ABSTRACT
In fission yeast, MBF-dependent transcription is required for cells to complete S phase. The MBF transcription factor is regulated through a complex feedback mechanism that involves the co-repressors Yox1 and Nrm1 that are loaded onto MBF at the end of S phase, while positive transactivation is achieved through the constitutive binding of the co-activator Rep2. Here we show that Rep2 is required to fully recruit the chromatin remodelers SWI/SNF and RSC to MBF-regulated promoters. On the contrary, Nrm1 and Yox1, when bound to the MBF complex, block the approximation of these chromatin remodelers to MBF-regulated promoters. We propose that SWI/SNF and RSC are recruited to MBF-regulated genes, and RSC together with SAGA complex are important to regulate the G1-to-S transcriptional wave. Mutants of these remodeler complexes are highly sensitive when cells are exposed to insults that challenge DNA synthesis.
KEYWORDS:
Introduction
During fission yeast life cycle, cells face with a critical decision between vegetative growth, sexual conjugation, or remain in stationary phase. This decision point, known as Start in yeasts and restriction point in metazoans, occurs in late G1 phase of the cell cycle. Genetic analyses have identified several proteins that are required for passage through Start in Schizosaccharomyces pombe [Citation1]. Among them, it was originally described the cyclin-dependent kinase CDK1Cdc2 [Citation2] and the transcription factor Cdc10 [Citation3]. While the activity of Cdc2 is required for Start and for the G2/M transition, Cdc10 is only required for the completion of Start. Cdc10 is an essential component of a multimeric complex known as MBF complex, whose core proteins are Cdc10 and the DNA binding proteins Res1 and Res2 [Citation4–8]. Additional proteins that also form part of the MBF complex are involved in its cell cycle-dependent regulation. Among them, the co-activator Rep2 [Citation9], the SAGA complex, which binds the MBF complex through Rep2 [Citation10] and the co-repressors Nrm1 and Yox1, which are involved in switching-off MBF-dependent transcription during the G2 phase of the cell cycle [Citation11–14].
MBF, which is the functional analog of mammalian pRB/E2F, is responsible for the G1-to-S transcriptional wave [Citation15], controlling the expression of a set of genes that are directly or indirectly required for DNA synthesis during the S phase of the cell cycle [Citation16]. Among them, cdc22 which codes for the large subunit of ribonucleotide reductase, the S phase cyclin cig2 and cdc18 and cdt1, which are an intrinsic part of the DNA replication machinery [Citation15–17]. Not surprisingly, it has been described that among the genes that are regulated by the MBF complex one can also find those that are involved in the negative feedback that represses MBF-dependent transcription at the end of S phase, such as nrm1, yox1, and cig2 [Citation11,Citation12,Citation14,Citation16].
The cell-cycle-dependent expression of the MBF operon is disrupted in strains with impaired MBF function and cells are unable to complete Start. On the contrary, hyperactivation of MBF induces extremely high levels of the MBF-regulated genes, leading to over-replication of DNA and replicative stress [Citation18]. Interestingly, the crosstalk between checkpoints and MBF works in both directions. Cells under replicative stress activate the DNA replication checkpoint through the effector kinase CHK2Cds1; one of the substrates of Cds1 is the co-repressor Yox1, which when phosphorylated is released from the MBF complex activating MBF-dependent transcription [Citation11,Citation12]. Interestingly, when there is severe damage to DNA and the DNA integrity checkpoint is activated, its effector kinase CHK1Chk1 phosphorylates Cdc10 repressing MBF-dependent transcription [Citation19,Citation20]. Therefore, tight control of MBF activity is essential to ensure normal cell-cycle progression.
The core of MBF (Cdc10, Res1, and Res2) has been found bound to its target promoters throughout the cell cycle [Citation21], implicating that MBF activity is not achieved by modulating its DNA-binding activity. We have recently shown that the activity of the histone acetyltransferase (HAT) Gcn5/SAGA is required to fully activate the transcription of MBF-regulated genes [Citation10]. The SAGA complex is recruited to the MBF complex through interaction to the co-activator Rep2. This recruitment is also cell cycle-regulated, achieving its maximum during S phase. Opposed to this recruitment, the co-repressors Nrm1 and Yox1 work as physical barriers for SAGA recruitment. Gcn5 is required at MBF promoters to acetylate-specific histone H3 residues (K9 and K18), facilitating nucleosome eviction and promoter clearance to load RNA polymerase. However, other large complexes are known to participate in the regulation of transcription. Chromatin remodelers use the energy of ATP hydrolysis to relocate DNA around nucleosomes, disrupting histone–DNA interaction. There are at least five families of chromatin-remodeling complexes based on structural or functional domains: SWI/SNF, ISWI, INO80, CHD, and Mi-2 [Citation22]. Fission yeast lacks ISWI‐type remodelers, and we showed in the past that the activity of the INO80 complex helps in the timing of transcriptional activation of MBF-regulated genes [Citation23]. Regarding the SWI/SNF family, there are two complexes (SWI/SNF and RSC), although both complexes function in the sliding and eviction of nucleosomes. The two complexes have ATP-dependent nucleosome-remodeling activity, but they contain different proteins (the core protein in SNI/SNF is Snf22, while in RSC is Snf21). But they also share some of them, like Ssr1, Ssr2, Ssr3, Ssr4, Arp9, and Arp42 [Citation24].
Here we investigate how the MBF factor recruits the activity of the SWI/SNF and RSC chromatin remodeling complexes when the transcription of MBF-regulated genes is induced during S phase. We show that this recruitment is also mediated by the MBF co-activator Rep2, which is reminiscent of what happens also with the SAGA complex. This points to Rep2 as a hub to bring enzymatic activities that modify chromatin to MBF-regulated genes facilitating the transcriptional activation during S phase.
Materials and methods
Strains and media
All S. pombe strains are isogenic to wild-type 972h- and are listed in . Media were prepared as previously described [Citation25]. Cultures were synchronized at the end of G2 phase using the temperature-sensitive strain cdc25-22. Briefly, cells were cultured in minimal media at the permissive temperature (25°C) before shifting to non-permissive temperature (36°C) for 4 h and then released at permissive temperature. Alternatively, cells were synchronized at metaphase using the cdc2-asM17 allele, as described elsewhere [Citation10]. Samples from synchronizations were analyzed on microscopy and septation index was measured from cell fixed in 70% ethanol and stained with 25 μg/ml calcofluor.
Table 1. Strains used in this work
Sensitivity analysis
Cells were grown in liquid YE5S media to an OD600 of 0.3. Cultures were 10-fold serially diluted in YE5S and spotted onto drug-free or HU-containing YE5S plates at different concentrations. Plates were incubated at 30°C or 25°C for 2–5 d. In liquid cultures, HU was used at a concentration of 12 mM.
Gene expression analysis
Total RNA was extracted from 40 ml of cells at an OD600 of 0.5 by standard hot-phenol method, as described before [Citation10]. 10 µg of purified RNA were incubated with DNase I (Roche) for 30 min at 37°C, followed by purification using acid phenol-chloroform extraction. Reverse transcription was performed on 4 µg of total RNA using Reverse Transcription System (Promega), as indicated in the manufacturer´s manual. The cDNA was diluted 1:2 prior to PCR amplification. cDNAs were quantified by real-time qPCR on Light Cycler II using Light Cycler 480 SYBR Green I Master (Roche). Values were normalized to tbf2. The error bars (SEM) were calculated from at least three biological triplicates, unless indicated otherwise. Primers used to quantify are listed in .
Table 2. Primers used in this work
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were carried out as previously described [Citation26] with minor modifications. Briefly, 50 ml of cells at an OD600 of 0.5 per sample were cross-linked adding 1% formaldehyde for 10 min at 25°C; cross-linking was stopped with 125 mM glycine. Pellets were lysed with a bead beater in lysis buffer (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM PMSF). Lysates were sonicated, yielding chromatin fragments of ≈400 bp average size. Chromatin lysate was immunoprecipitated with specific antibodies and protein G-Sepharose beads overnight at 4°C on a rot o-wheel. Beads were washed once in lysis buffer, twice in lysis buffer containing 0.5 M NaCl, twice in washing buffer (10 mM Tris pH 8.0, 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, and 1 mM PMSF) and once in TE (10 mM Tris pH 8.0, 1 mM EDTA). DNA was eluted by incubating the beads for 20 min at 65°C with elution buffer (50 mM Tris pH 7.5, 10 mM EDTA pH 8.0, 1% SDS), and a second incubation in TE with 0.67% SDS. Formaldehyde cross-linking was reversed by overnight incubation at 65°C. Proteins were digested by incubation for 2 h at 37°C with 0.3 mg/ml proteinase K and 0.04 mg/ml glycogen. 125 mM NaCl was added, chromatin was purified by phenol/chloroform extraction and precipitated with cold ethanol for at least 2 h at −20°C. DNA was precipitated, air-dried, and resuspended in TE buffer. Recovered DNA was amplified by quantitative real-time PCR using Light Cycler 480 SYBR Green I Master (Roche). A region corresponding to chromosome I (SPNCRNA.650) was used as negative control. The error bars (SEM) were calculated from at least three biological triplicates, unless indicated otherwise. The primers used are listed in .
Results and discussion
Screening of chromatin remodelers that increase hydroxyurea (HU) sensitivity in fission yeast
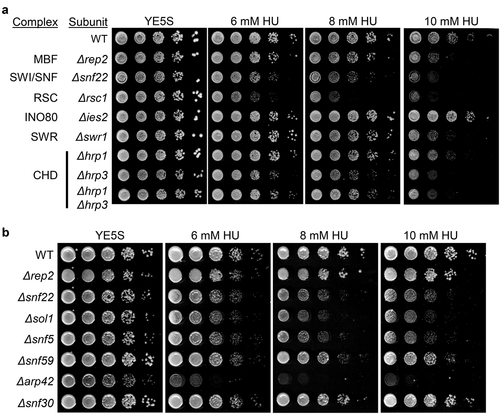
As a first approximation to determine chromatin remodelers that could modulate MBF activity, we decided to screen mutants of the different chromatin remodeling complexes for sensitivity to hydroxyurea (HU). As can be observed in , mutants in subunits of the SWI/SNF complex (Δsnf22), the RSC complex (Δrsc1), and CHD complex (Δhrp3) were sensitive to different HU concentrations. It is worth noting that some of them were even more sensitive to HU than Δrep2 cells (the co-activator of the MBF complex). On the other hand, the INO80 and the SWR1 mutants that we tested (Δies2 and Δswr1) showed no sensitivity in this assay since cells were as resistant to HU as wild-type cells. The results with the CHD complex were not conclusive since some mutants behave as wild-type cells (Δhrp1) and some were slightly sensitive (Δhrp1Δhrp3 double mutant). This result, in combination with the absence of effect on MBF-dependent transcription (not shown), led us to concentrate on the effect of the SWI/SNF and the RSC complexes on MBF regulation.
Figure 1. Genetic interaction between chromatin remodeler complexes and HU sensitivity. (a) Fivefold serial dilutions of the strains indicated on the left were spotted onto rich media or in media with 6 mM HU, 8 mM HU, or 10 mM HU. Plates were incubated at 30°C for 2–4 d. (b) Fivefold serial dilutions of the strains indicated on the left were spotted onto rich media or in media with 6 mM HU, 8 mM HU, or 10 mM HU. Plates were incubated at 30°C for 2–4 d
The SWI/SNF complex is recruited at the promoters of MBF-regulated genes
We further analyzed the sensitivity to HU of different mutant strains lacking nonessential components of the SWI/SNF complex. As shown in , not only Δsnf22 cells were sensitive to increasing concentrations of HU, but also Δsol1, Δsnf5, and Δarp42 were highly sensitive to HU, while Δsnf59 was mildly sensitive. This result confirmed that when SWI/SNF activity was compromised, cells were sensitive to HU, not circumscribing the effect to a specific mutation on snf22. Next, and to further determine if there was a physical connection between SWI/SNF complex and MBF-regulated genes, we determined if there was such interaction by ChIP. As shown in , we were able to detect the binding of Snf22 to cdc18 and cdc22, the two best characterized genes regulated by the MBF complex. Interestingly, we were able to detect a noticeable increase in the binding of Snf22 to the promoter region of these two genes after treatment with HU. This recruitment was negligible to the ORF or to the 3' region of any of the MBF-regulated genes that we tested.
Figure 2. Snf22 binds to the promoters of MBF-regulated genes. (a) Cells expressing Snf22-Myc under the control of its own promoter were treated or not with HU for 3 h. ChIP experiments were performed using primers covering promoter (prom), coding (ORF), and termination (term) sequences of the cdc18 and cdc22 genes (as indicated). Error bars (SEM) were calculated from biological triplicates. Significant differences between treated and untreated samples were determined by the Student´s t-test (*P < 0.05). (b) Wild type (WT) or Δgcn5 cells expressing Snf22-Myc were treated or not with HU for 3 h. ChIP experiments were performed using primers covering the promoter region of the cdc18 and cdc22 genes or the SPNCRNA.650 gene as a control. Error bars (SEM) were calculated from biological triplicates. (c) Wild type (WT), Δrep2, Δyox1, or Δnrm1 cells expressing Snf22-Myc were treated or not with HU for 3 h, as indicated. Error bars (SEM) were calculated from biological triplicates. Significant differences between treated and untreated samples were determined by the Student´s t-test (*P < 0.05; ** P < 0.01)
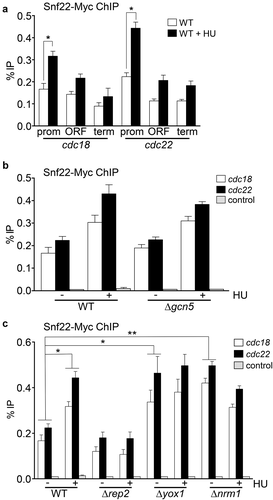
Snf22 has a bromo-domain at the carboxy-terminal region [Citation27]. These domains recognize acetylated lysine residues at histones. Since we have recently shown that SAGA/Gcn5 is required for the acetylation of several lysines of histone H3 (H3K9, H3K14, and H3K19) at the promoter region of MBF-regulated genes [Citation10], we wondered if Gcn5 was required for Snf22 recruitment at these genes. We determined the binding of Snf22 to the promoter region of cdc18 and cdc22 in a wild-type strain and in a Δgcn5 strain. As shown in , binding of Snf22 was unaffected by the absence of Gcn5, both in untreated or in cells treated with HU. This result pointed to the possibility that the recruitment of Snf22 could be dependent on transcription but independent on the status of acetylation at H3K9, H3K14, and H3K19 at the promoters of MBF-regulated genes. To confirm this hypothesis, we measured Snf22 binding to cdc18 or cdc22 promoters in different genetic backgrounds that keep MBF-dependent transcription constitutively repressed (Δrep2) or induced (Δyox1 or Δnrm1). As shown in , binding of Snf22 to either cdc18 or cdc22 promoters absolutely paralleled the transcription of both genes, pointing that Snf22 binding could be a consequence of the level of transcription and not being a regulator of MBF-dependent transcription.
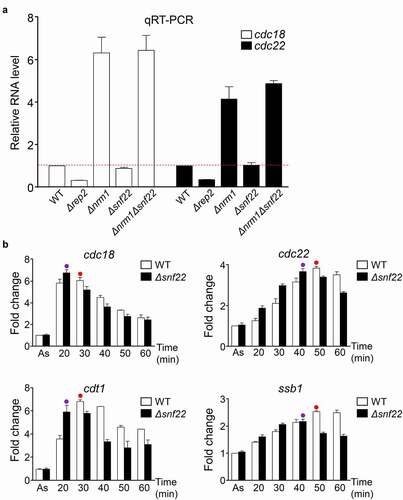
To determine if there was any effect of Snf22 (and SWI/SNF) on the regulation of MBF-dependent transcription, we measured the expression of cdc18 and cdc22 in different genetic backgrounds and in synchronized cultures (). First, we prepared RNA from asynchronous cultures of a wild-type strain and from Δrep2, Δnrm1, Δsnf22, and Δnrm1Δsnf22 strains. As can be observed in , deletion of snf22 does not affect the expression of cdc18 or cdc22 in either a wild-type background or in a Δnrm1 background. To fully discard any role of Snf22 in the regulation of MBF-dependent transcription, we synchronized cultures of wild-type cells or Δsnf22 using the temperature-sensitive cdc25-22 strain. This strain allows the synchronization of the cultures at the G2/M transition by inactivating the phosphatase Cdc25 at restrictive temperature. Upon shifting back the temperature of the culture to the permissive 25°C, cells are forced to progress synchronously into mitosis, G1 phase, S phase, and the next G2. As shown in , the deletion of snf22 has no major effect on the transcription of the four MBF-regulated genes that we analyzed (cdc18, cdc22, cdt1, and ssb1). The only noticeable change that we observed was a minor but reproducible advancement (10 min) at which the different RNAs reached its maximum in the Δsnf22 cells.
Figure 3. SWI/SNF mutant strains do not have impaired MBF-dependent transcription. (a) Expression of the MBF-dependent genes cdc18 and cdc22 was analyzed by RT-qPCR in the indicated strains, before (-) and after (+) treatment with HU for 3 h. tfb2 was used as a control for normalization. Each column represents the mean value and SEM, calculated from three biological replicates. (b) Fission yeast cdc25-22 cells or cdc25-22 Δsnf22 were synchronized at the G2/M transition. After release, samples were collected every 10 min, processed for RNA isolation, and analyzed by RT-qPCR for the expression of the genes indicated on top. Dots indicate the timepoint with the maximum value (in red, WT; in purple, Δsnf22). Error bars (SEM) were calculated from biological triplicates
RSC complex is recruited at MBF-regulated genes and is required to fully activate their transcription
Since we have shown that Δrsc1 cells were sensitive to HU (, we decided to determine if cells lacking other components of the RSC complex were also sensitive to HU. As shown in , Δrsc4 cells were as sensitive to HU as Δrsc1 cells. Snf21 is the catalytic subunit of the RSC complex. Contrary to what happens with the SWI/SNF complex whose catalytic subunit, Snf22, is not essential in fission yeast, Δsnf21 cells are inviable. However, it is described a point mutation in Snf21 (snf21-36) that renders a temperature-sensitive strain [Citation27]. This strain can grow at the permissive temperature (25°C), but upon shifting the culture to non-permissive temperature (37°C) Snf21 is inactivated and cells stop growing ()). Interestingly, snf21-36 cells are very sensitive to HU even at the permissive temperature ()). To further investigate the role of the RSC complex in the regulation of MBF-dependent transcription, we analyzed the binding of Snf21 to cdc18 and cdc22 by ChIP. We were able to detect binding of Snf21 not only to the promoter of both genes, but also to the ORFs and to the 3' regions (, and this binding was noticeably increased upon treatment with HU. It is worth noting an important difference with the binding of Snf22 to the MBF-regulated genes: while Snf21 bound equally well along each gene, Snf22 binding was restricted to the promoter region of cdc18 and cdc22.
Figure 4. RSC complex mutants are sensitive to HU. (a) Fivefold serial dilutions of the strains indicated on the left were spotted onto rich media or in media with 4 mM HU, 6 mM HU, 8 mM HU, or 10 mM HU. Plates were incubated at 30°C for 2–4 d. (b) Fivefold serial dilutions of the strains indicated on the left were spotted onto rich media or in media with 4 mM HU or 6 mM HU. Plates were incubated at 25°C or 37°C, as indicated in the top
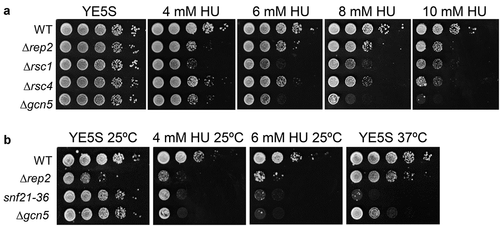
Figure 5. Snf21 binds to MBF-regulated genes. (a) Cells expressing Snf21-Myc under the control of its own promoter were treated or not with HU for 3 h. ChIP experiments were performed using primers covering promoter (prom), coding (ORF), and termination (term) sequences of the cdc18 and cdc22 genes (as indicated). Error bars (SEM) were calculated from biological triplicates. Significant differences between treated and untreated samples were determined by the Student´s t-test (*P < 0.05; **P < 0.01). (b) Wild type (WT) or Δgcn5 cells expressing Snf21-Myc were treated or not with HU for 3 h. ChIP experiments were performed using primers covering the promoter region of the cdc18 and cdc22 genes or the SPNCRNA.650 gene as a control. Error bars (SEM) were calculated from biological triplicates. Significant differences between treated and untreated samples were determined by the Student´s t-test (*P < 0.05). (c) Wild type (WT), Δrep2, Δyox1, or Δnrm1 cells expressing Snf21-Myc were treated or not with HU for 3 h, as indicated. Error bars (SEM) were calculated from biological triplicates. Significant differences between treated and untreated samples were determined by the Student´s t-test (*P < 0.05)
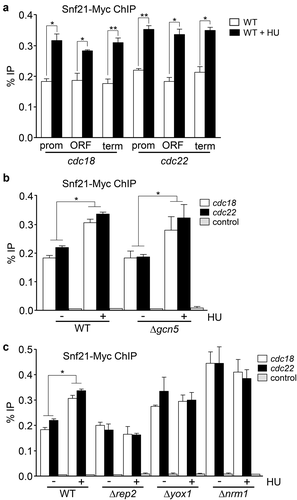
Snf21 also has a bromo-domain at the carboxy-terminal region, which is supposed to recognize histones with acetylated lysines. Thus, we tested if the binding of Snf21 was affected in a Δgcn5 strain. As shown in , binding of Snf21 to the promoter of cdc18 or cdc22 was unaltered in a Δgcn5 cells. In fact, Snf21 was also recruited when Δgcn5 cells were treated with HU, similarly to what happens to the wild-type cells. On the other hand, recruitment of Snf21 after treatment with HU was not increased in Δrep2 cells and was constitutively increased in Δyox1 or Δnrm1 cells (, which is reminiscent of our previous observation regarding the recruitment of Snf22 to the same promoters (.
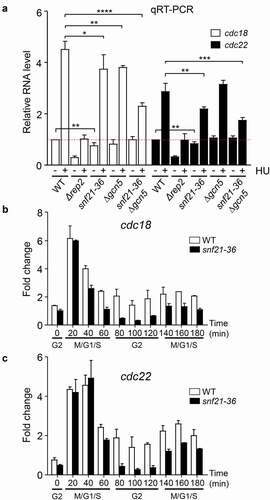
To determine if recruitment of Snf21 could affect the transcription of MBF-dependent genes, we decided to analyze the expression of cdc18 and cdc22 in different genetic backgrounds bearing mutant alleles of the RSC complex. First, we analyzed the effect on transcription in the strain bearing the temperature-sensitive allele snf21-36 (. Although the experiments were done at the permissive temperature (25°C), the levels of cdc18 and cdc22 mRNA were reduced, when compared with the wild-type strain, both in the presence or absence of HU. Interestingly, this effect was even more noticeable in the snf21-36 Δgcn5 strain when treated with HU (and not in untreated cells), pointing to an additive effect between the RSC complex and the SAGA complex in the regulation of the MBF-dependent genes. However, the mutation of other subunits (like Rsc1) had only a minor effect and not on all MBF-regulated genes. To confirm the effect of the RSC complex on MBF-dependent transcription, we synchronized cultures of wild-type cells or snf21-36 using the cdc2-asM17 allele, which in the presence of 1 µM 1-NM-PP1 arrests cells in metaphase. Upon release (after washing out the ATP analog), cells progress synchronously into the cell cycle. As shown in , while the induction of expression of cdc18 and cdc22 was similar in both strains, the repression during the next G2 was more profound in the snf21-36 strain, indicating that Snf21 had a role in maintaining a minimal expression of the MBF-regulated genes during the G2 phase of the cell cycle.
Figure 6. RSC mutant strains have impaired MBF-dependent transcription. (a) Expression of the MBF-dependent genes cdc18 and cdc22 was analyzed by RT-qPCR in the indicated strains, before (-) and after (+) treatment with HU for 3 h. tfb2 was used as a control for normalization. Each column represents the mean value and SD, calculated from three biological replicates. Significant differences between samples were determined by the Student´s t-test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). (b) Fission yeast cdc2-asM17 cells (WT) or cdc2-asM17 snf21-36 (snf21-36) were synchronized at metaphase. After release, samples were collected every 20 min, processed for RNA isolation, and analyzed by RT-qPCR for the expression of cdc18. Error bars (SD) were calculated from biological triplicates. The different phases of the cell cycle are indicated at the bottom. (c) Same samples from (b) were analyzed for the expression of cdc22.
In summary, we believe that SWI/SNF and RSC chromatin remodeler complexes are recruited to MBF-regulated genes in a Gcn5-independent manner. However, while SWI/SNF is recruited to the promoter region of these genes, the RSC complex is recruited along the whole gene. This difference might have an important impact in the outcome of this interaction since while we did not detect any effect on transcription that depended on the SWI/SNF complex, the recruitment of the RSC complex has clearly impacts, increasing MBF-dependent transcription: in the absence of RSC complex, MBF-dependent transcription is diminished when compared to a wild-type strain. Further work will be necessary to determine if a similar mechanism also applies in higher eukaryotes.
Acknowledgments
We thank Kunihiro Ohta for the snf21-36 strain (KYP176). We thank members of the Oxidative Stress and Cell Cycle Group for suggestions and comments and Mercè Carmona for her technical support.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Nurse P. Mutants of the fission yeast Schizosaccharomyces pombe which alter the shift between cell proliferation and sporulation. Mol Gen Genet. 1985;198(3):497–502.
- Nurse P, Bissett Y. Gene required in G1 for commitment to cell cycle and in G2 for control of mitosis in fission yeast. Nature. 1981;292(5823):558–560.
- Aves SJ, Durkacz BW, Carr A, et al. Cloning, sequencing and transcriptional control of the Schizosaccharomyces pombe cdc10 ‘start’ gene. EMBO J. 1985;4(2):457–463.
- Ayte J, Leis JF, DeCaprio JA. The fission yeast protein p73res2 is an essential component of the mitotic MBF complex and a master regulator of meiosis. Mol Cell Biol. 1997;17(11):6246–6254.
- Miyamoto M, Tanaka K, Okayama H. res2+, a new member of the cdc10+/SWI4 family, controls the ‘start’ of mitotic and meiotic cycles in fission yeast. EMBO J. 1994;13(8):1873–1880.
- Zhu Y, Takeda T, Nasmyth K, et al. pct1+, which encodes a new DNA-binding partner of p85cdc10, is required for meiosis in the fission yeast Schizosaccharomyces pombe. Genes Dev. 1994;8(8):885–898.
- Tanaka K, Okazaki K, Okazaki N, et al. A new cdc gene required for S phase entry of Schizosaccharomyces pombe encodes a protein similar to the cdc 10+ and SWI4 gene products. EMBO J. 1992;11(13):4923–4932.
- Ayte J, Leis JF, Herrera A, et al. The Schizosaccharomyces pombe MBF complex requires heterodimerization for entry into S phase. Mol Cell Biol. 1995;15(5):2589–2599.
- Nakashima N, Tanaka K, Sturm S, et al. Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle ‘start’ function of Res2-Cdc10. EMBO J. 1995;14(19):4794–4802.
- Gonzalez-Medina A, Hidalgo E, Ayte J. Gcn5-mediated acetylation at MBF-regulated promoters induces the G1/S transcriptional wave. Nucleic Acids Res. 2019;47(16):8439–8451.
- Caetano C, Klier S, de Bruin RA. Phosphorylation of the MBF repressor Yox1p by the DNA replication checkpoint keeps the G1/S cell-cycle transcriptional program active. PLoS One. 2011;6(2):e17211.
- Gómez-Escoda B, Ivanova T, Calvo IA, et al. Yox1 links MBF-dependent transcription to completion of DNA synthesis. EMBO Rep. 2011;12(1):84–89.
- Purtill FS, Whitehall SK, Williams ES, et al. A homeodomain transcription factor regulates the DNA replication checkpoint in yeast. Cell Cycle. 2011;10(4):664–670.
- de Bruin RA, Kalashnikova TI, Aslanian A, et al. DNA replication checkpoint promotes G1-S transcription by inactivating the MBF repressor Nrm1. Proc Natl Acad Sci U S A. 2008;105(32):11230–11235.
- Lowndes NF, McInerny CJ, Johnson AL, et al. Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10+. Nature. 1992;355(6359):449–453.
- Ayté J, Schweitzer C, Zarzov P, et al. Feedback regulation of the MBF transcription factor by cyclin Cig2. Nat Cell Biol. 2001;3(12):1043–1050.
- Kelly TJ, Martin GS, Forsburg SL, et al. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74(2):371–382.
- Caetano C, Limbo O, Farmer S, et al. Tolerance of deregulated G1/S transcription depends on critical G1/S regulon genes to prevent catastrophic genome instability. Cell Rep. 2014;9(6):2279–2289.
- Ivanova T, Alves-Rodrigues I, Gomez-Escoda B, et al. DNA damage and the DNA replication checkpoints converge at the MBF transcription factor. Mol Biol Cell. 2013;24(21):3350–3357.
- Ivanova T, Gomez-Escoda B, Hidalgo E, et al. G1/S transcription and the DNA synthesis checkpoint: common regulatory mechanisms. Cell Cycle. 2011;10(6):912–915.
- Wuarin J, Buck V, Nurse P, et al. Stable association of mitotic cyclin B/Cdc2 to replication origins prevents endoreduplication. Cell. 2002;111(3):419–431.
- Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim Biophys Acta. 2005;1681(2–3):59–73.
- Knezevic I, Gonzalez-Medina A, Gaspa L, et al. INO80 complex activates the transcription of S-phase genes in a cell cycle-regulated manner. FEBS J. 2018;285(20):3870–3881.
- Monahan BJ, Villen J, Marguerat S, et al. Fission yeast SWI/SNF and RSC complexes show compositional and functional differences from budding yeast. Nat Struct Mol Biol. 2008;15(8):873–880.
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823.
- Moldon A, Malapeira J, Gabrielli N, et al. Promoter-driven splicing regulation in fission yeast. Nature. 2008;455(7215):997–1000.
- Yamada K, Hirota K, Mizuno K, et al. Essential roles of Snf21, a Swi2/Snf2 family chromatin remodeler, in fission yeast mitosis. Genes Genet Syst. 2008;83(5):361–372.
