ABSTRACT
Lung adenocarcinoma (LUAD) is originated from the mucus-producing glands of the lungs. The involvement of long noncoding RNAs (lncRNAs) has been discovered in multiple diseases. In the present research, we aimed to unmask the role of C2CD4D and THEM5 antisense RNA 1 (C2CD4D-AS1) in LUAD. RT-qPCR or western blot analysis was respectively applied in the detection of RNA or protein expressions. The function of C2CD4D-AS1 in LUAD was assessed by functional assays. Through ChIP, RNA pull down, DNA pull down, RIP and luciferase reporter assays, the in-depth regulatory mechanism of C2CD4D-AS1 in LUAD was explored. C2CD4D-AS1 was dramatically overexpressed in LUAD tissues and cell lines. As a result, depletion of C2CD4D-AS1 significantly repressed cell proliferation, migration, invasion and stimulated cell apoptosis in LUAD. Mechanistically, ETS variant transcription factor 4 (ETV4) activated the transcription of C2CD4D-AS1 and stimulated its up-regulation in LUAD cells, thus affecting LUAD cell biological functions. Furthermore, C2CD4D-AS1 sponged microRNA-3681-3p (miR-3681-3p) and regulated NIMA-related kinase 2 (NEK2), thus participating in modulating LUAD cell biological behaviors. To conclude, C2CD4D-AS1 up-regulation induced by ETV4 enhanced NEK2 expression by sequestering miR-3681-3p to contribute to the malignant behaviors of LUAD cells.
Introduction
As the major subtype of lung cancer, lung adenocarcinoma (LUAD) is characterized by aggression and rapid fatality. Although many efforts have been made for the treatment of LUAD in recent years, improvements in poor diagnosis and tumor metastasis remain few [Citation1]. Therefore, understandings of factors involved in tumor progression are urgently needed.
Long noncoding RNAs (lncRNAs) are classified as a family of transcripts with more than 200 nucleotides [Citation2]. Aberrant expression of lncRNAs confers their different roles in tumor growth, metastasis and so on [Citation3,Citation4]. For example, Yang et al. have demonstrated the oncogenicity of lncRNA PVT1 in prostate cancer. PVT1 was highly expressed in prostate cancer tissues and cells, and the overexpression of PVT1 would lead to poorer survival [Citation5]. Wang et al. have found that HOXD-AS1 mediated by STAT3 promotes liver cancer metastasis [Citation6]. Zhang et al. have uncovered that lncRNA XIAT may exist as a biomarker for colorectal cancer [Citation7]. As a novel lncRNA, C2CD4D-AS1 has been reported to exert functions on autoimmune diseases [Citation8]. However, it still remains obscure in LUAD.
Transcription factors account a lot for dysregulation of lncRNAs. In brief, transcription factors always control the transcription of genes via recognizing DNA sequences [Citation9]. Moreover, targeting transcription factors may prevent the transcription of oncogenic genes, thereby inhibiting cancer process [Citation10]. For instance, lncRNA SNHG15 could promote colon cancer progression by stabilizing Slug [Citation11]. LINC00312 has been verified to induce migration and invasion of LUAD cells by direct binding to the transcription factor Y-Box Binding Protein 1 (YBX1) [Citation12]. ETV4, a transcription factor, has been discovered as a crucial element in estrogen signaling and growth in endometrial cancer [Citation13]. Hence, the interaction between transcription factors and C2CD4D-AS1 needs more investigations.
Competing endogenous RNA (ceRNA) network has been widely reported to be implicated in the pathogenesis of cancers [Citation14]. To be specific, lncRNAs indirectly regulate mRNAs by competitively binding with miRNAs [Citation15]. For instance, Yang et al. have proposed that LINC01133 could sponge miR-106a-3p to regulate APC expression and the Wnt/ꞵ-catenin pathway, thus inhibiting progression of gastric cancer [Citation16]. LncRNA FAL1 regulates the proliferative and migratory capacities of hepatocellular cells through sequestering miR-1236 [Citation17]. LncRNA NEAT1 could sponge miR-193a-3p as a ceRNA to accelerate LUAD [Citation18]. However, how C2CD4D-AS1 serves as a ceRNA in modulating LUAD has not been analyzed.
In this paper, we aimed to get a better understanding of the functional role and underlying mechanism of C2CD4D-AS1 in LUAD.
Methods
Cell culture
Human LUAD cell lines (H1975, A549, PC-9, and H1299) and human normal lung epithelial cell line (BEAS-2B) were chosen for this study. Among them, H1975, A549, H1299, and BEAS-2B cell lines were acquired from American Type Culture Collection (ATCC; Manassas, VA, USA) while PC-9 cell lines were procured from Huatuo Biotechnology Co., Ltd. (Shenzhen, China). H1975, PC-9, H1299 cell lines were cultured in RPMI-1640 Medium. A549 cell line was cultured in F-12 K Medium and BEAS-2B cell line in BEGM medium. All mediums were added with 10% fetal bovine serum (FBS) and cultivated at room temperature with 5% CO2.
Cell transfection
The designed shRNAs targeting C2CD4D-AS1 synthesized by GenePharma (Shanghai, China) were obtained to knock down C2CD4D-AS1. Moreover, for overexpression of NEK2, the sequences of NEK2 were cloned into pcDNA3.1 vectors to construct pcDNA3.1/NEK2. MiR-3681-3p mimics/inhibitor and respective negative controls were all bought from RiboBio (Shanghai, China). All cell transfections were used by utilizing Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Reverse transcription quantitative real-time PCR (RT-qPCR) analysis
TRIzol Reagent (Introgen, Carlsbad, CA, USA) was utilized for extracting total RNA, which was then reversely transcribed into cDNA with the application of RevertAid First-Strand cDNA Synthesis Kit (Thermo Fisher, IL, USA). SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) was utilized to perform PCR analysis. The quantitative analysis of relative expression levels was determined by 2−ΔΔCt method [Citation19]. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 snRNA (U6) was used as an internal control.
Cell counting kit 8 (CCK8) assay
CCK8 assay was carried out to evaluate the proliferation of PC-9 and H1299 cells. Cells were firstly placed into 96-well plates (5 × 103 cells per well). 24 h, 48 h and 72 h after transfection, each well was supplemented with CCK8 reagent (10 μl per well). After an additional incubation for 2 h, the absorbance at 450 nm was measured.
Colony formation assay
In brief, 20000–80000 stably transfected cells were added into 6-well plates for 14-day incubation. Cells were then fixed in 4% paraformaldehyde, followed by staining with 0.5% crystal violet. The stained cell colonies were counted manually.
Transwell assay
3 × 104 cells were seeded onto the membrane in the upper chambers of transwell plates. Then, the bottom chamber was added with 100% complete culture medium. Matrigel (BD Biosciences San Diego, CA, USA) was added into the chambers for invasion assay and no Matrigel for migration assay. After 24 h, invaded and migrated cells were stained using crystal violet.
Transferase-mediated dUTP nick end labeling (TUNEL) assay
Transfected LUAD cells were treated with 4% paraformaldehyde (PFA) and TUNEL reagent (Merck KGaA, Darmstadt, Germany) was added. After being stained with DAPI, labeled samples were analyzed with the help of optical microscopy (Olympus).
Subcellular fractionation
Via cytoplasmic and Nuclear RNA Purification Kit (Norgen, Thorold, ON, Canada), cytoplasmic and nuclear elements were isolated from 5 × 104 cells in 24-well plates. Expression levels of C2CD4D-AS1 were analyzed by RT-qPCR. GAPDH or U6 was respectively regarded as cytoplasmic or nuclear control.
Fluorescence in situ hybridization (FISH)
Cells were permeabilized for 1 hour, followed by incubation with digoxigenin (DIG)-labeled C2CD4D-AS1 probe in the dark at 37°C overnight. Hoechst was applied to stain nucleus. Finally, images were acquired under the fluorescence microscope (Leica Microsystems, Mannheim, Germany).
RNA binding protein immunoprecipitation (RIP)
RIP assay was undertaken with the help of RNA-binding protein immunoprecipitation kit (Millipore, Burlington, MA, USA). In brief, 6 × 107 PC-7 and H1299 cells were lysed using RIP buffer and incubated with Ago2 antibody (1:50) or immunoglobulin G (IgG) antibody (ab172730, Abcam; 1:20) for 2 h. Finally, RT-qPCR analysis was conducted.
RNA pull down assay
2 × 107 PC-9 or H1299 cells were treated with biotinylated C2CD4D-AS1 (Bio-C2CD4D-AS1) or biotinylated negative control probes (Bio-NC) (PCR; 50 ng/μl). Magnetic beads were then added into cells for cultivation overnight at 4°C. The pull-downs collected by beads were purified for RT-qPCR.
DNA pull down assay
DNA pull down assay was done to explore the potential binding correlation between ETV4 and C2CD4D-AS1 promoter as previously described [Citation20]. Briefly, biotinylated promoter probes including Bio-C2CD4D-AS1 promoter, Bio-C2CD4D-AS1 promoter Mut), and the negative control (Bio-NC) were co-cultured with cell lysates. The input group was utilized as the positive control. Streptavidin magnetic beads were then added for further cultivation with the DNA-protein complex, and the proteins eluted from DNA were measured via western blot.
Chromatin immunoprecipitation (ChIP)
The EZ ChIP™ Chromatin Immunoprecipitation Kit (Millipore, Burlington, MA, USA) was used for implementation of ChIP assay. Briefly, 2 × 107 LUAD cells fixed with 1% formaldehyde were sonicated to be fragments (200–500 bp). Rabbit anti-ETV4 (Abcam) antibody and IgG antibody (Abcam) were precipitated with chromatin. The immunoprecipitated DNA was purified and subjected to RT-qPCR analysis.
Luciferase reporter assay
The binding sites (wild type or mutant type) of C2CD4D-AS1 promoter region were sub-cloned into the pGL3 vector (Promega, Madison, WI) to construct luciferase reporter genes (Wt-luc or Mut-luc). The constructed luciferase reporter genes were co-transfected along with ETV4 overexpression plasmids or the empty vector into LUAD cells. Similarly, the sequence of C2CD4D-AS1 or NEK2 mRNA 3ʹuntranslated region (3ʹUTR) containing wild-type (Wt) and mutant type (Mut) of miR-3681-3p binding site was inserted into pmirGLO dual-luciferase vector to form pmirGLO-C2CD4D-AS1-Wt/Mut and pmirGLO-NEK2 3ʹUTR-Wt/Mut respectively. Later, we co-transfected miR-3681-3p mimics or NC mimics with the reporter gene into PC-9 and H1299 cells (1 × 104). After 48 h, the Dual-Luciferase Reporter Gene Assay Kit (Yeasen, Shanghai, China) was applied to measure luciferase activity.
Western blot analysis
RIPA buffer was firstly utilized to obtain the protein lysates. Total protein samples were separated with 12.5% SDS-PAGE and later transferred to PVDF membranes (Thermo fisher, IL, USA). Afterward, membranes went through incubation with primary antibodies at 4°C overnight after being blocked in 5% milk without fat. After being washed with PBS for three times, the blots were then incubated with secondary antibodies against NEK2 and GAPDH (Abcam) for 1 h at dark room. Chemiluminescence system (GE Healthcare, Chicago, USA) was utilized to perform the quantification analysis of protein.
Statistical analysis
Experimental data were analyzed by SPSS 22.0 statistical software package. Representative data were expressed as mean ± standard deviation (SD). The differences between two groups or more groups were analyzed with the employment of Student’s t-test or analysis of variance (ANOVA). All the experiments were independently conducted at least three times. Differences were considered statistically significant when P < 0.05.
Results
Silence of C2CD4D-AS1 hinders cell proliferation, migration, invasion, and promotes cell apoptosis in LUAD
Firstly, we searched for Ensembl gene ID of C2CD4D-AS1 on Ensembl (http://asia.ensembl.org/index.html/) for subsequent data screening (Fig. S1A). Through GEPIA database (http://gepia2.cancer-pku.cn/), C2CD4D-AS1 was discovered to be overexpressed in LUAD, cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) and thymoma (THYM) (Fig. S1B). Furthermore, based on the searching results of overexpressed genes in LUAD from GEPIA, C2CD4D-AS1 was found (Fig. S1C). Also through GEPIA database, we found C2CD4D-AS1 was highly expressed in LUAD tissues compared with normal tissues (*P < 0.05) (). In comparison with normal cell line (BEAS-2B), the up-regulation of C2CD4D-AS1 in LUAD cell lines (H1975, A549, PC-9, and H1299), especially in PC-9 and H1299, was also discovered by RT-qPCR analysis (*P < 0.05, **P < 0.01) (). The two cell lines, PC-9 and H1299, were selected for following experiments. Subsequently, we conducted loss-of-function assays to assess the role of C2CD4D-AS1 in LUAD cells. Sh-C2CD4D-AS1#1 and sh-C2CD4D-AS1#2 were selected for subsequent studies for the excellent knockdown efficiencies (**P < 0.01) (). CCK8 and colony formation assays demonstrated that down-regulation of C2CD4D-AS1 obviously suppressed the proliferation of LUAD cells (**P < 0.01) (). The findings of transwell assay indicated that C2CD4D-AS1 silence inhibited LUAD cell migration and invasion (**P < 0.01) (). TUNEL assay suggested that cell apoptosis was promoted after C2CD4D-AS1 was depleted (**P < 0.01) (). Taken together, C2CD4D-AS1 facilitated growth of LUAD cells.
Figure 1. Silence of C2CD4D-AS1 hinders cell malignant phenotype in LUAD a. The overexpression of C2CD4D-AS1 in LUAD tissues was showed in GEPIA database. b. Differences of C2CD4D-AS1 expression between LUAD cell lines and normal cell line were evaluated with help of RT-qPCR analysis. c. The knockdown efficiency of sh-C2CD4D-AS1#1/2/3 was detected. d-e. Cell proliferation capacity was examined by CCK8 and colony formation assays. F-G. Transwell assays were applied to measure the migration and invasion of PC-9 and H1299 cells. h. Cell apoptosis was observed with the employment of TUNEL assay. *P < 0.05, **P < 0.01
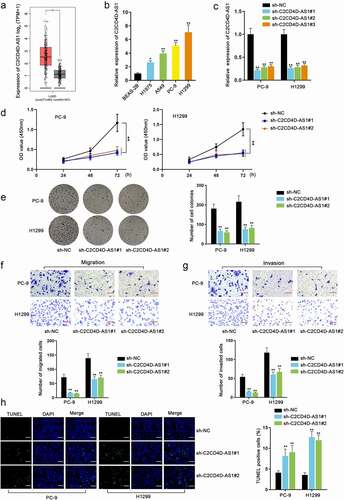
ETV4 stimulates the up-regulation of C2CD4D-AS1
Transcription factors have been considered to regulate genes positively or negatively [Citation9]. In view of the notable up-regulation of C2CD4D-AS1 in LUAD cells, we conjectured that C2CD4D-AS1 was also activated by certain transcription factors. Through HumanTFDB database (http://bioinfo.life.hust.edu.cn/HumanTFDB#!/) (Score ≥ 25) and GEPIA database (Log2 Fold change ≥ 2), we discovered that ETV4 was dramatically high expressed in LUAD cell lines and had a high binding score with C2CD4D-AS1 promoter, and thus ETV4 was screened out as the potential transcription factor of C2CD4D-AS1 (). RT-qPCR analysis discovered that ETV4 expression could be inhibited by sh-ETV4 transfection and promoted by pcDNA3.1/ETV4 transfection, and the expression level of C2CD4D-AS1 markedly decreased or increased in consistence with ETV4 down-regulation or up-regulation (**P < 0.01) (). It indicated that C2CD4D-AS1 was positively regulated by ETV4. The transcriptional response element for ETV4 in the promoter of C2CD4D-AS1 is identified in . Based on the detection of C2CD4C-AS1 promoter enrichment in ChIP assay, the strong binding affinity between C2CD4D-AS1 promoter and ETV4 protein was ascertained (**P < 0.01) (). In DNA pull down assay, ETV4 could be abundantly pulled down by Bio-C2CD4D-AS1 promoter, further confirming the cohesion between C2CD4D-AS1 promoter and ETV4. Luciferase reporter assay uncovered that ETV4 overexpression increased the luciferase activity of wild-type C2CD4D-AS1 promoter reporter (**P < 0.01) instead of the mutant type (). We further overexpressed ETV4 after silencing C2CD4D-AS1 and it was clearly shown that pcDNA3.1/ETV4 could recover the reduced C2CD4D-AS1 expression induced by sh-C2CD4D-AS1#1 transfection (**P < 0.01) (Fig. S1D). Furthermore, with the aim to explore whether ETV4 could affect LUAD cell biological function through C2CD4D-AS1, functional assays in a rescue manner were carried out. The overexpression efficiency of pcDNA3.1/C2CD4D-AS1 was first detected and it was suggested that the transfection of pcDNA3.1/C2CD4D-AS1 led to an overt increment in C2CD4D-AS1 level (**P < 0.01) (Fig. S2A). As presented in Fig. S2B-C, ETV4 depletion could overtly weaken the proliferation of LUAD cells, which was then discovered to be reversed on account of C2CD4D-AS1 up-regulation (**P < 0.01). Similarly, the suppressive influences of ETV4 knockdown on LUAD cell migration and invasion were also countervailed by C2CD4D-AS1 overexpression (**P < 0.01) (Fig. S2D-E). Moreover, the changes of LUAD cell apoptosis under the influence of ETV4/C2CD4D-AS1 were also evaluated. The results of TUNEL assay suggested that the strengthened cell apoptosis, resulting from ETV4 deficiency, could be rescued by C2CD4D-AS1 overexpression (**P < 0.01) (Fig. S2F). Overall, ETV4 activated the transcription of C2CD4D-AS1, and mediated the biological functions of LUAD cells through C2CD4D-AS1.
Figure 2. ETV4 stimulates the up-regulation of C2CD4D-AS1 a. Venn diagram demonstrated transcription factors with high binding scores in HumanTFDB overlapped with up-regulated transcription factors in LUAD. b-c. RT-qPCR tested the efficiency of ETV4 inhibition or overexpression, as well as the variations in C2CD4D-AS1 expression caused by ETV4 down-regulation or up-regulation. d. The binding sequence between ETV4 and C2CD4D-AS1 promoter was presented. e. The affinity of ETV4 and C2CD4D-AS1 promoter was examined through ChIP assay. f. In DNA pull down assay, the enrichment of ETV4 in Bio-C2CD4D-AS1 promoter was measured by western blot. g. The binding of ETV4 and C2CD4D-AS1 promoter was further confirmed by luciferase reporter assay. **P < 0.01, n.s.: no significance
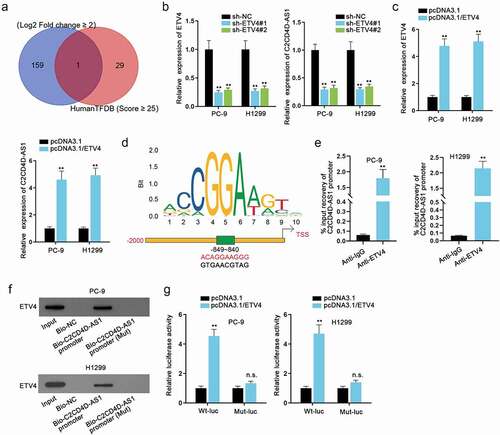
C2CD4D-AS1 serves as a miR-3681-3p sponge
Subcellular fractionation and FISH assays were carried out and the results indicated the main location of C2CD4D-AS1 in cytoplasm rather than in nucleus (). In consideration of that post-transcriptional regulation often occurs in cytoplasm, ceRNA mechanism was hypothesized for it was a classic event in post-transcriptional regulation [Citation21]. According to starBase (http://starbase.sysu.edu.cn/), we found 6 potential downstream miRNAs (miR-128-3p, miR-3681-3p, miR-216a-3p, miR-27a-3p, miR-27b-3p and miR-218-5p) of C2CD4D-AS1. RNA pull down assay identified miR-3681-3p as the putative miRNA for C2CD4D-AS1 for the reason that only miR-3681-3p could be notably pulled down by biotinylated C2CD4D-AS1 probe (**P < 0.01) (). The binding site of C2CD4D-AS1 and miR-3681-3p was identified (). Furthermore, miR-3681-3p expression was enhanced after transfection of miR-3681-3p mimics (**P < 0.01) (). Then we found that the luciferase activity was observably decreased in the C2CD4D-AS1-Wt group (**P < 0.01) while remaining almost unchanged in the C2CD4D-AS1-Mut group after co-transfection with miR-3681-3p mimics (). In summary, miR-3681-3p was sponged by C2CD4D-AS1.
Figure 3. C2CD4D-AS1 serves as a miR–3681–3p sponge a. The distribution of C2CD4D-AS1 in nucleus and cytoplasm of LUAD cells was detected via subcellular fractionation. b. FISH assay was done for the confirmation of C2CD4D-AS1 distribution in LUAD cells. C. RNA pull down revealed the strong interaction between C2CD4D-AS1 and miR–3681–3p. d. Binding sequences between C2CD4D-AS1 and miR–3681–3p, along with mutated C2CD4D-AS1 sequence were displayed. e. MiR–3681–3p expression in LUAD cells with miR–3681–3p mimics transfection was quantified via RT-qPCR. f. The luciferase activity of C2CD4D-AS1-Wt/Mut was assessed by luciferase reporter assay in response to the transfection of miR–3681–3p mimics. **P < 0.01, n.s.: no significance
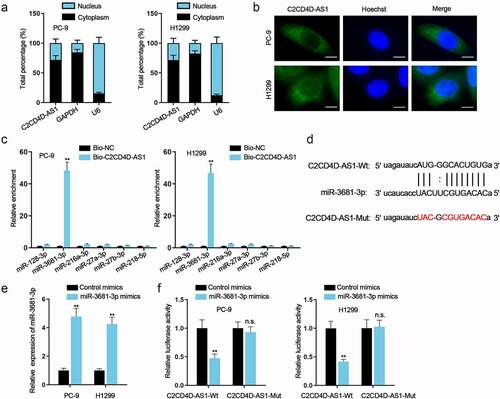
NEK2 is negatively regulated by miR-3681-3p
Based on starBase (CLIP Data ≥ 5), high-mobility group box 3 (HMGB3), Gremlin 1 (GREM1) and NEK2 were discovered by overlapping with obviously up-regulated mRNAs in GEPIA database (log2 fold change ≥ 2) (). RT-qPCR analysis manifested that only NEK2 expression was prominently down-regulated (**P < 0.01) when miR-3681-3p was up-regulated (). The hypothetic miR-3681-3p binding site was identified in the NEK2 3ʹ UTR region (). Data of RIP assay demonstrated the high enrichment of C2CD4D-AS1, miR-3681-3p and NEK2 in Ago2-containing beads (**P < 0.01) (). Moreover, an overt augment in the enrichment of NEK2 and decline in the enrichment of C2CD4D-AS1 were discovered on account of C2CD4D-AS1 inhibition, while no obvious variation was found in that of miR-3681-3p (**P < 0.01) (Fig. S2G). Additionally, from luciferase reporter assays, we observed decreased luciferase activity of NEK2 3ʹ UTR-Wt (**P < 0.01), while no apparent change was observed when the putative binding site was mutated (). According to RT-qPCR and western blot results, miR-3681-3p inhibitor could restore the reduced RNA level and protein level of NEK2 caused by knockdown of C2CD4D-AS1 (**P < 0.01) (). All these data supported that NEK2 was a target gene of miR-3681-3p and negatively regulated by miR-3681-3p.
Figure 4. NEK2 is negatively regulated by miR–3681–3p a. 3 potential mRNAs were predicted by utilizing starBase (CLIP Data ≥ 5) and GEPIA (log2 fold change ≥ 2). b. The expression of these mRNAs was examined after overexpressing miR–3681–3p. c. Bioinformatics tools analyzed the binding sequence between miR–3681–3p and NEK2 3ʹUTR. d. RIP assay was performed for investigation into the combination among C2CD4D-AS1, miR–3681–3p and NEK2. e. The binding of miR–3681–3p and NEK2 3ʹUTR was validated by luciferase reporter assay. f-g. The mRNA and protein levels of NEK2 in sh-NC group, sh-C2CD4D-AS1#1 group and sh-C2CD4D-AS1#1+ miR–3681–3p inhibitor group was measured via RT-qPCR and western blot. **P < 0.01, n.s.: no significance
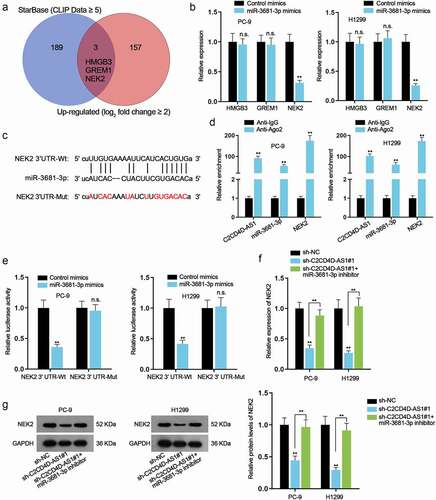
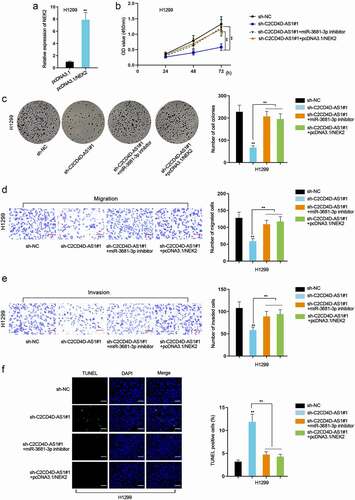
C2CD4D-AS1 accelerates LUAD cell proliferation, migration, invasion, and suppresses cell apoptosis via miR-3681-3p/NEK2 axis
Rescue assays were performed to investigate the impacts of C2CD4D-AS1/miR-3681-3p/NEK2 axis on LUAD cell behaviors. Firstly, we enhanced the expression of NEK2 in H1299 by transfection of pcDNA3.1/NEK2 plasmids (**P < 0.01) (). CCK8 and colony formation assays substantiated that cell proliferative capacity was repressed by depletion of C2CD4D-AS1, which was completely rescued by miR-3681-3p inhibition or up-regulation of NEK2 (**P < 0.01) (). Knockdown of C2CD4D-AS1 led to a decrease in migrated and invaded cell number, while the influence could be countervailed by down-regulation of miR-3681-3p or NEK2 overexpression (**P < 0.01) (). Additionally, TUNEL assay showed that miR-3681-3p down-regulation or NEK2 up-regulation counteracted the strengthened apoptotic ability of LUAD cells induced by C2CD4D-AS1 silencing (**P < 0.01) (). To be concluded, C2CD4D-AS1 exacerbated the malignant behaviors of LUAD cells via modulation of miR-3681-3p/NEK2 axis.
Figure 5. C2CD4D-AS1 accelerates LUAD cell proliferation, migration, invasion and suppresses cell apoptosis by modulating miR–3681–3p/NEK2 axis a. RT-qPCR analyzed NEK2 expression in response to NEK2 augment. b-c. Cell proliferation in sh-NC group, sh-C2CD4D-AS1#1 group, sh-C2CD4D-AS1#1+ miR-3681–3p inhibitor group and sh-C2CD4D-AS1#1+ pcDNA3.1/NEK2 group was detected by CCK8 and colony formation assays. d-e. Transwell assay was implemented for observation of cell migration and invasion in different groups. f. The apoptotic capacity of LUAD cells transfected with indicated plasmids was examined by TUNEL assay. **P < 0.01
Discussion
In view of the fact that the overall 5-year survival rate of LUAD patients remains unsatisfactory in spite of advancements in the diagnosis, treatment, and prognosis, exploring vital biomarkers in LUAD is extremely urgent.
More and more lncRNAs have been discovered to play the role of biomarkers and have multiple functions in LUAD tumorigenesis and metastasis [Citation22]. As a novel lncRNA, C2CD4D-AS1 is scarce reported. C2CD4D-AS1 has been only investigated in autoimmune diseases and identified to be a potential therapeutic target [Citation8]. On this basis, we explored the role of C2CD4D-AS1 in LUAD in our study. Bioinformatics analysis and RT-qPCR have exhibited that C2CD4D-AS1 was conspicuously up-regulated in LUAD. Moreover, loss-of-function assays proved that C2CD4D-AS1 silencing exerted a suppressive role on cell proliferation, migration, and invasion while promoted cell apoptosis in LUAD.
Transcription factors are often recruited to modulate the transcription and regulate gene expression [Citation23]. Additionally, ETV4 has been reported to facilitate cell proliferation and invasion of non-small cell lung cancer and LUAD [Citation24,Citation25]. Likewise, our experimental data verified that ETV4 had a strong binding force with C2CD4D-AS1 promoter and positively modulated the expression of C2CD4D-AS1 to influence LUAD cell malignant behaviors.
LncRNA-miRNA-mRNA ceRNA network is universal in the regulatory mechanism of the occurrence and development of LUAD [Citation26]. This paper revealed C2CD4D-AS1 was predominantly localized in the cytoplasm of LUAD cells. Logically, we applied bioinformatics tools and RNA pull down assay to identify miR-3681-3p as the possible downstream of C2CD4D-AS1 in LUAD cells. The interaction between C2CD4D-AS1 and miR-3681-3p was also supported by mechanism research. Furthermore, inhibition of miR-3681-3p completely rescued C2CD4D-AS1 knockdown mediated suppression on LUAD cell proliferation, migration and, invasion and boost on apoptosis.
NEK2 has been extensively investigated in lung cancer and the results all elucidate that NEK2 can be regarded as a marker in lung cancer. Chen et al. have testified that high expression of NEK2 regulated by EGFR, could promote lung cancer progression [Citation27]. Consistent with this finding, we found that NEK2 was targeted by miR-3681-3p and had a negative correlation with miR–3681–3p. Moreover, rescue assays showed that NEK2 overexpression countervailed the influence of C2CD4D-AS1 silencing on LUAD cell proliferation, migration, invasion and apoptosis. Nevertheless, our study did not prove that the axis of ETV4/C2CD4D-AS1/miR–3681–3p/NEK2 functioned as the dominant regulatory mechanism driving LUAD initiation and development. It still requires substantial efforts to make clear the comprehensive regulatory pathways involved in LUAD.
Conclusions
To be concluded, C2CD4D-AS1 was determined to accelerate growth of LUAD cells. From the perspective of mechanism, C2CD4D-AS1 expression transcriptionally modulated by ETV4 could sponge miR–3681–3p and consequently regulate NEK2 expression. Finally, rescue experiments testified the validity of ETV4/C2CD4D-AS1/miR–3681–3p/NEK2 axis in LUAD cells, which might contribute to a better understanding of LUAD.
Supplemental Material
Download Zip (2.4 MB)Acknowledgments
We appreciate the supports of our experimenters.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Kuhn E, Morbini P, Cancellieri A, et al. Adenocarcinoma classification: patterns and prognosis. Pathologica. 2018;110(1):5–11.
- Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46.
- Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109.
- Weidle UH, Birzele F, Kollmorgen G, et al. Long Non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics. 2017;14(3):143–160.
- Yang J, Li C, Mudd A, et al. LncRNA PVT1 predicts prognosis and regulates tumor growth in prostate cancer. Biosci Biotechnol Biochem. 2017;81(12):2301–2306.
- Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136.
- Zhang XT, Pan SX, Wang AH, et al. Long Non-Coding RNA (lncRNA) X-Inactive Specific Transcript (XIST) plays a critical role in predicting clinical prognosis and progression of colorectal cancer. Med Sci Monit. 2019;25:6429–6435.
- Teimuri S, Hosseini A, Rezaenasab A, et al. Integrative Analysis of lncRNAs in Th17 cell lineage to discover new potential biomarkers and therapeutic targets in autoimmune diseases. Mol Ther Nucleic Acids. 2018;12:393–404.
- Bouhlel MA, Lambert M, David-Cordonnier MH. Targeting Transcription Factor Binding to DNA by competing with DNA binders as an approach for controlling gene expression. Curr Top Med Chem. 2015;15(14):143–160.
- Lambert M, Jambon S, and Depauw S, et al. Targeting transcription factors for cancer treatment. Molecules. 2018;23(6):1479 .
- Jiang H, Li T, Qu Y, et al. Long non-coding RNA SNHG15 interacts with and stabilizes transcription factor Slug and promotes colon cancer progression. Cancer Lett. 2018;425:78–87.
- Peng Z, Wang J, Shan B, et al. The long noncoding RNA LINC00312 induces lung adenocarcinoma migration and vasculogenic mimicry through directly binding YBX1. Mol Cancer. 2018;17(1):167.
- Rodriguez AC, Vahrenkamp JM, Berrett KC, et al. ETV4 is necessary for estrogen signaling and growth in endometrial cancer cells. Cancer Res. 2020;80(6):1234–1245.
- Qi X, Zhang DH, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718.
- Zhang Y, Xu Y, Feng L, et al. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7(39):64148–64167.
- Yang XZ, Cheng TT, He QJ, et al. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer. 2018;17(1):126.
- Li B, Mao R, Liu C, et al. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018;197:122–129.
- Xiong DD, Li ZY, Liang L, et al. The LncRNA NEAT1 accelerates lung adenocarcinoma deterioration and binds to Mir-193a-3p as a competitive endogenous RNA. Cell Physiol Biochem. 2018;48(3):905–918.
- Arocho A, Chen B, Ladanyi M, et al. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathology Am J Surg Pathol Part B. 2006;15(1):56–61
- Wu W, Tan W, Ye S, et al. Analysis of the promoter region of the human miR-32 gene in colorectal cancer. Oncol Lett. 2019;17(4):3743–3750.
- Smillie CL, Sirey T, Ponting CP. Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Crit Rev Biochem Mol Biol. 2018;53(3):231–245.
- Wang Y, Fu J, Wang Z, et al. Screening key lncRNAs for human lung adenocarcinoma based on machine learning and weighted gene co-expression network analysis. Cancer Biomarkers. 2019;25(4):24–231.
- Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455.
- Cheng T, Zhang Z, Cheng Y, et al. ETV4 promotes proliferation and invasion of lung adenocarcinoma by transcriptionally upregulating MSI2. Biochem Biophys Res Commun. 2019;516(1):278–284.
- Wang Y, Ding X, Liu B, et al. ETV4 overexpression promotes progression of non-small cell lung cancer by upregulating PXN and MMP1 transcriptionally. Mol Carcinog. 2020;59(1):73–86.
- Li DS, Ainiwaer JL, Sheyhiding I, et al. Identification of key long non-coding RNAs as competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Eur Rev Med Pharmacol Sci. 2016;20:2285–2295.
- Chen C, Peng S, Li P, et al. High expression of NEK2 promotes lung cancer progression and drug resistance and is regulated by mutant EGFR. Mol Cell Biochem. 2020;475(1–2):15–25.
