ABSTRACT
Accumulating evidence demonstrate that long non-coding RNAs (lncRNAs) play an important role in regulating the biological function of cervical cancer cells. However, the regulatory role of lncRNA Wilms tumor 1 homolog antisense RNA (WT1-AS) in cervical cancer cells remains uncertain. In this study, we explored the participation of WT1-AS in cervical cancer by first using the reverse transcription quantitative polymerase-chain reaction (RT-qPCR) was to analyze the expression of WT1-AS and phosphoinositide-3-kinase adaptor protein 1 (PIK3AP1) in cervical cancer tissues and cells. Dual-luciferase reporter gene assay, RNA pull-down/RNA immunoprecipitation (RIP) assays and Chromatin Immunoprecipitation (ChIP) assay were conducted to explore the interactions among WT1-AS, PIK3AP1, and SPI1. Gain- and loss-of-function approaches were carried out to determine the effects of lncRNA WT1-AS, PIK3AP1 on cell biological characteristics, followed by assays of cell proliferation, autophagy, and apoptosis abilities using, respectively, EdU, monodansylcadaverine (MDC) staining, and flow cytometry. Finally, we measured growth of xenograft tumors in nude mice. We found decreased expression of lncRNA WT1-AS and increased expression of PIK3AP1 in cervical cancer samples. Moreover, PIK3AP1 was negatively regulated by WT1-AS, which promoted apoptosis, but inhibited cell proliferation and autophagy of cervical cancer cells. Furthermore, WT1-AS inhibited PIK3AP1 expression by recruiting SPI1, and inhibited the progression of cervical cancer through the SPI1/PIK3AP1 axis in vivo and in vitro. In summary, lncRNA WT1-AS repressed the development of cervical cancer by reducing PIK3AP1 expression through an interaction with SPI1, which may suggest new therapeutic approaches for treating cervical cancer.
Abbreviations: HPV, human papillomavirus; lncRNAs, Long non-coding RNAs; WT1-AS, wilms tumor 1 antisense RNA; HCC, hepatocellular carcinoma; SFFV, Spleen focus forming virus; SPI1, Spleen focus forming virus proviral integration oncogene 1; TF, transcription factor; PIK3AP1, Phosphoinositide-3-kinase adaptor protein 1; NCBI, National Center for Biotechnology Information; oe, overexpressed; sh-PIK3AP1, short hairpin RNA against PIK3AP1; RIPA, radioimmunoprecipitation; PMSF, phenylmethylsulfonyl fluoride; HRP, horseradish peroxidase; IgG, immunoglobulin G; GAPDH, Glyceraldehyde-3-phosphate dehydrogenase; PCR, polymerase chain reaction; EP, Eppendorf; RIP, RNA-binding protein immunoprecipitation; CHIP, Chromatin immunoprecipitation; EdU, 5-ethynyl-2ʹ-deoxyuridine; PI, propidium iodide; MDC, Monodansylcadaverine; PFA, paraformaldehyde; SPF, specific pathogen free; TV, tumor volume; DLG1-AS1, discs large MAGUK scaffold protein 1 antisense RNA 1; TOB1-AS1, transducer of epidermal growth factor receptor-2.1 antisense RNA 1; LC3II, light chain 3 type II; LC3I, light chain 3 type I; IRF4, interferon regulatory factor 4
Introduction
Cervical cancer remains the fourth leading cause of gynecological cancer-related deaths, with an approximate mortality rate of 311,000 out of 570,000 cases in 2018 worldwide [Citation1]. Chronic human papilloma virus (HPV) infection is the major cause of cervical cancer and, therefore, risk factors of cervical cancer relate to history of HPV infection and/or impaired immunity against HPV infection impairment [Citation2]. Hence, cervical cancer prevention is based on screening and vaccination, and the treatments include radical hysterectomy, chemoradiotherapy, and radiotherapy [Citation3]. Given the inadequate responses to these treatments, we undertook this study aiming to expand our knowledge of cervical cancer at the molecular level and explore new therapeutic targets for its treatment.
Long non-coding RNAs (lncRNAs) are a large class of transcribed RNA molecules with a length of more than 200 nucleotides by which are without capacity of protein-encoding [Citation4]. LncRNAs can play oncogenic roles or act as tumor suppressors in different malignancies through their various interactions with DNA, RNA, or protein [Citation5]. LncRNA Wilms tumor 1 antisense RNA (WT1-AS) has been identified as a suppressor of hepatocellular carcinoma (HCC) through promoting cell apoptosis by binding to WT1, which otherwise exerts an oncogenic role in HCC progression [Citation6]. Furthermore, the lncRNA WT1-AS reportedly negatively regulates the progression of cervical cancer by mediating the miR-203a-5p/FOXN2 axis [Citation7]. Spleen focus forming virus (SFFV) proviral integration oncogene 1 (SPI1), also known as PU.1, is a crucial transcription factor (TF) for the T-cell and hematopoietic lineages [Citation8,Citation9]. Meanwhile, accumulating evidence suggest that SPI1 also has the potential to regulate the progression of malignancies, such as myeloma, glioma, and breast cancer [Citation10–12]. Phosphoinositide-3-kinase adaptor protein 1 (PIK3AP1), also known as B-cell adaptor for PI3K (BCAP), is a specific protein adaptor of B-cells that is expressed in hematopoietic cells [Citation13,Citation14]. Besides, PIK3AP1 has been implicated in the development of gastric cancer [Citation15].
Given this background, we hypothesized that the lncRNA WT1-AS may influence cervical cancer progression through effects on SPI1 and PIK3AP1. We undertook a series of experiments in vivo and in vitro to test this hypothesis, aiming to understand better the molecular mechanisms underlying cervical cancer and provide novel therapeutic targets.
Materials and methods
Ethics statement
All experimental procedures were approved by the Ethics Committee of The First Affiliated Hospital, Hengyang Medical School, University of South China. Written informed consent was obtained from each participant. All animal experiments were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and conformed to relevant national provisions.
Bioinformatics analysis
Cervical cancer-related microarrays were downloaded from the GEO database using cervical cancer as the key word. The affy package of R language was used to standardize the microarray expression data, and the limma package was used to screen the differentially expressed lncRNAs and genes. The corrected p value was represented by adj.P.Val, where the genes with |log2FC| > 1.5 and adj.P.Val < 0.05 were considered to be differentially expressed, and these thresholds were used to draw expression heat maps of lncRNAs and genes. The LncMAP website was further utilized to investigate the possible regulatory function of the lncRNAs in cervical cancer.
Tissue samples
Cervical cancer tissues were collected from 56 patients who had undergone gynecologic surgery in The First Affiliated Hospital of University of South China from January 2015 to January 2018, and had a confirmed diagnosis of cervical cancer by the pathology department of The First Affiliated Hospital of University of South China. The clinical and pathological data of all cases were reviewed by pathologists. The patients aged from 27 to 83 years old with an average age of 45, among which there were 22 cases > 45 years old and 34 cases ≦ 45 years old. In addition, healthy tissues 5 cm away from the cervical cancer margins were also collected. The clinical data are shown in .
Table 1. The relations between WT1-AS expression and clinical pathology factors
Cell culture and transfection
Human cervical cancer cell lines Hela, HCE1, SiHa, and human cervical immortalized epithelial cell line H8 were selected for study. The cells were all obtained from the cell bank of the Cancer Institute of Xiangya School of Medicine (Central South University, Changsha, Hunan, China). RT-qPCR was used to detect expression of lncRNA WT1-AS in these four cell lines, and the cell line with the lowest expression was chosen for further study. Cells were cultured in RPMI 1640 medium supplemented with 10% serum, and incubated at 37°C with 5% CO2. The culture medium was changed every 2 − 3 days based on the growth of the cells, and subculture was performed when the cells reached 80–90% confluence.
According to the known sequences of lncRNA WT1-AS, SPI1, and PIK3AP1 in the National Center for Biotechnology Information (NCBI) database, Shanghai Sangon Biotech Co., Ltd. (Shanghai, China) was commissioned to construct silencing and overexpression plasmids of lncRNA WT1-AS. The Hela cervical cancer cells in logarithmic phase were transfected with overexpressed (oe)-WT1-AS, short hairpin RNA against PIK3AP1 (sh-PIK3AP1), or their negative controls (NC), respectively, or co-transfected with oe-WT1-AS and oe-PIK3AP1 using Lipofectamine 2000 (11,668–019, Invitrogen, Carlsbad, CA, USA).
RT-qPCR
The total RNA of tissues was extracted using a TRIzol kit and was extracted after transfection using a miRNeasy Mini Kit (217,004, QIAGEN, Duesseldorf, Germany). LncRNA WT1-AS and PIK3AP1 primers were designed and synthesized by Takara (Supplementary Table 1). Then, reverse transcription of RNA into cDNA was performed using the PrimeScript RT kit (RR036A, Takara, Tokyo, Japan). The RT-qPCR was carried out following the instructions of the SYBR® Premix Ex TaqTM II Kit (RR820A, TaKaRa, Tokyo, Japan) on an ABI7500 RT-qPCR instrument (7500, ABI, USA). GAPDH was used as an internal reference, and the relative transcription levels of lncRNA WT1-AS and PIK3AP1 were calculated using 2−ΔΔCT method.
Western blot assay
The total protein in the cells was extracted with radioimmunoprecipitation (RIPA) lysate containing phenylmethylsulfonyl fluoride (PMSF) (R0010, Beijing Solarbio Science & Technology Co., Ltd., Beijing, China), and portions were transferred to a polyvinylidene fluoride membrane after sodium dodecyl sulfate polyacrylamide gel electrophoresis. The membrane was blocked and then incubated with diluted mouse anti-human primary antibody PIK3AP1 (0.05 mg/μL, ab165145), Beclin1 (1: 2000, ab207612), LC3B (1.0 μg/mL, ab38394), Caspase 3 (1: 500, ab13847), Bax (1: 5000, ab32503), and Bcl-2 (1: 1000, ab32124) overnight at 4°C, all of the antibodies used above were purchased from Abcam (Cambridge, UK). The membrane was then washed 3 times with TBST for 5 min each time, and incubated with horseradish peroxidase (HRP)-labeled secondary goat anti-mouse immunoglobulin G (IgG) antibody (1: 100, (HA1003, Shanghai Yanhui Biotechnology Co., Ltd., Shanghai, China) for 1 h. The membrane was reacted with electrochemiluminescence solution (ECL 808–25, Biomiga, San Diego, CA, USA) for 1 min, and observed using X-ray screens (36209ES01, Shanghai QCbio Science & Technologies Co. Ltd., Shanghai, China). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was taken as an internal reference, and the relative expression of target protein was analyzed by comparing the gray value of the target band to that of the internal reference.
Bioinformatics prediction and dual luciferase reporter gene assay
The promoter sequences of lncRNA WT1-AS and PIK3AP1 were amplified by PCR. The target fragment was digested with Xho I and Not I endonucleases and then cloned into the downstream region of the luciferase reporter gene pmirGLO (3,577,193, Promega, Madison, WI, USA) to construct pPIK3AP1-Wt. Site-directed mutagenesis was conducted at the binding sites of lncRNA WT1-AS and PIK3AP1 to generate pPIK3AP1-Mut. The two reporter plasmids were co-transfected with WT1-AS mimic and the NC into HEK-293 cells, respectively. After 24 h of transfection, the cells were lysed and centrifuged at 12,000 rpm for 1 min, and the supernatant was collected. Luciferase activity was measured using a dual luciferase reporter system (Dual-Luciferase® Reporter Assay System, E1910, Promega, Madison, WI, USA).
Nuclear and cytoplasmic extraction
The nuclear and cytoplasmic fraction were isolated according to the manufacturer’s instructions of the PARIS kit (Life Technologies corporation, Gaithersburg, MD, USA).
RNA binding protein immunoprecipitation (RIP)
The binding of lncRNA WT1-AS with SPI1 was detected using a RIP kit (Merck Millipore, Billerica, MA, USA). In brief, the cells were lysed with RIPA lysis buffer (P0013B, Beyotime, Shanghai, China) on ice for 5 min, and centrifuged at 14,000 rpm for 10 min at 4°C. Part of the cell extract was taken as an input, and the rest was used for co-precipitation with antibody. Detailed procedures were as follows: 50 μL of magnetic beads were washed and then resuspended in 100 μL of RIP wash buffer, and 5 μg of antibody was added for binding. The magnetic bead-antibody complex was washed and resuspended in 900 μL of RIP wash buffer and incubated with 100 μL of cell extract overnight at 4°C. The sample was then placed on a magnetic stand to collect the magnetic bead-protein complex. The RNA was extracted from sample and input after digestion with proteinase K. LncRNA WT1-AS and PIK3AP1 levels were detected by RT-qPCR. The antibody used for RIP was SPI1 (ab76543, 1: 5000), with immunoglobulin G (IgG, ab109489, 1: 100) serving as NC.
Chromatin immunoprecipitation (CHIP)
CHIP was conducted to verify the binding ability of SPI1 to the PIK3AP1 promoter. Hela cells were plated in a 6-well plate. After fixation with 16% formaldehyde, the cells were lysed, sonicated, and incubated with SPI1 (ab76543, 1: 5000) overnight. Magnetic beads were added to capture the protein DNA-binding complexes, which was then washed with washing solution, and added with 5 mM NaCl for decrosslinking. After completion of decrosslinking, DNA was recovered, and the relative expression of PIK3AP1 in the complex was detected by RT-qPCR (Primer sequences: Left: TAATGGAGGGTTCTGCCTTG; Right: AGAGAGCCAAAGGACAGCAG).
5-ethynyl-2ʹ-deoxyuridine (EdU) assay
EdU solution was added to each group of cells, and the cells were incubated for 2 h before being fixed with 40 g/L paraformaldehyde for 30 min, incubated in a glycine solution for 5 min, and then rinsed with PBS containing 0.5% Triton X-100. Next, Apollo® staining reaction solution was added and incubated at room temperature for 30 min in the dark, and the cells were then washed twice with methanol and PBS. Finally, Hoechst 33,342 reaction solution was added, incubated with cells at room temperature for 30 min in the dark, and the stained cells were observed under a fluorescence microscope. Three fields of view were selected in which EdU-stained cells (proliferating cells) and Hoechst 33,342-stained cells (total cells) were counted, and the cell proliferation rate was calculated as the number of proliferating cells/number of total cells × 100%.
Flow cytometry
Each group of cells was seeded in a 96-well plate at a density of 2.0 × 103 cells/well. After inoculation for 16 h, the cells were replaced with freshly prepared culture solutions containing different concentrations of cisplatin (0, 0.5, 1, 2, 3 μg/mL, 5 wells each). After 5 h of drug treatment, the cells were collected and then resuspended in 200 μL of binding buffer. Next, 10 μL of Annexin-V-fluorescein isothiocyanate (Annexin V-FITC) (ab14085, Abcam) was added into the cells and mixed gently along with 5 μL of propidium iodide (PI). After and reaction at room temperature for 15 min in the dark, 300 μL of binding buffer was added into the cells, and the apoptosis was evaluated by flow cytometry at an excitation wavelength of 488 nm.
Monodansylcadaverine (MDC) staining
Pre-treated sterile coverslips were placed in a 6-well plate, and the prepared cell suspension was added to a 6-well plate (1 × 105/2 mL of medium), and incubated at 37°C with 5% CO2 for 24 h. 2 μL of MDC staining solution was added to each well and incubated at 37°C with 5% CO2 for 20 min. The cells were fixed with 2 mL of 4% paraformaldehyde (PFA) for 15 min, and the PFA was then removed. The MDC staining was observed under a fluorescence microscope. Autophagy rate = number of autophagic vacuoles/total number of cells × 100%.
Xenograft tumor in nude mice
Forty BALB/c-nu/nu nude male mice of specific pathogen-free (SPF) grade, aged 5 weeks, were purchased from Shanghai Slack Laboratory Animal Co., Ltd. (Shanghai, China). Mice were transfected with Hela cells infected with NC (negative control plasmids), oe-WT1-AS, sh-PIK3AP1 or co-transfected with oe-WT1-AS + oe-PIK3AP1, 10 mice each. The cervical cancer cell-line Hela was adjusted to a cell concentration of 2.5 × 107 cells/mL, and 200 μL was subcutaneously inoculated into the neck of the nude mice, which were then reared in the same environment. The tumor formation was observed weekly, and the maximum diameter (a) and the minimum diameter (b) of the tumor nodules of the nude mice were measured for calculation of tumor volume (TV) according to the TV = 1/2 × a × b2, where a and b are the long and short axes. On the 35th day, the nude mice were euthanized, the tumors were dissected, and three tumors were collected from different groups mice to make paraffin sections.
Statistical analysis
All experimental data were analyzed using SPSS 21.0 statistical software (IBM Corp, Armonk, NY, USA). The measurement data were expressed as mean ± standard deviation. Normal distribution and homogeneity of variance were tested. All data were in line with normal distribution and homogeneity of variance. The comparison between two groups in paired design was analyzed by paired t-test. The comparison between two groups in unpaired design was analyzed by unpaired t-test. Data comparisons among multiple groups were performed using one-way analysis of variance (ANOVA) with Tukey’s post hoc test. Data comparison among groups at different time points was performed by repeated measurement ANOVA with Bonferroni’s post hoc test. p < 0.05 was considered statistically significant.
Results
LncRNA WT1-AS is poorly expressed in cervical cancer
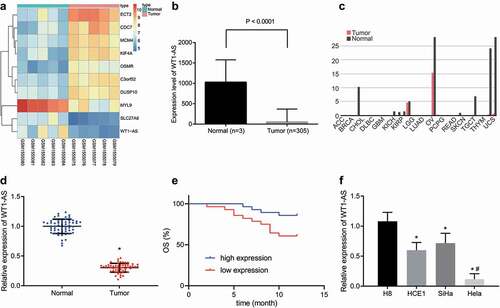
Data analysis of the microarray GSE63678 revealed that the lncRNA WT1-AS was down-regulated in cervical cancer (). By analyzing the cervical cancer data in the TCGA database, we found that expression of lncRNA WT1-AS was also decreased in cervical cancer tissues compared to normal samples (). The expression of lncRNA WT1-AS in other tumors is shown in . RT-qPCR showed that, compared with the adjacent tissues, the expression of lncRNA WT1-AS was significantly lower in human cervical cancer tissues (). As shown in , the expression of lncRNA WT1-AS had no significant correlation with age and lymph node metastasis of cervical cancer patients (p > 0.05) but was closely related to tumor size, differentiation degree, and FIGO classification (p < 0.05). Then, to analyze the relationship between lncRNA WT1-AS expression and overall survival, we used the Kaplan Meier method after dividing the samples into high and low expression groups according to the group mean value of lncRNA WT1-AS expression in the cervical cancer tissues. The results suggested that patients with high lncRNA WT1-AS expression had a longer survival time than patients with low lncRNA WT1-AS expression (). Meanwhile, compared with human immortalized cervical epithelium cell line H8, the expression of lncRNA WT1-AS was also significantly lower in the three human cervical cancer cell lines, among which the expression of lncRNA WT1-AS was lowest in Hela cells ().
Figure 1. LncRNA WT1-AS shows low expression in cervical cancer. a, The screening heatmap of microarray GSE63678. b, The expression of lncRNA WT1-AS in cervical cancer tissues and adjacent tissues analyzed in the TCGA database. c, The expression of lncRNA WT1-AS in several other cancers analyzed in the TCGA database. d, The expression of lncRNA WT1-AS in cancer tissues and adjacent tissues, * p < 0.05 vs. the normal tissues. E, Kaplan Meier method of lncRNA WT1-AS expression and survival in patients with cervical cancer. F, The expression of lncRNA WT1-AS in human immortalized cervical epithelial cells H8 and cervical cancer cells HCE1, SiHa and Hela, * p < 0.05 vs. the H8 cells, # p < 0.05 vs. the HCE1 and SiHa cells. Figure D is analyzed using the paired t test, and figure E using one-way ANOVA with Tukey’s post hoc test. The cell experiments were repeated three times. lncRNA WT1-AS, long non-coding RNA Wilms tumor 1 homolog antisense RNA; PIK3AP1, phosphoinositide-3-kinase adaptor protein 1; SPI1, Spleen focus forming virus proviral integration oncogene 1
LncRNA WT1-AS promotes apoptosis, inhibits cell proliferation and autophagy of cervical cancer cells
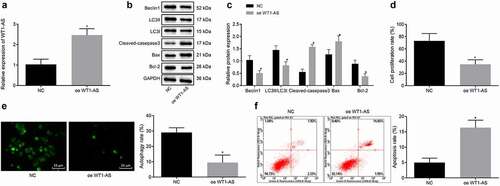
Previous studies have shown that autophagy and apoptosis affect the development of cervical cancer [Citation16,Citation17]. Therefore, we detected the autophagy and apoptosis of cervical cancer cells after overexpression of lncRNA WT1-AS. The results of RT-qPCR and Western blot assay showed that, compared with that of the cells treated with NC, expression of lncRNA WT1-AS was up-regulated, Beclin1, Bcl-1, and LC3II/LC3I expression was down-regulated and Cleaved-caspase3 and Bax were up-regulated in the cells transfected with oe-WT1-AS (p < 0.05) (). The proliferation assay performed using EdU showed that, compared with NC cells, the proliferation rate of the cells transfected with oe-WT1-AS was significantly lower (). The autophagy determined by MDC staining showed that when the cells were under nutritional stress, the autophagy rate of the cells transfected with oe-WT1-AS was significantly decreased (). To investigate the effect of lncRNA WT1-AS overexpression on cell apoptosis, we performed flow cytometry of cells double-stained with Annexin V and PI. Our results showed that the apoptosis rate of the cells transfected with oe-WT1-AS was significantly increased compared with that of the cells treated with NC (). These findings indicate that lncRNA WT1-AS overexpression can increase apoptosis rate while decreasing the proliferation and autophagy of Hela cells.
Figure 2. LncRNA WT1-AS suppressed proliferation and autophagy while promoted apoptosis of cervical cancer cells. a, The expression of lncRNA WT1-AS measured by RT-qPCR. b-c, The protein expression of Beclin1, Bcl-1, LC3II, LC3I, Cleaved-caspase3 and Bax determined by Western blot assay. d, The proliferation rate of cancer cells transfected with oe-WT1-AS detected by EdU staining. e, The autophagy of cancer cells transfected with oe-WT1-AS detected by MDC staining. f, Analysis of cell apoptosis in cancer cells transfected with oe-WT1-AS analyzed by flow cytometry. * p < 0.05 compared with negative control (NC) plasmids transfected cells. The above results are all measurement data, expressed as mean ± standard derivation. The unpaired t-test was used and the cell experiment was repeated three times. EdU, 5-ethynyl-2ʹ-deoxyuridine; MDC, Monodansylcadaverine; Bax, Bcl2 associated X, apoptosis regulator; Bcl-2, B cell leukemia/lymphoma 2; LC3I/II, microtubule-associated protein 1 light chain 3; ANOVA, analysis of variance; NC, negative control
LncRNA WT1-AS may inhibit PIK3AP1 expression by recruiting SPI1 by bioinformatics analysis
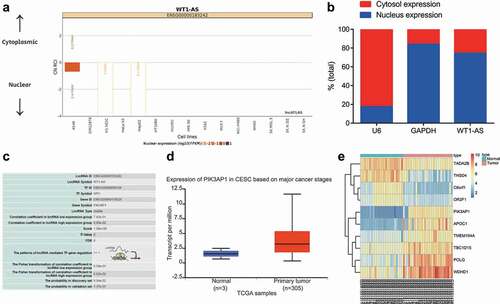
The subcellular fractionation experiment showed that expression of lncRNA WT1-AS was greatest in the nucleus (). The effect of transcriptional regulation is important in many diseases. In addition to studying the direct regulation between transcription factors and TF, researchers have found that lncRNA can also participate in transcriptional regulation through the lncRNA-TF-gene regulatory network [Citation18,Citation19]. So, our interrogation of the LncMAP database (http://bio-bigdata.hrbmu.edu.cn/LncMAP/) suggested that lncRNA WT1-AS may inhibit PIK3AP1 expression by the transcription factor SPI1 (). Analysis of the microarray GSE63514 revealed that expression of PIK3AP1 in cervical cancer was higher than that in normal control tissues ().
Figure 3. LncRNA WT1-AS might regulate PIK3AP1 expression via SPI1. a, Location of lncRNA WT1-AS in cells determined by cell fractionation experiments; b, The expression localization of lncRNA WT1-AS detected by nuclear separation; c, The predicted regulating mechanism of lncRNA WT1-AS analyzed on the LncMAP website. d, The expression of PIK3AP1 in cervical cancer tissues and adjacent tissues analyzed in TCGA database; e, The screening heatmap of microarray GSE63514. TCGA, The cancer genome atlas
LncRNA WT1-AS regulates PIK3AP1 expression through transcription factor SPI1
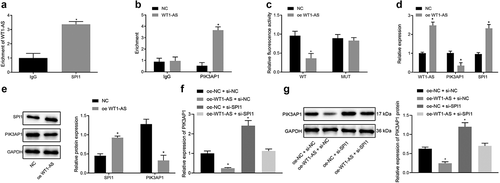
Moreover, a previous study has shown that SPI1 regulates the occurrence of cervical cancer as a transcription factor [Citation20]. To investigate whether lncRNA WT1-AS regulated PIK3AP1 expression through transcription factor SPI1, we undertook RIP assays, which showed that, compared with cells treated with IgG, the lncRNA WT1-AS enrichment was higher in the cells treated with SPI1 (), indicating that lncRNA WT1-AS can bind to SPI1. CHIP assay results showed that, compared with IgG-treated cells, the SPI1 enrichment in the PIK3AP1 promoter region was significantly increased (), indicating that SPI1 can bind to the promoter region of PIK3AP1. The results of the dual luciferase reporter gene assay showed that, compared with NC cells treated, the fluorescence intensity of the cells treated with PIK3AP1-Wt and oe-WT1-AS was decreased significantly, but there was no such significant change in the cells treated with PIK3AP1-Mut (). RT-qPCR and Western blot assays showed that, compared with NC cells, the expression of lncRNA WT1-AS and SPI1 was significantly up-regulated and PIK3AP1 expression was significantly down-regulated in cells treated with oe-WT1-AS (). The rescue experiment showed that, compared with cells treated with oe-NC and si-NC, PIK3AP1 expression was down-regulated by treatment with oe-WT1-AS and si-NC, but was up-regulated by oe-NC and si-SPI1 (). In addition, the interference of SPI1 reversed the down-regulation of PIK3AP1 induced by oe-WT1-AS. Similarly, Western blot analysis showed that PIK3AP1 protein level was down-regulated in oe-WT1-AS and si-NC treated cells but up-regulated in oe-NC and si-SPI1 treated cells (). Our results suggest that lncRNA WT1-AS inhibits PIK3AP1 expression by recruiting SPI1.
Figure 4. LncRNA WT1-AS inhibited PIK3AP1 expression by recruiting SPI1. a, The enrichment of lncRNA WT1-AS and SPI1 detected by RIP assay; b, The enrichment of SPI1 in PIK3AP1 promoter region detected by ChIP assay, where * p < 0.05 compared with that of the cells treated by IgG; c, The binding fluorescence intensity of lncRNA WT1-AS and PIK3AP1 promoter region detected by dual luciferase reporter gene assay; d, The relative expression of lncRNA WT1-AS and PIK3AP1 in cells transfected with oe-WT1-AS; e, The relative protein expression of PIK3AP1 and lncRNA WT1-AS in cells after transfected with oe-WT1-AS, where * p < 0.05 compared with that of the NC treated cells. f, The relative mRNA expression of PIK3AP1. g, The relative protein expression of PIK3AP1, where * p < 0.05 compared with that of cells treated with oe-NC and si-NC, # p < 0.05 compared with that of cells treated with oe-WT1-AS and si-NC. The above results are all measurement data, expressed as mean ± standard derivation and analyzed using an unpaired t-test. The cell experiments were repeated three times. IgG, immunoglobulin G; ChIP, chromatin Immunoprecipitation; FISH, Fluorescence In Situ Hybridization; RIP, RNA binding protein immunoprecipitation; WT, wild type; MUT, mutant
LncRNA WT1-AS inhibits the progression of cervical cancer through mediating SPI1/PIK3AP1
We further investigated whether lncRNA WT1-AS affected cervical cancer through SPI1/PIK3AP1. The results of RT-qPCR and Western blot assay showed that, compared with NC cells, the expression of lncRNA WT1-AS was upregulated in cells transfected with oe-WT1-AS, while expression of PIK3AP1 were down-regulated; expression of lncRNA WT1-AS in cells transfected with sh-PIK3AP1 showed no significant changes, while the expression of PIK3AP1 was significantly interfered (). Further, compared with NC cells, overexpression of lncRNA WT1-AS or silencing PIK3AP1 downregulated the expression of Beclin1, Bcl-1, and LC3II/LC3I proteins, but upregulated the expression of cleaved-caspase3 and Bax proteins, and that overexpression of lncRNA WT1-AS together with PIK3AP1 neutralized the effects of overexpression of lncRNA WT1-AS alone (). Therefore, in Hela cells transfected with oe-WT1-AS, PIK3AP1 expression was down-regulated, the expression of proliferation and autophagy-related proteins decreased, and apoptosis-related genes were up-regulated.
Figure 5. LncRNA WT1-AS inhibited cervical cancer by inhibiting PIK3AP1 through recruiting SPI1. a, The expression of lncRNA WT1-AS and PIK3AP1 mRNA in different cells lines; b-c, The expression of Beclin1, Bcl-1, LC3II, LC3I, cleaved-caspase3, and Bax protein; d, The proliferation rate of cancer cells transfected with NC, oe-WT1-AS, sh-PIK3AP1 or co-transfected with oe-WT1-AS and sh-PIK3AP1 detected by EdU staining (× 200); e, The proliferation rate of cancer cells transfected with NC, oe-WT1-AS, sh-PIK3AP1 or co-transfected with oe-WT1-AS and oe-PIK3AP1 detected by RT-qPCR and western blot assay; f, The autophagy of cancer cells transfected with NC, oe-WT1-AS, sh-PIK3AP1 or co-transfected with oe-WT1-AS and oe-PIK3AP1 detected by MDC staining; G, The apoptosis of cancer cells transfected with NC, oe-WT1-AS, sh-PIK3AP1 or co-transfected with oe-WT1-AS and oe-PIK3AP1 detected by flow cytometry. * p < 0.05 compared with the NC treated cells, # p < 0.05 compared with that of the cells transfected with oe-WT1-AS. The above results are measurement data, expressed as mean ± standard derivation, analyzed by an unpaired t-test, and the cell experiments were repeated three times. mRNA, message RNA; RT-qPCR, quantitative Reverse transcription polymerase chain reaction; EDU, 5-ethynyl-2’-deoxyuridine; MDC staining, Monodansylcadaverine staining
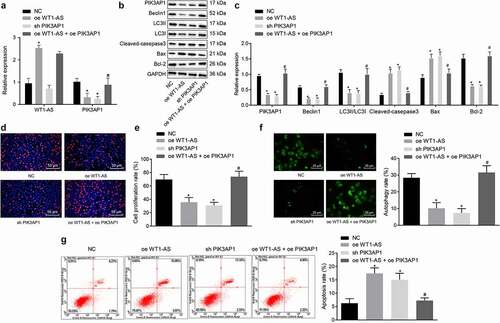
The proliferation rate determined by EdU assay showed that, compared with the NC group, the proliferation rates of the cells transfected with oe-WT1-AS or sh-PIK3AP1 were significantly lower; compared with the cells transfected with oe-WT1-AS, the proliferation rate of cells co-transfected with oe-WT1-AS and oe-PIK3AP1 was significantly increased ().
MDC staining showed that when Hela cells were under nutritional stress, the autophagy rate of the cells transfected with oe-WT1-AS or sh-PIK3AP1 was lower than that of the NC cells. Compared with the cells transfected with oe-WT1-AS, the autophagy rate of the cells co-transfected with oe-WT1-AS + oe-PIK3AP1 was increased ().
Flow cytometry showed that, compared with NC cells, the apoptosis rate of cells transfected with oe-WT1-AS and cells transfected with sh-PIK3AP1 increased significantly. Compared with the cells transfected with oe-WT1-AS, the apoptosis rate of cells co-transfected with oe-WT1-AS + oe-PIK3AP1 was significantly decreased ().
LncRNA WT1-AS inhibits cervical cancer growth in vivo
Subsequently, the results of xenograft tumors in nude mice showed that, compared with that of the NC treated mice, tumor growth was significantly attenuated in mice treated with oe-WT1-AS or sh-PIK3AP1, while concomitant overexpression of lncRNA WT1-AS and PIK3AP1 neutralized the effects of overexpression of lncRNA WT1-AS or silencing PIK3AP1 alone (). Thus, overexpression of lncRNA WT1-AS inhibits tumor growth in vivo.
Figure 6. LncRNA WT1-AS suppressed cervical cancer development in vivo. a, Representative images of xenograft tumors in mice implanted with cells transfected with NC, oe-WT1-AS, h-PIK3AP1 or co-transfected with oe-WT1-AS and oe-PIK3AP1; b, The volume change of tumor in mice transfected with NC, oe-WT1-AS, sh-PIK3AP1 or co-transfected with oe-WT1-AS and oe-PIK3AP1; c-d, The expression of PIK3AP1 protein in mice transfected with NC, oe-WT1-AS, sh-PIK3AP1 or co-transfected with oe-WT1-AS and oe-PIK3AP1; * p < 0.05 compared with that treated with NC, # p < 0.05 compared with that treated with oe-WT1-AS. n = 10. The above results are all measurement data, expressed as mean ± standard derivation. The data comparison between multiple groups is performed by one-way ANOVA with Tukey’s post hoc test; the data comparison among groups at different time points is performed by repeated measurement ANOVA with Bonferroni’s post hoc test. ANOVA, analysis of variance; N, number of samples
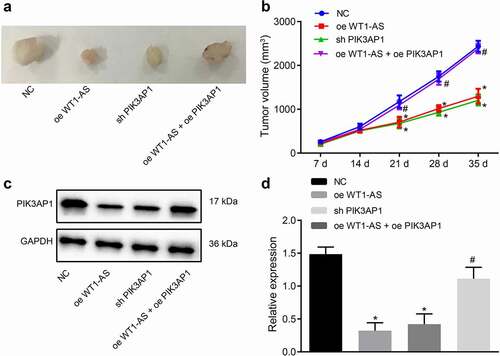
Western blot assay showed that, compared with NC treated mice, the expression of PIK3AP1 protein was down-regulated in mice treated by oe-WT1-AS or sh-PIK3AP1, but there was no such significant change in mice treated with oe-WT1-AS and oePIK3AP1 together (). Hence, overexpression of lncRNA WT1-AS inhibited PIK3AP1 protein expression in vivo, confirming that t lncRNA WT1-AS inhibits the development of cervical cancer xenografts.
Discussion
Cervical cancer affects women all over the world, often in the aftermath of a chronic HPV infection [Citation21,Citation22]. Clinical methods like excision and radiation are useful for treating primary cervical cancer, while metastatic cervical cancer remains largely incurable, thus calling for new therapies [Citation23]. Accumulating evidence demonstrates that lncRNAs play an important role in regulating the biological function of cervical cancer cells, as in the cases of lncRNA discs large MAGUK scaffold protein 1 antisense RNA 1 (DLG1-AS1), LINC00675, and others [Citation24–26]. However, the regulatory role of lncRNA WT1-AS in cervical cancer cells remained uncertain. Therefore, we designed this study to expand the knowledge of lncRNA WT1-AS, and collected evidence that lncRNA WT1-AS inhibited the expression of PIK3AP1 by recruiting SPI1, thus interfering in the progression of cervical cancer. As such, this study provides a promising new therapeutic target for cervical cancer and gives insight into the underlying molecular mechanisms of lncRNA WT1-AS.
In this study, we first showed that lncRNA WT1-AS expression was decreased in cervical cancer tissues and cells. This lncRNA is also downregulated in liver cancer tissues, and acts as an inhibitor of the development of liver cancer [Citation6]. Interestingly, previous work showed that the lncRNA transducer of epidermal growth factor receptor-2.1 antisense RNA 1 (TOB1-AS1) was also poorly expressed in cervical cancer cells, suggesting that its overexpression could be a tumor suppressor in cervical cancer [Citation27]. To further elucidate the function of lncRNAs in cervical cancer, we transfected cancer cells with oe-WT1-AS and found that expression of Beclin1, Bcl-1 and the ratio of microtubule associated protein 1 light chain 3 type II (LC3II)/microtubule associated protein 1 light chain 3 type I (LC3I) decreased, while the expression of cleaved-caspase3 and Bax increased in cervical cancer cells. Beclin1 is a vital autophagic protein and its repression was observed to attenuate the autophagy and metastasis of osteosarcoma cells [Citation28]. Previous studies have shown that Beclin1 overexpression can inhibit the occurrence of cervical cancer. In recent years, studies have shown that circRNA can promote the occurrence of cervical cancer by promoting the expression of Beclin1 [Citation29,Citation30]. In addition, the anti-cervical cancer effect of ferulic acid may be related to the decrease of autophagy-related proteins LC3-II, Beclin1 and ATG12-ATG5 [Citation31]. Further, it has been observed that lncRNA JPX promotes gastric cancer progression by regulating Beclin1 via inhibiting miR-197 [Citation32]. And lncRNA GAS5 inhibits cell migration and invasion and promotes autophagy by targeting miR-222-3p via Beclin1 in colorectal cancer [Citation33]. These studies suggest that Beclin1 may be regulated by lncRNAs and affect the occurrence of cervical cancer. Downregulation of Bcl-1 has been shown to be an antioncogene in colorectal cancer cells [Citation34,Citation35]. The ratio of LC3II/LC3I is also correlated with autophagy, and a decrease of this ratio suggested mitigation of autophagic injury in bronchial epithelial cells [Citation36]. The upregulation of cleaved-caspase3 and Bax expression after knockdown of lncRNA HAGLROS was permissive to increased apoptosis of colorectal cancer cells [Citation37]. Hence, these consistent findings in various cell types support our conclusion that the up-regulation of lncRNA WT1-AS in cervical cancer cells potentially plays a functional inhibitory role in relation to tumor progression.
Further, we observed that lncRNA WT1-AS could bind to SPI1 and further inhibit expression of PIK3AP1, which ultimately suppressed the progression of cervical cancer. SPI1, also known as or PU.1, is a crucial transcriptional regulator [Citation38]. A recent study has reported that SPI1 acted as a suppressor of myeloma by repressing the transcription of interferon regulatory factor 4 (IRF4) [Citation39] and others showed that SPI1 restoration suppressed the proliferation of cancer cells, thus presenting it as a novel therapeutic target for primary effusion lymphoma [Citation40]. Meanwhile, SPI1 was reported to be able to bind to the regulatory region of lncRNA HOTAIRM1, thus further increasing the expression of HOTAIRM1 [Citation41], and suggesting that it may have a generally negative effect on the progression of cancers. At the same time, we found in the current study that SPI1 can bind to the promoter region of PIK3AP1. PIK3AP1, which is also known as BCAP, is a signaling adaptor for B-cells in hematopoietic cells [Citation13,Citation14]. In addition, PIK3AP1 could be inhibited by miR-486 and played a suppressing role in the progression of lung cancer [Citation42]. Taken together, our results suggest that lncRNA WT1-AS has suppressive roles of elevation of SPI1 and consequent inhibition of PIK3AP1 in cervical cancer progression.
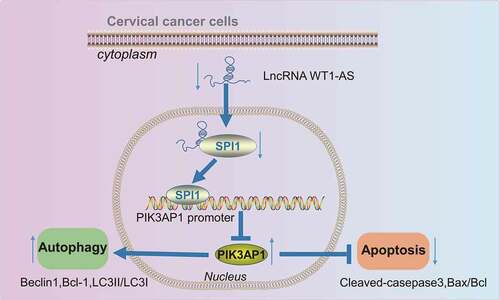
In summary, this study has demonstrated that overexpression of lncRNA WT1-AS suppressed the proliferation, autophagy and promoted the apoptosis of cervical cancer cells by inhibiting PIK3AP1 through recruitment of SPI1, suggesting that the WT1-AS/SPI1/PIK3AP1 axis might be a novel therapeutic target for treating cervical cancer (). Although lncRNA WT1-AS may prove useful in cervical cancer management, the underlying mechanisms of lncRNA WT1-AS still require further investigation.
Supplemental Material
Download MS Word (19.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Cohen PA, Jhingran A, Oaknin A, et al. Cervical cancer. Lancet. 2019;393(10167):169–182.
- Small W Jr., Ma B, Bajaj A, et al. Cervical cancer: a global health crisis. Cancer. 2017;123(13):2404–2412.
- Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer. 2016;15(1):39.
- Bach DH, Lee SK. Long noncoding RNAs in cancer cells. Cancer Lett. 2018;419:152–166.
- Lv L, Chen G, Zhou J, et al. WT1-AS promotes cell apoptosis in hepatocellular carcinoma through down-regulating of WT1. J Exp Clin Cancer Res. 2015;34(1):119.
- Dai SG, Guo LL, Xia X, et al. Long non-coding RNA WT1-AS inhibits cell aggressiveness via miR-203a-5p/FOXN2 axis and is associated with prognosis in cervical cancer. Eur Rev Med Pharmacol Sci. 2019;23:486–495.
- Ungerback J, Hosokawa H, Wang X, et al. Pioneering, chromatin remodeling, and epigenetic constraint in early T-cell gene regulation by SPI1 (PU.1). Genome Res. 2018;28(10):1508–1519.
- Goyal S, Suzuki T, Li JR, et al. RUNX1 induces DNA replication independent active DNA demethylation at SPI1 regulatory regions. BMC Mol Biol. 2017;18(1):9.
- Nishimura N, Endo S, Ueno S, et al. A xenograft model reveals that PU.1 functions as a tumor suppressor for multiple myeloma in vivo. Biochem Biophys Res Commun. 2017;486(4):916–922.
- Xu Y, Gu S, Bi Y, et al. Transcription factor PU.1 is involved in the progression of glioma. Oncol Lett. 2018;15:3753–3759.
- Lin J, Liu W, Luan T, et al. High expression of PU.1 is associated with Her-2 and shorter survival in patients with breast cancer. Oncol Lett. 2017;14:8220–8226.
- Pimienta G, Fok V, Haslip M, et al. Proteomics and transcriptomics of BJAB cells expressing the epstein-barr virus noncoding RNAs EBER1 and EBER2. PLoS One. 2015;10(6):e0124638.
- Duggan JM, Buechler MB, Olson RM, et al. BCAP inhibits proliferation and differentiation of myeloid progenitors in the steady state and during demand situations. Blood. 2017;129(11):1503–1513.
- Zhang F, Li K, Yao X, et al. A miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop regulates tumour growth and chemoresistance in gastric cancer. EBioMedicine. 2019;44:311–321.
- Zhang L, Wei Z, and Wang Y, et al. Long noncoding RNA ROR1-AS1 enhances STC2-mediated cell growth and autophagy in cervical cancer through miR-670-3p. J Recept Signal Transduct Res. 2020;41(6):582–592 .
- Xiang W, Zhang RJ, Jin GL, et al. RCE4, a potential anticervical cancer drug isolated from Reineckia carnea, induces autophagy via the dual blockade of PI3K and ERK pathways in cervical cancer CaSki cells. Int J Mol Med. 2020;45:245–254.
- Ren F, Ren JH, Song CL, et al. LncRNA HOTAIR modulates hepatitis B virus transcription and replication by enhancing SP1 transcription factor. Clin Sci (Lond). 2020;134(22):3007–3022.
- Gong J, Fan H, Deng J, et al. LncRNA HAND2-AS1 represses cervical cancer progression by interaction with transcription factor E2F4 at the promoter of C16orf74. J Cell Mol Med. 2020;24(11):6015–6027.
- Tao L, Wang X, Zhou Q. Long noncoding RNA SNHG16 promotes the tumorigenicity of cervical cancer cells by recruiting transcriptional factor SPI1 to upregulate PARP9. Cell Biol Int. 2020;44(3):773–784.
- Boussios S, Seraj E, Zarkavelis G, et al. Management of patients with recurrent/advanced cervical cancer beyond first line platinum regimens: where do we stand? A literature review. Crit Rev Oncol Hematol. 2016;108:164–174.
- Castle PE, Kinney WK, and Xue X, et al. Role of screening history in clinical meaning and optimal management of positive cervical screening results. J Natl Cancer Inst. 2018;11: 820–827 .
- The Cancer Genome Atlas Research Network. Smith Genome Sciences C, Harvard Medical S. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–384.
- Rui X, Xu Y, Huang Y, et al. lncRNA DLG1-AS1 promotes cell proliferation by competitively binding with miR-107 and up-regulating ZHX1 expression in cervical cancer. Cell Physiol Biochem. 2018;49(5):1792–1803.
- Ma S, Deng X, Yang Y, et al. The lncRNA LINC00675 regulates cell proliferation, migration, and invasion by affecting Wnt/beta-catenin signaling in cervical cancer. Biomed Pharmacother. 2018;108:1686–1693.
- Aalijahan H, Ghorbian S. Long non-coding RNAs and cervical cancer. Exp Mol Pathol. 2019;106:7–16.
- Yao J, Li Z, Yang Z, et al. Long noncoding RNA TOB1-AS1, an epigenetically silenced gene, functioned as a novel tumor suppressor by sponging miR-27b in cervical cancer. Am J Cancer Res. 2018;8:1483–1498.
- Zhang F, Yan T, Guo W, et al. Novel oncogene COPS3 interacts with Beclin1 and Raf-1 to regulate metastasis of osteosarcoma through autophagy. J Exp Clin Cancer Res. 2018;37(1):135.
- Chen M, Ai G, Zhou J, et al. circMTO1 promotes tumorigenesis and chemoresistance of cervical cancer via regulating miR-6893. Biomed Pharmacother. 2019;117:109064.
- Guo J, Chen M, Ai G, et al. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed Pharmacother. 2019;115:108957.
- Gao J, Yu H, Guo W, et al. The anticancer effects of ferulic acid is associated with induction of cell cycle arrest and autophagy in cervical cancer cells. Cancer Cell Int. 2018;18(1):102.
- Han X, and Liu Z. Long noncoding RNA JPX promotes gastric cancer progression by regulating CXCR6 and autophagy via inhibiting miR197. Mol Med Rep. 2021;23(1): 60 .
- Liu L, Wang HJ, Meng T, et al. lncRNA GAS5 inhibits cell migration and invasion and promotes autophagy by targeting miR-222-3p via the GAS5/PTEN-signaling pathway in CRC. Mol Ther Nucleic Acids. 2019;17:644–656.
- Qin A, Yu Q, Gao Y, et al. Inhibition of STAT3/cyclinD1 pathway promotes chemotherapeutic sensitivity of colorectal caner. Biochem Biophys Res Commun. 2015;457(4):681–687.
- Cao L, Liu Y, Wang D, et al. MiR-760 suppresses human colorectal cancer growth by targeting BATF3/AP-1/cyclinD1 signaling. J Exp Clin Cancer Res. 2018;37(1):83.
- Ding S, Hou X, Wang F, et al. Regulation of Eclipta prostrata L. components on cigarette smoking-induced autophagy of bronchial epithelial cells via keap1-Nrf2 pathway. Environ Toxicol. 2018;33(8):811–820.
- Zheng Y, Tan K, Huang H. Retracted: long noncoding RNA HAGLROS regulates apoptosis and autophagy in colorectal cancer cells via sponging miR-100 to target ATG5 expression. J Cell Biochem. 2019;120(3):3922–3933.
- Delestre L, Cui H, Esposito M, et al. Senescence is a Spi1-induced anti-proliferative mechanism in primary hematopoietic cells. Haematologica. 2017;102(11):1850–1860.
- Ueno N, Nishimura N, Ueno S, et al. PU.1 acts as tumor suppressor for myeloma cells through direct transcriptional repression of IRF4. Oncogene. 2017;36(31):4481–4497.
- Goto H, Kariya R, Kudo E, et al. Restoring PU.1 induces apoptosis and modulates viral transactivation via interferon-stimulated genes in primary effusion lymphoma. Oncogene. 2017;36(37):5252–5262.
- Wei S, Zhao M, Wang X, et al. PU.1 controls the expression of long noncoding RNA HOTAIRM1 during granulocytic differentiation. J Hematol Oncol. 2016;9(1):44.
- Xu Y, Wang Y, Yao A, et al. Low frequency magnetic fields induce autophagy-associated cell death in lung cancer through miR-486-mediated inhibition of Akt/mTOR signaling pathway. Sci Rep. 2017;7(1):11776.