ABSTRACT
Circular RNA (circRNA) deregulation impacts on normal cell physiology leading to malignant phenotypic changes. Here, we determined the function of the circRNA, hsa_circ_0065217 in malignant renal cell carcinoma (RCC). Hsa_circ_0065217 was abundantly expressed in RCC tissue and cell lines, and its expression linked to advanced TNM stages, large tumor sizes, and lymph-node metastasis. Hsa_circ_0065217 silencing reduced in vitro RCC cell-line growth and aggressiveness. Mechanistically, hsa_circ_0065217 promoted alpha protein kinase 2 (ALPK2) expression via its competing endogenous RNA (ceRNA) activity toward miR-214-3p. Moreover, ALPK2 overexpression reversed hsa_circ_0065217 knockdown effects on RCC cell-line malignancy. Thus, hsa_circ_0065217/miR-214-3p/ALPK2 signaling putatively promotes RCC tumorigenesis and is a putative RCC treatment target.
Introduction
Globally, renal cell carcinoma (RCC) is a highly lethal malignancy, and in 2020, accounted for approximately 14,830 cancer deaths in America [Citation1,Citation2]. Although technological developments have made significant contributions to diagnostics and treatment, the 5-year survival rate for advanced RCC is limited due to therapy resistance [Citation3,Citation4]. Therefore, underlying RCC mechanisms must be comprehensively investigated to develop new treatment strategies.
Circular RNAs (circRNAs) are non-coding RNAs (ncRNA) with covalently closed loop structures, but with no 5’ cap and 3’ polyadenylated tail. Their deregulation is frequently associated with tumorigenesis and malignant phenotypic changes [Citation5,Citation6]. Increasingly, studies have shown circRNAs exert crucial roles in disease evolution by serving as microRNA (miRNA) sponges, including RCC [Citation7,Citation8]. For example, Chen et al. found that hsa_circ_001895 functioned as a miR‐296‐5p sponge to promote clear cell RCC proliferation and invasion by affecting SOX12 expression [Citation9]. Xue et al. found that circ-AKT3 reduced clear RCC metastasis by altering miR-296-3p/E-cadherin signaling [Citation10]. Li et al. found that circPRRC2A promoted tumor epithelial-to-mesenchymal transition and aggression in patients with RCC by regulating circPRRC2A-miR-514a-5p/miR-6776-5p/TRPM3 signaling [Citation11]. Nonetheless, underlying circRNA mechanisms in RCC remain relatively unclear, therefore, we analyzed hsa_circ_0065217 roles in RCC.
We used GSE137836 and GSE100186 datasets from the Gene Expression Omnibus database and predicted the hitherto unknown hsa_circ_0065217 as a significantly up-regulated circRNA in RCC. Furthermore, our experimental studies suggested hsa_circ_0065217 promoted RCC progression by functioning as a competing endogenous RNA (ceRNA) toward Alpha Protein Kinase 2 (ALPK2) and up-regulated its expression by competing with miR-214-3p. Combined, our data indicated hsa_circ_0065217 potentially functioned as a viable target for RCC therapy.
Materials and methods
Clinical specimens
RCC tissues and adjacent non-cancerous tissues (NC) (N = 45) were collected from patients in The First Affiliated Hospital of Zhejiang Chinese Medicine University. Tissues were immediately placed in liquid nitrogen and maintained at −80°C. Studies were conducted with reference to the Declaration of Helsinki, and the study was approved by the Ethics Committee of The First Affiliated Hospital of Zhejiang Chinese Medicine University. Patients provided signed informed consent and did not undergo chemotherapy or radiotherapy before surgery.
Cell culture and transfection
ACHN, SW839, OS-RC-2, A498, 786-O, and Caki-1 RCC cell lines and kidney tubular epithelial cells, HK-2 were used in this study (American Type Culture Collection, Manassas, VA, USA). Cells were grown in Dulbecco’s modified Eagle medium (DMEM, Gibco, NY, USA) plus 10% fetal bovine serum (FBS, Gibco) and grown at 37°C in saturated humidity at 5% CO2.
Small interfering RNAs (si-RNAs) against hsa_circ_0065217 (si-circ_0065217), miR-214-3p mimics, miR-214-3p inhibitors as well as control groups (NC) were synthesized by GenePharma (Shanghai, China). Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used for transfections. After 48 h, cells were processed for downstream analyses.
Cell viability assay
We used the Cell Counting Kit-8 (CCK-8) (Beyotime, Shanghai, China) to assay viability. Transfected RCC cells (1 × 104/well) were added to 96-well plates and grown in complete medium. Then, cell supernatants were removed at 24 h, 48 h, 72 h, and 96 h and plates incubated with CCK-8 solution for 4 h at 37°C. Absorbance was recorded at 450 nm on a microplate reader (Thermo Fisher, Waltham, MA, USA).
Colony formation assay
Transfected RCC cells (1000 cells/well) were added to 6-well plates and grown at 37°C in 5% CO2 for 2 weeks to generate colonies. Colonies were fixed in 95% ethanol, stained in 0.5% crystal violet for 20 min, and enumerated using inverted microscopy (Nikon, Tokyo, Japan).
Invasion assays
Cell invasion was examined using a 24-well Transwell system (Corning, NY, USA). Briefly, 200 μL transfected RCC cells (1 × 105/chamber) in serum-free medium were added to matrigel pre-coated upper Transwell chambers. To lower chambers, 600 μL complete medium was added. After 1-day, invaded cells were stained in 0.5% crystal violet and enumerated using inverted microscopy (Nikon).
Quantitative real-time polymerase chain reaction (qRT-PCR)
To isolate RNA, RCC tissues and cells were lysed in TRIzol (Invitrogen, Carlsbad, CA, USA). RNAs were reverse transcribed to cDNA using HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China), and RNAs expression was detected in qPCR assay using specific primers and ChamQ SYBR Color qPCR Master Mix (Vazyme). The expression of circRNAs, mRNAs and miRNAs were calculated normalized to internal control GAPDH or U6 using the 2−ΔΔCt method.
Dual-luciferase reporter assay
Hsa_circ_0065217 wild-type (WT) or mutant (MUT) DNAs were cloned into the pmir-GLO dual-luciferase plasmid (Promega, Madison, WI, USA) and co-transfected into RCC cells plus NC or miR-214-3p mimics. PmirGLO-ALPK2-WT or pmirGLO-ALPK2-MUT plasmids were also generated and transfected with miR-214-3p or NC mimics into RCC cells. After 48 h, luciferase activity was assayed by the Dual Luciferase Reporter Assay System (Promega).
Western blot assay
Protein was extracted using RIPA buffer plus protease and phosphatase inhibitors. Protein concentrations were assayed using the bicinchoninic acid protein assay. Approximately 20 μg protein was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and proteins transferred to polyvinylidene fluoride membranes. They were blocked in 5% skim milk and incubated with primary antibodies overnight at 4°C (anti-E-cadherin; anti-vimentin; anti-ALPK2, and anti-GAPDH, 1:1000; Abcam, Cambridge, MA, USA). Membranes were then washed in TBST and a secondary HRP-IgG antibody (Abcam) added and incubated at for 1 h at 37°C. Blots were developed by enhanced chemiluminescence (Millipore, Bedford, MA, USA) and images viewed in Image Lab™ (Bio-Rad).
RNA pull-down assay
Biotinylated hsa_circ_0065217 (Bio-hsa_circ_0065217) and control (Bio-NC) were transfected into RCC cells for 48 h, lysed in lysis buffer (Sigma) for 10 min, after which lysates were incubated with streptavidin-coupled Dynabeads (Invitrogen) for 1 h at room temperature. Proteinase K (Sigma) was then added to digest eluent proteins. RNA was isolated and miRNAs assayed.
RNA immunoprecipitation (RIP) assay
Briefly, RCC cell lysates were added to immunoprecipitation buffer containing magnetic beads conjugated to IgG or Ago2 antibodies (Abcam). Hsa_circ_0065217 and miR-214-3p enrichment was then investigated using qRT-PCR. The assay was performed using the Magna RIP kit (Millipore) based on manufacturer’s instructions.
Statistical analysis
Data from three independent experiments were processed by SPSS (IBM, NY, USA). Results were presented as the mean ± standard deviation (SD) and statistical significance determined by one-way analysis of variance or Student’s t-test. Differences were statistically significant at P < 0.05.
Results
Hsa_circ_0065217 is overexpressed in RCC
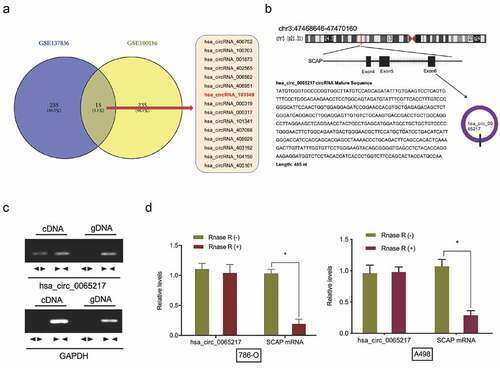
To investigate circRNA expression profiles in RCC, the microarray gene profiling datasets, GSE137836 and GSE100186 were used. Fifteen circRNAs were deregulated in both datasets (). We therefore focused on the most significantly up-regulated circRNA, hsa_circ_0065217 (has_circRNA_103349), which mapped to chromosome 3 and consisted of three exons (exons 4–6) from its host gene, SCAP (). Then, cDNA and genomic DNA were used as templates to amplify hsa_circ_0065217 from cDNA using divergent primers. Importantly, amplification products were not found from genomic DNA (). Our RNase R assay indicated hsa_circ_0065217 was highly stable in the presence of RNase R, however, linear SCAP was not; it was digested by RNase R ().
Figure 1. Hsa_circ_0065217 characterization in RCC. (a) Venn diagrams demonstrate aberrant circRNAs in GSE137836 and GSE100186 datasets. (b) The information of hsa_circ_0065217 (has_circRNA_103349). (c) Hsa_circ_0065217 validation by expression assays. Divergent primers amplified hsa_circ_0065217 from cDNA but not genomic DNA. (d) QRT-PCR was used to determine hsa_circ_0065217 and linear SCAP mRNA abundance in RCC cells incubated with RNase R. *P < 0.05
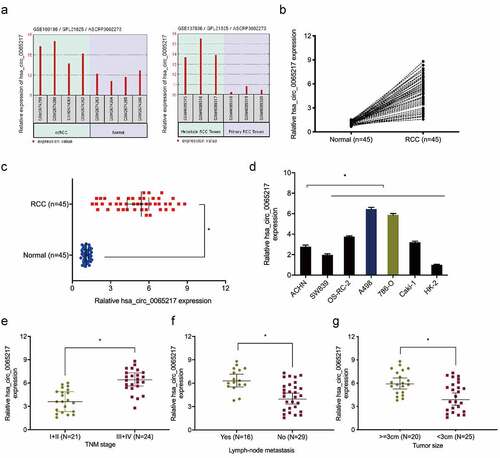
Next, we showed hsa_circ_0065217 expression was upregulated in GSE137836 and GSE100186 datasets (). Hsa_circ_0065217 expression was further confirmed in RCC. The results exhibited that hsa_circ_0065217 expression was observably higher in RCC tissues and cell lines (). Moreover, correlation analysis showed RCC patients with advanced features (advanced TNM stage, lymph-node metastasis, and larger tumor size) showed significant increased expression of hsa_circ_0065217 (–g)).
Figure 2. Hsa_circ_0065217 is elevated in RCC. (a) Expression of hsa_circ_0065217 in GSE100186 and GSE137836 databsets. (b-d) Hsa_circ_0065217 expression in RCC tissue and cell lines by qRT-PCR. (e-g) Elevated hsa_circ_0065217 levels were associated with advanced TNM stages, lymph-node metastasis and larger tumor sizes in RCC patients. *P < 0.05
Hsa_circ_0065217 promotes RCC cell proliferation and invasion
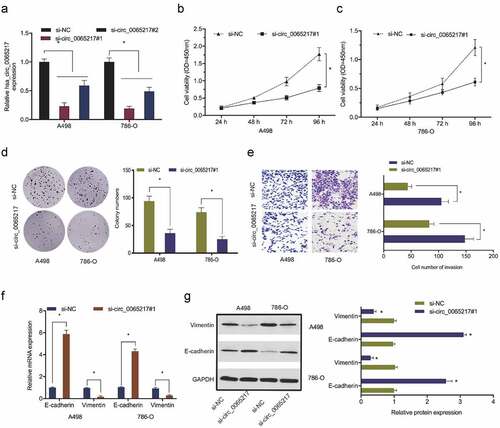
To explore hsa_circ_0065217 functions in RCC, si-circ_0065217 and si-NC reagents were transfected into 786-O and A498 cells. Transfection efficiencies are shown in . Cell viability assays showed that hsa_circ_0065217 silencing lowered in vitro 786-O and A498 proliferation (). Also, hsa_circ_0065217 silencing repressed the colony formation capabilities of RCC cells (). Transwell assays indicated that 786-O and A498 cell invasion was reduced after hsa_circ_0065217 silencing (). Moreover, hsa_circ_0065217 silencing increased E-cadherin and inhibited vimentin levels in RCC cells (). These data indicated a suppressive role for hsa_circ_0065217 inhibition toward in vitro RCC growth and metastasis.
Figure 3. Hsa_circ_0065217 silencing reduces RCC growth phenotypes. (a) Hsa_circ_0065217 silencing in RCC cells. (b-d) Hsa_circ_0065217 inhibits RCC proliferation. (e) The impact of hsa_circ_0065217 silencing on RCC by Transwell assay. (f, g) E-cadherin and vimentin levels in RCC cells transfected with si-circ_0065217 were explored by qRT-PCR and Western blot. *P < 0.05
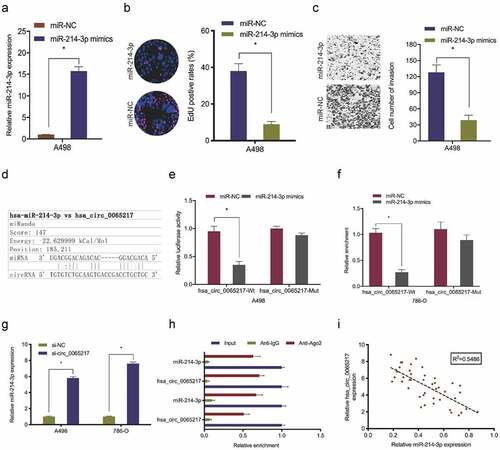
Hsa_circ_0065217 acts as a sponge of miR-214-3p in RCC
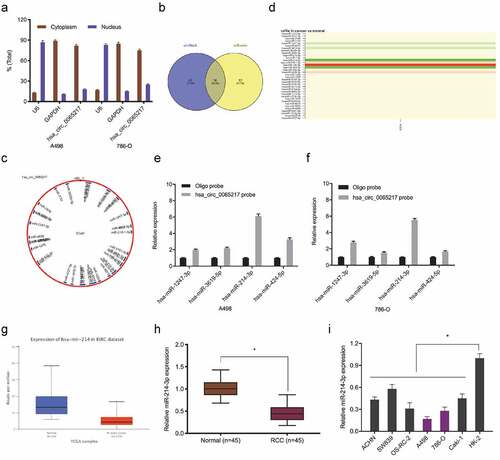
It was previously indicated that circRNAs, which are primarily cytoplasmic in nature, function as miRNA sponges [Citation7]. Therefore, we investigated hsa_circ_0065217 localization in RCC cells using nuclear and cytoplasmic separation assays. These data showed hsa_circ_0065217 was primarily localized to the cytoplasm in RCC cells (). Next, hsa_circ_0065217 potential target miRNAs were predicted using circBank and miRanda databases, with 36 miRNAs identified (). TCGA database showed hsa-miR-1247-3p, hsa-miR-3619-5p, hsa-miR-214-3p and hsa-miR-424-5p were significantly decreased in renal cancer tissues (). RNA-pull down assay showed all 4 miRNAs were abundantly pulled-down by the hsa_circ_0065217 probe compared with the NC probe, and miR-214-3p was the most enriched (), and importantly, the Cancer Genome Atlas (TCGA) database showed miR-214-3p was significantly reduced in RCC samples (). These observations were also confirmed in RCC tissues and cell lines ().
Figure 4. MiR-214-3p is a hsa_circ_0065217 target. (a) Hsa_circ_0065217 cellular distribution was analyzed using a cellular RNA fractionation assay. (b, c) Potential binding targets of hsa_circ_0065217 predicted by circBank and miRanda. (d) Potential target miRNAs expression in TCGA datasets. (e, f) miR-214-3p was pulled down by using a hsa_circ_0065217 specific probe. (g) MiR-214-3p expression in the Cancer Genome Atlas (TCGA) database. (h, i) MiR-214-3p expression in RCC tissue and cell lines. *P < 0.05
Next, we explored miR-214-3p roles in RCC. Functional assays revealed miR-214-3p up-regulation significantly lowered in vitro A498 cell proliferation and invasion (). Luciferase reporter assays showed miR-214-3p mimics attenuated hsa_circ_0065217‐WT luciferase activity, but not MUT activity (). Our qRT-PCR data revealed that hsa_circ_0065217 silencing significantly evaluated miR-214-3p levels in RCC cell lines (). RIP assay showed hsa_circ_0065217 and miR-214-3p were both enriched in the Ago2 group, but not the IgG group (). Correlation analysis identified a negative link between hsa_circ_0065217 and miR-214-3p levels in RCC ()). Combined, hsa_circ_0065217 functioned as a sponge for miR-214-3p in RCC cells.
Figure 5. Hsa_circ_0065217 is a miR-214-3p sponge. (a-c) MiR-214-3p overexpression impacts RCC growth phenotypes. (d-f) Luciferase data shows miR-214-3p mimics inhibited hsa_circ_0065217-WT activity in RCC cells. (g) Hsa_circ_0065217 silencing evaluated miR-214-3p in RCC cells. (h) Hsa_circ_0065217 and miR-214-3p enrichment on Ago2-beads in RCC cells. (i) Hsa_circ_0065217 was inversely linked with miR-214-3p levels in RCC tissue. *P < 0.05
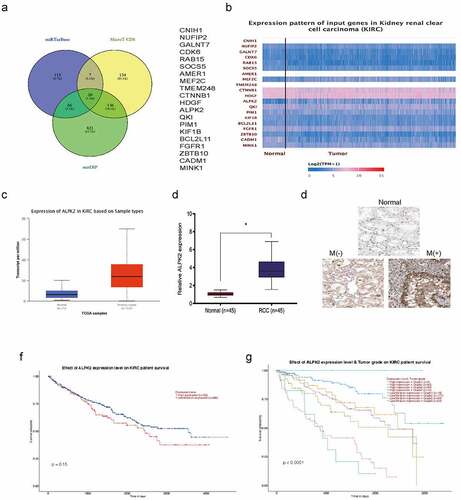
MiR-214-3p promotes RCC progression by targeting ALPK2
To understand miR-214-3p function in RCC progression, miRTarBase, MicroT-CDS, and miDIP databases were investigated, and predicted 20 downstream target genes (). Candidate gene expression was further explored in TCGA database, and ALPK2 was chosen to further study (). QRT-PCR showed that ALPK2 was significantly increased RCC tissues (). These data were further confirmed by immunohistochemistry (IHC) (). Moreover, overall survival analysis revealed elevated ALPK2 was linked with bad prognoses in patients with RCC ().
Figure 6. ALPK2 is highly expressed in RCC. (a) Potential binding targets for miR-214-3p as predicted by miRTarBase, MicroT-CDS, and miDIP databases. (b) Potential target gene expression in the Cancer Genome Atlas (TCGA). (c) ALPK2 levels in TCGA samples. (d, e) ALPK2 expression in RCC tissue was explored by qRT-PCR and IHC. (f, g) High ALPK2 expression was linked with poor prognoses in patients with RCC. *P < 0.05
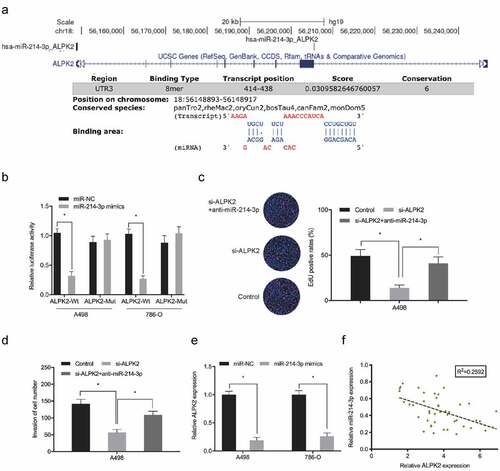
Dual-luciferase reporter assay showed that miR-214-3p overexpression significantly lowered luciferase levels in ALPK2 3’UTR-WT cells, but not MUT group (). The inhibitory effects on RCC cells proliferation and invasion induced by si-ALPK2 could be abolished by downregulation of miR-214-3p (). Expression data revealed that ALPK2 was down-regulated concomitant with miR-214-3p overexpression in RCC cells (). Correlation analyses revealed that high ALPK2 levels were negatively correlated with miR-214-3p in RCC (). These data suggested miR-214-3p directly interacted with ALPK2 in RCC.
Figure 7. ALPK2 is a direct target of miR-214-3p. (a, b) Luciferase data shows miR-214-3p mimics inhibited ALPK2-WT activity in RCC cells. (c, d) miR-214-3p suppression abolished the roles of ALPK2 inhibition on RCC cells proliferation and invasion. (e) MiR-214-3p mimics reduced ALPK2 expression in RCC cells. (f) MiR-214-3p was inversely linked with ALPK2 levels in RCC tissues. *P < 0.05
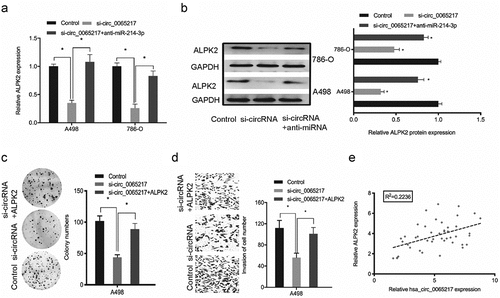
Hsa_circ_0065217 facilitates RCC cells progression by targeting miR-214-3p/ALPK2 axis
To assess if hsa_circ_0065217 regulated the miR-214-3p/ALPK2 axis, rescue assays were performed. Our qRT-PCR and western blotting data revealed ALPK2 levels were positively modulated by hsa_circ_0065217, whereas miR-214-3p silencing diminished the hsa_circ_0065217 knockdown effects on ALPK2 expression (). Next, colony formation assay revealed that ALPK2 overexpression significantly reversed the hsa_circ_0065217 suppression-induced growth inhibition in cells (). Transwell data indicated that ALPK2 forced expression reversed hsa_circ_0065217 silencing-induced cell invasion inhibition (). Also, elevated hsa_circ_0065217 levels were positively correlated with ALPK2 levels in RCC (). Thus, hsa_circ_0065217 promoted RCC progression by targeting miR-214-3p/ALPK2 signaling.
Figure 8. Oncogenic hsa_circ_0065217/miR-214-3p/ALPK2 signaling in RCC cells. (a, b) ALPK2 expression levels in RCC cells transfected with si-circ_0065217 or miR-214-3p inhibitors. (c, d) Forced ALPK2 expression reversed hsa_circ_0065217 silencing effects on RCC cells colony formation and invasion. (e) High ALPK2 levels were positively correlated with hsa_circ_0065217 levels in RCC tissue. *P < 0.05
Discussion
In recent decade, the involvement of circRNAs in oncogenesis and progression has attracted considerable attention, and generated progress toward disease etiology [Citation12]. Increasingly, the evidence suggests circRNAs operate as ceRNAs to sponge miRNAs, thereby regulating target gene expression [Citation13,Citation14]. For example, Su et al showed that hsa_circ_0070269 inhibited hepatocellular carcinoma by affecting miR-182/NPTX1 signaling [Citation15]. Wang et al suggested that circRNA-002178 acted as a ceRNA to affect PDL1/PD1 levels in lung adenocarcinoma [Citation16]. Lu et al revealed that circSLC8A1 sponged miR-130b/miR-494 to suppress bladder cancer via PTEN regulation [Citation17]. However, circRNA functions and mechanisms in tumor progression remain relatively unknown.
Here, hsa_circ_0065217 was increased in RCC tissue and cell lines. Elevated hsa_circ_0065217 levels were linked with advanced clinical characteristics in RCC patients. Hsa_circ_0065217 inhibition appeared to exert reducing effects toward in vitro RCC proliferation and invasion. Thus, we propose hsa_circ_0065217 may participate in RCC development by affecting RCC proliferation and invasion.
MiR-214-3p is a tumor suppressor molecule implicated in several tumor processes. For example, Fang et al showed that miR-214-3p inhibited endometrial cancer epithelial-to-mesenchymal transition and metastasis by targeting TWIST1 [Citation18]. Han et al found that miR-214-3p is a downstream target of long non-coding RNA (lncRNA) DNAJC3-AS1 in colon cancer, with DNAJC3-AS1 reportedly acting during tumorigenesis via miR-214-3p/LIVIN signaling [Citation19]. Huang et al showed that hsa_circ_0029589 knockdown suppressed vascular smooth cell growth phenotypes via miR-214-3p and STIM1 regulation [Citation20]. In our study, our nuclear/cytoplasmic fractionation assay confirmed that hsa_circ_0065217 was primarily cytoplasmic in RCC cells. Also, bioinformatics analyses inferred an interactive link between hsa_circ_0065217 and miR-214-3p. A comprehensive series of verification assays confirmed hsa_circ_0065217 specifically and competitively bound to miR-214-3p. Accordingly, we propose a novel regulatory network, hsa_circ_0065217/miR-214-3p is implicated in RCC.
ALPK2 is part of an atypical alpha protein kinase family known to regulate cell cycle and DNA repair genes in cancer [Citation21]. For example. Zhu et al showed that ALPK2 knockdown inhibited ovarian cancer cell development and progression [Citation22]. Moreover, Jiang et al showed that ALPK2 knockdown inhibited RCC progression by targeting Akt signaling [Citation23]. Here, ALPK2 levels were upregulated in RCC and linked with poor prognoses in patients with RCC. We then confirmed that miR-214-3p interacted with ALPK2 in RCC cells. Furthermore, hsa_circ_0065217 silencing lowered ALPK2 expression, while miR-214-3p inhibition abrogated these effects. Additionally, rescue assays indicated that forced ALPK2 expression reversed hsa_circ_0065217 silencing induced RCC cell proliferation and invasion inhibition. Therefore, we hypothesize hsa_circ_0065217 promotes cancer growth phenotypes via miR-214-3p/ALPK2 signaling in RCC.
Conclusion
In the present study, we report a new hsa_circ_0065217/miR-214-3p/ALPK2 axis which promotes malignant progression in RCC cells. Our study provides novel insights into a potential RCC strategy where hsa_circ_0065217 putatively functions as a promising therapeutic target.
Ethics approval and consent participate
Written informed consent was obtained from patients with approval by the Institutional Review Board in The First Affiliated Hospital of Zhejiang Chinese Medicine University.
Disclosure statement
The authors declare that they have no financial conflicts of interest.
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30.
- Capitanio U, Bensalah K, Bex A, et al. Epidemiology of renal cell carcinoma. Eur Urol. 2019;75(1):74–84.
- Sun M, De Velasco G, Brastianos PK, et al. The development of brain metastases in patients with renal cell carcinoma: epidemiologic trends, survival, and clinical risk factors using a population-based cohort. Eur Urol Focus. 2019;5(3):474–481.
- Wu J, Zhang P, Zhang G, et al. Renal cell carcinoma histological subtype distribution differs by age, gender, and tumor size in coastal Chinese patients. Oncotarget. 2017;8(42):71797.
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461.
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211.
- Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):1–11.
- Wang Y, Zhang Y, Wang P, et al. Circular RNAs in renal cell carcinoma: implications for tumorigenesis, diagnosis, and therapy. Mol Cancer. 2020;19(1):1–10.
- Chen Z, Xiao K, Chen S, et al. Circular RNA hsa_circ_001895 serves as a sponge of microRNA‐296‐5p to promote clear cell renal cell carcinoma progression by regulating SOX12. Cancer Sci. 2020;111(2):713.
- Xue D, Wang H, Chen Y, et al. Circ-AKT3 inhibits clear cell renal cell carcinoma metastasis via altering miR-296-3p/E-cadherin signals. Mol Cancer. 2019;18(1):1–13.
- LiW, YangFQ, SunCM, etal. circPRRC2A promotes angiogenesis and metastasis through epithelial-mesenchymal transition and upregulates TRPM3 in renal cell carcinoma. Theranostics. 2020;10(10):4395.
- Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165(2):289–302.
- Verduci L, Strano S, Yarden Y, et al. The circRNA–microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669–680.
- Wang J, Zhang JQ, Zhao XL, et al. Circular RNA DHX33 promotes malignant behavior in ccRCC by targeting miR-489-3p/MEK1 axis. Aging (Albany NY). 2020;12(14):14885.
- Su X, Su J, He H, et al. Hsa_circ_0070269 inhibits hepatocellular carcinoma progression through modulating miR-182/NPTX1 axis. Biomed Pharmacother. 2019;120:109497.
- Wang JF, Zhao XH, Wang YB, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11(1):1–11.
- Lu Q, Liu T, Feng H, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18(1):1–13.
- Fang YY, Tan MR, Zhou J, et al. miR-214-3p inhibits epithelial-to-mesenchymal transition and metastasis of endometrial cancer cells by targeting TWIST1. Onco Targets Ther. 2019;12:9449.
- Han B, Ge Y, Cui J, et al. Down-regulation of lncRNA DNAJC3-AS1 inhibits colon cancer via regulating miR-214-3p/LIVIN axis. Bioengineered. 2020;11(1):524–535.
- Huang Z, Li P, Wu L, et al. Hsa_circ_0029589 knockdown inhibits the proliferation, migration and invasion of vascular smooth muscle cells via regulating miR-214-3p and STIM1. Life Sci. 2020;259:118251.
- Yoshida Y, Tsunoda T, Doi K, et al. ALPK2 is crucial for luminal apoptosis and DNA repair-related gene expression in a three-dimensional colonic-crypt model. Anticancer Res. 2012;32(6):2301–2308.
- Zhu X, Yan S, Xiao S, et al. Knockdown of ALPK2 inhibits the development and progression of Ovarian Cancer. Cancer Cell Int. 2020;20(1):1–10.
- Jiang J, Han P, Qian J, et al. Knockdown of ALPK2 blocks development and progression of renal cell carcinoma. Exp Cell Res. 2020;392(2):112029.
