferase reporter gene assay; C, binding relation between MCM3AP-AS1 and miR–24–3p was confirmed by RNA pull-down assay; D, binding sites of miR–24–3p and EIF4G2 were predicted at http://starbase.sysu.edu.cn/; E, binding relation between miR–24–3p and EIF4G2 was confirmed by dual luciferase reporter gene assay; * P < 0.05 vs the mimic NC group, # P < 0.05 vs the Bio-NC group; N = 3; the data were expressed as mean ± standard deviation, the unpaired t-test was performed for comparisons between two groups, one-way ANOVA was used for comparisons among multiple groups and Tukey’s post hoc test was used for pairwise comparisons after one-way ANOVA.
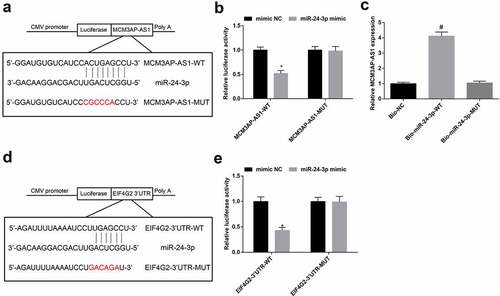
RNA pull-down assay revealed that () MCM3AP-AS1 was enriched by Bio-miR-24-3p-WT, suggesting that MCM3AP-AS1 decreased dissociation degree of miR-24-3p by binding miR-24-3p.
Existence of targeting relationship between miR-24-3p and EIF4G2 was predicted by a bioinformatic software (). It was further confirmed that () the co-transfection of EIF4G2-3ʹUTR-WT and miR-24-3p mimic into VECs repressed luciferase activity, implying that EIF4G2 was targeted by miR-24-3p.
Discussion
MI remains the main cause of cardiac morbidity and mortality worldwide [Citation21]. This research was designed to investigate the impact of the MCM3AP-AS1/miR-24-3p/EIF4G2 axis in the progression of MI, and our study revealed that the knockdown of MCM3AP-AS1 could enhance miR-24-3p expression to promote proliferation and migration of VECs in MI by inhibiting EIF4G2.
We established both animal and cellular models to observe the role of altered expression of MCM3AP-AS1, miR-24-3p and EIF4G2 during MI development. To begin with, MCM3AP-AS1, miR-24-3p and EIF4G2 expression in rat myocardial tissues was assessed. The results of our experiments showed that MCM3AP-AS1 and EIF4G2 were highly expressed while miR-24-3p was poorly expressed in MI rat myocardial tissues. It has also been validated that MCM3AP-AS1 expression was increased in lipopolysaccharide-stimulated chondrocytes, indicating an ectopic expression of MCM3AP-AS1 in osteoarthritis [Citation22]. A recent document has verified that miR-24-3p was lowly expressed in I/R mice [Citation12], and Pan et al. have found that miR-24 was decreased in myocardium after acute MI [Citation23]. We also confirmed that MCM3AP-AS1 acted as a ceRNA to sponge miR-24-3p. A research has revealed that MCM3AP-AS1 played as a ceRNA of miR–194–5p in hepatocellular carcinoma [Citation24], and also sponge miR-93 in cervical squamous cell carcinoma [Citation25]. Nevertheless, the binding relationship between MCM3AP-AS1 and miR-24-3p has not been uncovered yet. As for the abnormal expression of EIF4G2, it has been unraveled that EIF4G2 was upregulated in glioblastoma cells [Citation15]. Furthermore, the target relation between miR–24–3p and EIF4G2 has been confirmed in our study, which has not been unveiled yet.
In our animal experiments, MI rats were treated with altered MCM3AP-AS1 or miR–24–3p to explore their roles during MI in vivo. We found that silenced MCM3AP-AS1 or elevated miR–24–3p improved cardiac function and attenuated myocardial injury and apoptosis in the MI rats. Consistently, Tan et al. have illuminated that miR–24–3p attenuated the infarct area and impairment of cardiac function in a myocardial I/R injury mouse model [Citation12]. A publication has also suggested that miR–24–3p protected cardiomyocytes from I/R injury [Citation26]. The limited literatures showed that the effect of MCM3AP-AS1 on the progression of MI was rarely studied and there needs further explorations. The in vitro cellular experiment has also been performed in our research and the VECs were transfected, respectively. The results indicated that MCM3AP-AS1 knockdown or miR–24–3p overexpression promoted proliferation and migration of the VECs. Similarly, it has been recently identified that the overexpression of MCM3AP-AS1 promoted malignant behaviors of papillary thyroid cancer cells [Citation27], and Li et al. have figured out that MCM3AP-AS1 knockdown constrained lung cancer cell growth [Citation28]. In a study focusing on the role of miR-24 in diabetes, downregulation of miR-24 was found to suppress endothelial cell migration, while upregulation of miR-24 rescued the migration in diabetic endothelial cells [Citation29]. Additionally, Maegdefessel et al. have discovered that miR-24 accelerated migration of aortic smooth muscle cells and promoted adhesion molecule expression in aortic endothelial cells [Citation30]. Moreover, the animal and cellular models were both treated with overexpressed EIF4G2 after treatment of miR–24–3p agomir/mimic to explore its reverse effect. We found that the overexpression of EIF4G2 reversed the protective role of elevated miR–24–3p in MI. A similar finding in a publication implied that inhibition of EIF4G2 reversed effects of miR-139 inhibitor on chondrocyte proliferation and migration [Citation31]. These data are helpful for exploring the roles of MCM3AP-AS1, miR–24–3p and EIF4G2 in MI progression.
In conclusion, we found that MCM3AP-AS1 silencing upregulated miR–24–3p to decelerate MI development and promote proliferation and migration of VECs. Our study may provide novel biomarkers for MI treatment. Nevertheless, the molecular mechanisms remain further investigated.
Supplemental Material
Download MS Word (11.5 KB)Supplemental Material
Download MS Word (173 KB)Acknowledgments
We would like to acknowledge the reviewers for their helpful comments on this paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Mortality GBDC. Causes of death, global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the global burden of disease study 2013. Lancet. 2015;385(9963):117–171.
- Curley D, Lavin Plaza B, Shah AM, et al. Molecular imaging of cardiac remodelling after myocardial infarction. Basic Res Cardiol. 2018;113(2):10.
- Swirski FK, Nahrendorf M. Leukocyte behavior in atherosclerosis, myocardial infarction, and heart failure. Science. 2013;339(6116):161–166.
- Montrief T, Davis WT, Koyfman A, et al. Mechanical, inflammatory, and embolic complications of myocardial infarction: an emergency medicine review. Am J Emerg Med. 2019;37(6):1175–1183.
- McCarthy CP, Vaduganathan M, McCarthy KJ, et al. Left ventricular thrombus after acute myocardial infarction: screening, prevention, and treatment. JAMA Cardiol. 2018;3(7):642–649.
- Wang H, Zheng X, Jin J, et al. LncRNA MALAT1 silencing protects against cerebral ischemia-reperfusion injury through miR-145 to regulate AQP4. J Biomed Sci. 2020;27(1):40.
- Yang C, Zheng J, Xue Y, et al. The effect of MCM3AP-AS1/miR-211/KLF5/AGGF1 axis regulating glioblastoma angiogenesis. Front Mol Neurosci. 2017;10:437.
- Wang H, Xu, T, Wu, L, et al. Molecular mechanisms of MCM3AP-AS1 targeted the regulation of miR–708–5p on cell proliferation and apoptosis in gastric cancer cells. Eur Rev Med Pharmacol Sci. 2020;24(5):2452–2461.
- Li X, Lv J, Liu S. MCM3AP-AS1 KD inhibits proliferation, invasion, and migration of PCa cells via DNMT1/DNMT3 (A/B) methylation-mediated upregulation of NPY1R. Mol Ther Nucleic Acids. 2020;20:265–278.
- Yang B, Gao G, Wang Z, et al. Long non-coding RNA HOTTIP promotes prostate cancer cells proliferation and migration by sponging miR-216a-5p. Biosci Rep. 2018;38(5):5.
- Fang YY, Tan M-R, Zhou J, et al. miR–214–3p inhibits epithelial-to-mesenchymal transition and metastasis of endometrial cancer cells by targeting TWIST1. Onco Targets Ther. 2019;12:9449–9458.
- Tan H, Qi J, Fan B-Y, et al. MicroRNA–24–3p attenuates myocardial ischemia/reperfusion injury by suppressing ripk1 expression in mice. Cell Physiol Biochem. 2018;51(1):46–62.
- Fan PC, Chen -C-C, Peng -C-C, et al. A circulating miRNA signature for early diagnosis of acute kidney injury following acute myocardial infarction. J Transl Med. 2019;17(1):139.
- Hao GJ, Hao H-J, Ding Y-H, et al. Suppression of EIF4G2 by miR-379 potentiates the cisplatin chemosensitivity in nonsmall cell lung cancer cells. FEBS Lett. 2017;591(4):636–645.
- Chai Y, Xie M. LINC01579 promotes cell proliferation by acting as a ceRNA of miR–139–5p to upregulate EIF4G2 expression in glioblastoma. J Cell Physiol. 2019;234(12):23658–23666.
- Xie X, Li Y-S, XiaoW-F, et al. MicroRNA-379 inhibits the proliferation, migration and invasion of human osteosarcoma cells by targeting EIF4G2. Biosci Rep. 2017;37(3):3.
- Gao S, Liu, L, Zhu , S, et al. MicroRNA-197 regulates chondrocyte proliferation, migration, and inflammation in pathogenesis of osteoarthritis by targeting EIF4G2. Biosci Rep. 2020;40(9):9.
- Luo ZR, Li H, Xiao Z-X, et al. Taohong siwu decoction exerts a beneficial effect on cardiac function by possibly improving the microenvironment and decreasing mitochondrial fission after myocardial infarction. Cardiol Res Pract. 2019;2019:5198278.
- Yang Y, Yang J, Liu X-W, et al. Down-regulation of mir-327 alleviates ischemia/reperfusion-induced myocardial damage by targeting RP105. Cell Physiol Biochem. 2018;49(3):1049–1063.
- Lu D, Liao Y, Zhu S-H, et al. Bone-derived Nestin-positive mesenchymal stem cells improve cardiac function via recruiting cardiac endothelial cells after myocardial infarction. Stem Cell Res Ther. 2019;10(1):127.
- Xiao Y, Zhang Y, Chen Y, et al. Inhibition of MicroRNA–9–5p protects against cardiac remodeling following myocardial infarction in mice. Hum Gene Ther. 2019;30(3):286–301.
- Gao Y, ZhaoH, Li Y. LncRNA MCM3AP-AS1 regulates miR–142–3p/HMGB1 to promote LPS-induced chondrocyte apoptosis. BMC Musculoskelet Disord. 2019;20(1):605.
- Pan LJ, Wang, X, Ling, Y, et al. MiR-24 alleviates cardiomyocyte apoptosis after myocardial infarction via targeting BIM. Eur Rev Med Pharmacol Sci. 2017;21(13):3088–3097.
- Wang Y, Yang L, Chen T, et al. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR–194–5p/FOXA1 axis. Mol Cancer. 2019;18(1):28.
- Lan L, Liang Z, Zhao Y, et al. LncRNA MCM3AP-AS1 inhibits cell proliferation in cervical squamous cell carcinoma by down-regulating miRNA-93. Biosci Rep. 2020;40(2):2.
- Xiao X, Lu Z, Lin V, et al. MicroRNA miR–24–3p reduces apoptosis and regulates Keap1-Nrf2 pathway in mouse cardiomyocytes responding to ischemia/reperfusion injury. Oxid Med Cell Longev. 2018;2018:7042105.
- Liang M, Jia J, Chen L, et al. LncRNA MCM3AP-AS1 promotes proliferation and invasion through regulating miR–211–5p/SPARC axis in papillary thyroid cancer. Endocrine. 2019;65(2):318–326.
- Li X, Yu M, Yang C. YY1-mediated overexpression of long noncoding RNA MCM3AP-AS1 accelerates angiogenesis and progression in lung cancer by targeting miR–340–5p/KPNA4 axis. J Cell Biochem. 2020;121(3):2258–2267.
- Cui YX, Hua Y-Z, Wang N, et al. miR-24 suppression of POZ/BTB and AT-hook-containing zinc finger protein 1 (PATZ1) protects endothelial cell from diabetic damage. Biochem Biophys Res Commun. 2016;480(4):682–689.
- Maegdefessel L, Spin JM, Raaz U, et al. miR-24 limits aortic vascular inflammation and murine abdominal aneurysm development. Nat Commun. 2014;5(1):5214.
- Hu W, Zhang W, Li F, et al. miR-139 is up-regulated in osteoarthritis and inhibits chondrocyte proliferation and migration possibly via suppressing EIF4G2 and IGF1R. Biochem Biophys Res Commun. 2016;474(2):296–302.
