ABSTRACT
Hsa_circ_0001756 was reported to be upregulated in serum samples of ovarian cancer (OC) patients and may serve as a potential OC biomarker. This study aimed to investigate the role and molecular mechanisms of hsa_circ_0001756 in OC procession. Herein, we detected the expression of hsa_circ_0001756 in OC tissues and cell lines with RT-qPCR assay, which showed that hsa_circ_0001756 was upregulated in OC tissues and cell lines. Then small interfering RNA targeting hsa_circ_0001756 (si-hsa_circ_0001756) was transfected into SKOV3 and A2780 cells, and the proliferation, invasion, and expression of epithelial-mesenchymal transition (EMT) marker proteins were determined with CCK-8, Transwell and Western blotting assays, respectively. We found that hsa_circ_0001756 knockdown inhibited OC cell proliferation, invasion and EMT. Moreover, RNA pull-down assay verified the binding between hsa_circ_0001756 and IGF2 mRNA binding protein 2 (IGF2BP2), and rescue experiments indicated that IGF2BP2 overexpression reversed the effects of has_circ_0001756 knockdown on OC cell functions. Co-IP assay verified IGF2BP2 could interact with RAB GTPase 5A (RAB5A) protein. Then SKOV3 cells were transfected with si-IGF2BP2 alone or together with pcDNA-RAB5A, followed by the detection of SKOV3 cell functions. We found that IGF2BP2 knockdown inhibited OC cell proliferation, invasion, and EMT, while RAB5A overexpression reversed these effects. Finally, SKOV3 cells transfected with si-hsa_circ_0001756 were injected into nude mice through tail vein. Hsa_circ_0001756 knockdown significantly inhibited the xenograft tumor growth of OC in vivo. In conclusion, hsa_circ_0001756 knockdown inhibits OC cell proliferation, invasion, and EMT, and reduces xenograft tumor growth by suppressing IGF2BP2-mediated RAB5A expression and blocking the EGFR/MAPK signaling pathway.
1. Introduction
Ovarian cancer (OC) is one of the three major malignant tumors of the female genitalia. Due to its high malignancy and poor prognosis, the mortality rate of OC is higher than that of the sum of cervical and endometrial cancers [Citation1]. OC includes epithelial ovarian cancer (EOC) and non-epithelial ovarian cancer (NEOC). The five-year survival rate of EOC patients is poor because most of them are diagnosed with an advanced age and even accompanied with metastasis [Citation2]. NEOC is a heterogenous group of rare tumors with an incidence of approximately 10% of all ovarian cancers, affecting mainly young patients. The two most frequently diagnosed NEOCs are germ cell tumors (GCTs) and sex cord-stromal cell tumors (SCSTs). GCTs have an excellent prognosis, with cure rates approaching 100% for those with early-stage disease, whereas SCSTs are generally more indolent with an excellent short-term prognosis, but carry a significant risk of late relapse [Citation3]. Significant advances have been rapidly made on the OC treatment in the past decade. It was identified that defective DNA damage response is a defining hallmark of high-grade EOC, and poly (ADP-ribose) polymerase (PARP) inhibitors exploit this deficiency through synthetic lethality and have emerged as promising anticancer therapies, especially in breast cancer gene (BRCA1 or BRCA2) mutation carriers [Citation4]. However, the molecular mechanisms of OC progression have not been fully explored and the outcomes of OC patients remain seriously unsatisfactory. Thus, more efforts are urgently required to clarify the molecular mechanisms of OC progression and develop effective therapeutic targets.
The identification and study of cancer biomarkers is an ever-expanding field with promising recent findings. Accumulated studies have identified a large number of biomarkers including long coding RNAs (lncRNAs), microRNAS (miRNAs), circular RNAs (circRNAs) and proteins, which provide an effective insight into tumor genetics and help with understanding of the pathophysiology of cancers [Citation5,Citation6]. CircRNAs are a special type of endogenous non-coding RNA with closed loop without 5ʹ cap and 3ʹ poly A tail structure, which is different from traditional linear RNA [Citation7]. CircRNAs are not affected by RNA exonuclease and their expression is more stable, which have been proved to be widely expressed in a variety of eukaryotic organisms [Citation8]. Increasing evidence has revealed circRNAs are involved in a series of pathological processes, including human cancers [Citation9,Citation10]. Recently, a large number of circRNAs have been reported to serve as novel promising biomarkers for diagnosis and therapeutic targets for OC treatment, such as hsa_circ_0051240 [Citation11] and circPUM1 [Citation12]. More notably, a recent bioinformatics analysis revealed that hsa_circ_0001756 was upregulated in serum samples of OC patients, indicating that hsa_circ_0001756 may be involved in OC development [Citation13]. However, the underlying role of hsa_circ_0001756 in OC progression has rarely been explored to date.
Emerging evidence has suggested that circRNAs play important roles in human diseases by acting as miRNA sponges or interacting with RNA-binding protein (RBP) [Citation14]. IGF2 mRNA binding protein 2 (IGF2BP2) is an RBP, which have been reported to play oncogenic roles in various cancers. For example, the upregulated IGF2BP2 protein level in pancreatic cancer is correlated with poor survival of pancreatic cancer patients, which may be a prognostic marker and therapeutic target for the treatment of pancreatic cancer [Citation15]. Moreover, the mRNA expression of IGF2 mRNA binding proteins (IGF2BP1, IGF2BP2, and IGF2BP3) was significantly higher in epithelial ovarian tumors compared to normal ovarian samples [Citation16]. Therefore, IGF2BP2 may be involved in OC progression. RAB GTPase 5A (RAB5A) has been identified to be upregulated and serve as an oncogene in several human cancers, which is associated with cancer cell proliferation and metastasis [Citation17,Citation18]. It was previously reported that RAB5A was upregulated in OC samples, and RAB5A overexpression promoted OC cell proliferation [Citation19]. However, the mechanism related to RAB5A in OC progression has been little explored.
In this study, we found that hsa_circ_0001756 was upregulated in OC tissues and OC cell lines. Then we further explored the role of hsa_circ_0001756 in OC progression in vivo and in vitro. In addition, the regulatory mechanisms of hsa_circ_0001756 in OC progression were further explored.
2 Materials and methods
2.1 Tissue samples
A total of 35 OC tissues and matched adjacent tissues were collected from OC patients at the First Affiliated Hospital of Xi’an Jiaotong University (Xi’an, China) from 2018 April to 2019 October. The inclusion criteria were as follows: (i) diagnosis as OC confirmed by pathological and clinical examinations; (ii) age >18 years; (iii) completed clinical and pathological information, and clinical tissue samples are accessible for experimental use. (iv) patients who received treatment for the first time, without any medications such as hormones within the past 6 months. Exclusion criteria: patients with other ovarian diseases, other malignant tumors, severe complicated diseases of heart, lung, kidney, and other organs or severe infectious diseases. No patients obtained chemotherapy or radiotherapy before surgery. Samples were frozen in liquid nitrogen and stored at −80°C. This study was approved by the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University (XJTU-2018-023) and each patient involved provided written informed consent.
2.2 Cell lines and culture
The human ovarian cancer cell lines (A2780, SKOV3, CAOV3, and OVCAR3) and normal human ovarian epithelial cell line (HOSEpiC) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA), 100 U/mL penicillin, and 100 μg /mL streptomycin (Sigma, St. Louis, MO, USA), and maintained with 5% CO2 at 37°C.
2.3 Cell transfection
Overexpression plasmids of circ_0001756 (pLO-circ_0001756), IGF2BP2 (pcDNA-IGF2BP2) and RAB5A (pcDNA-RAB5A), small interfering RNAs targeting circ_0001756 (si-hsa_circ_0001756) and IGF2BP2 (si-IGF2BP2), and their negative controls (vector and si-NC) were obtained from RiboBio (Guangzhou, China). Cell transfection was performed by using the Lipofectamine 3000 Transfection Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. After transfection for 48 h, cells were collected for further experiments.
2.4 RNA extraction and RT-qPCR
Total RNA was extracted from tissues or cell lines by using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocols. Reverse transcription was carried out by using a Prime Script RT reagent Kit (Takara, Dalian, China) to synthesize cDNA. Before RT-qPCR, we confirmed that the high-quality cDNA was obtained by agarose gel electrophoresis. Real-time PCR analyses were conducted with the SYBR Premix Ex Taq II (Takara, Dalian, China) on an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) under the following conditions: 95°C for 1 min, and then 95°C for 20 s, 56°C for 10 s and 72°C for 15 s for 35 cycles. PCR reaction system contained 12.5 μL of SYBR Premix Ex Taq II, 1.0 μL of RT primer, 1 μL of cDNA sample, and 10.5 μL of double distilled H2O. The relative expression of circ_0001756 was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and calculated by 2−ΔΔCT method. The primers are as follows: has_circ_0001756 (forward: 5ʹ-GTG CTG GCT GAG ACC CTA AC-3ʹ, reverse: 5ʹ-AGC AGC ATC TGG AAC AAG GT-3ʹ). GAPDH (forward: 5ʹ-CTG GGC TAC ACT GAG CAC C-3ʹ, reverse: 5ʹ-AAG TGG TCG TTG AGG GCA ATG-3ʹ).
2.5 Cell proliferation assay
Cell proliferation was determined by using Cell Counting Kit-8 (CCK-8, Dojindo, Japan) assay. Briefly, SKOV3/A2780 cells (1 × 104 cells/well) were seeded into 96-well plates. After incubation for 0, 24, 48 and 72 h, 10 µL of CCK-8 solution was added and incubated for 2 h at 37°C. The absorbance at 450 nm of each well was measured by using a micro-plate reader (Molecular Devices, Shanghai, China).
2.6 Transwell invasion assay
Transwell assay was performed to detect cell invasion. Transwell chamber (8 µm pore size; Millipore Corporation, USA) was coated with Matrigel (BD Biosciences, USA). And a total of 1 × 105 SKOV3/A2780 cells in culture medium were seeded into the upper chamber, while DMEM medium containing 10% FBS was added to the lower chamber. After culturing for 24 h at 37°C, cells in the upper chamber were removed by using a cotton swab, and the cells in the bottom chamber were fixed with 70% ethanol for 10 min and stained with 0.1% crystal violet for 15 min. The number of invasive cells was counted with a light microscope (Olympus, Tokyo, Japan).
2.7 RNA pull-down assay
RNA pull-down assay was performed to confirm the binding relationship between circ_0001756 and IGF2BP2. In detail, the biotinylated wild-type has_circ_0001756 (Bio-has_circ_0001756-WT), mutant-type has_circ_0001756 (Bio-has_circ_0001756-MUT), and negative control (Bio-NC) were transfected into OC cells. After 48 h of transfection, the cells were collected and lysed in lysis buffer, and then the supernatant was collected and incubated with streptavidin magnetic beads (Invitrogen, Carlsbad, CA, USA) at 4°C for 3 h. The purified RNA-protein complex was then analyzed by Western blot assay.
2.8 Western blot analysis
Proteins were extracted from tissues or cell lines by using RIPA lysis buffer (Beyotime, Shanghai, China). The protein concentration was measured by BCA analysis (Millipore, Billerica, MA, USA), and then proteins were separated on SDS-PAGE under the following conditions: 70 V for 30 min, followed by 120 V for 90 min. And then the protein bands were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA, USA) at 300 mA for 2 h. The membranes were then blocked with 5% nonfat milk for 1 h at room temperature and incubated overnight at 4°C with the following primary antibodies obtained from Abcam: rabbit monoclonal anti-IGF2BP2 antibody (1:2000, ab124930), rabbit polyclonal anti-RAB5A antibody (1:1000, ab18211), rabbit monoclonal anti-EGFR antibody (1:1000, ab52894), rabbit monoclonal anti-p-EGFR antibody (1:1000, ab40815), anti-p38 antibody (1:1000, ab170099), rabbit monoclonal anti-p-p38 antibody (1:1000, ab178867) and rabbit polyclonal anti-GAPDH antibody (1:2500, ab9485). The membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:2000, Abcam, ab6721) for 1 h. Subsequently, the protein bands were visualized with ECL detection reagents and analyzed with ImageJ software (National Institutes of Health, Bethesda, MD, USA).
2.9 Co-immunoprecipitation (Co-IP) assay
SKOV3 cells were lyzed with lysis buffer, and the supernatant was collected and incubated with IGF2BP2 antibody at 4°C overnight. Then the mixture was captured by protein A/G agarose beads (Takara Biotechnology, Dalian, China). Subsequently, the beads were washed and eluted. The supernatant was collected by centrifugation and analyzed by using Western blot assay to detect the expression of binding proteins.
2.10 In vivo xenograft assay
Female BALB/c nude mice (aged 4 weeks) weighing 20 ± 2 g were provided by the animal experimental center of the First Affiliated Hospital of Xi’an Jiaotong University, which were randomly divided into two groups including si-NC group and si-hsa_circ_0001756 group (n = 8 per group). All mice were maintained in sterile cages under 22–25°C temperature, 55–60% humidity and 12 h light/dark cycle conditions with free access to food and water. SKOV3 cells (2 × 106) transfected with si-NC or si-hsa_circ_0001756 were injected into nude mice through tail vein (n = 8 per group). Then si-NC or si-hsa_circ_0001756 (50 μg/kg) was injected into nude mice through tail vein every 2 days. Tumor volumes were measured every 7 days for 28 consecutive days. The nude mice were euthanized on day 28, and tumor tissues were collected and tumor weights were determined. Then the expression of circ_0001756, IGF2BP2, RAB5A and EMT-associated proteins in tumor tissues was detected. Animal experiments were approved by the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University (XJTU-2018-023).
2.11 Statistical analysis
Data analysis was performed by SPSS version 22.0 software. Experimental results from triplicate independent experiments were presented as mean ± standard deviation (SD). The normality distribution of data was checked by the Shapiro–Wilk test. Comparisons between two groups or multiple groups were performed by using Student’s t-test or one-way ANOVA, respectively. P < 0.05 was considered to be statistically significant.
3. Results
3.1 Hsa_circ_0001756 is upregulated in OC tissues and cell lines
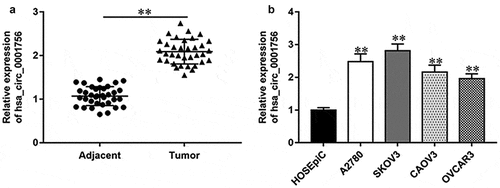
To investigate whether hsa_circ_0001756 is involved in OC progression, we firstly detected the expression of hsa_circ_0001756 in OC tissues and matched adjacent tissues by using RT-qPCR. As shown in , hsa_circ_0001756 was upregulated in OC tissues compared with matched adjacent tissues (P < 0.0001). Meanwhile, we also found that hsa_circ_0001756 was upregulated in OC cell lines (A2780, SKOV3, CAOV3 and OVCAR3) compared with the human ovarian epithelial cell line HOSEpiC (P < 0.0001; P < 0.0001; P = 0.0001; P = 0.0003) ().
Figure 1. Hsa_circ_0001756 is upregulated in OC tissues and cell lines. (a) Relative expression of hsa_circ_0001756 in OC tissues and matched adjacent tissues was measured by using RT-qPCR (n = 35 per group). (b) Relative expression of hsa_circ_0001756 in OC cell lines (A2780, SKOV3, CAOV3 and OVCAR3) and human ovarian epithelial cell line HOSEpiC was measured by using RT-qPCR. Data were presented as mean ± SD. **P < 0.01.
3.2 Has_circ_0001756 knockdown inhibits OC cell proliferation, invasion and EMT
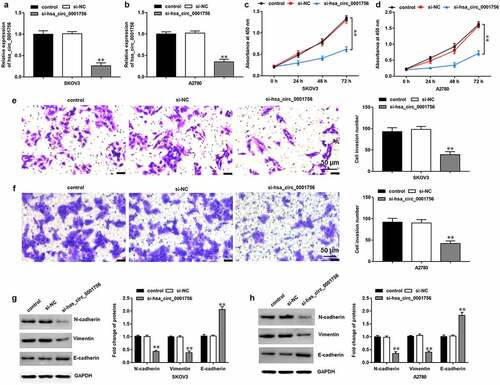
To further explore the effects of hsa_circ_0001756 on OC progression, SKOV3 and A2780 cells were transfected with si-hsa_circ_0001756, and RT-qPCR assay showed that hsa_circ_0001756 expression was downregulated by transfection of si-hsa_circ_0001756 (P < 0.0001; P < 0.0001) (). Then we found that hsa_circ_0001756 knockdown significantly inhibited proliferation (P < 0.0001; P < 0.0001) () and invasion (P < 0.0001; P < 0.0001) () in SKOV3 and A2780 cells. Moreover, Western blotting was performed to detect the expression of epithelial-mesenchymal transition (EMT) marker proteins, which showed that hsa_circ_0001756 knockdown increased E-cadherin expression (P < 0.0001; P = 0.0007) and decreased the expression of N-cadherin (P = 0.0004; P = 0.0005) and Vimentin (P < 0.0001; P = 0.0002) ().
Figure 2. Hsa_circ_0001756 knockdown inhibits SKOV3 cell proliferation, invasion and EMT. si-hsa_circ_0001756 (50 nM) and its negative control (si-NC) (50 nM) were transfected into SKOV3 and A2780 cells, respectively. (a, b) The expression of hsa_circ_0001756 was measured by RT-qPCR after transfection for 48 h in SKOV3 and A2780 cells. (c, d) CCK-8 assay was used to detect proliferation in SKOV3 and A2780 cells. (e, f) Transwell assay was performed to detect invasion in SKOV3 and A2780 cells. (g, h) Western blotting was conducted to detect the expression of EMT-related proteins (E-cadherin, N-cadherin and Vimentin). Data were presented as mean ± SD. **P < 0.01.
3.3 IGF2BP2 is an RNA binding protein of has_circ_0001756
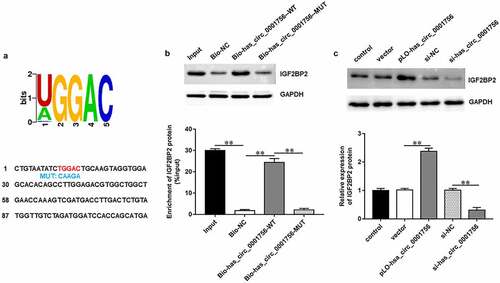
We obtained IGF2BP2 binding motifs from the online bioinformatics tool StarBase 2.0 (http://starbase.sysu.edu.cn/). As we expected, the motif was aligned with the hsa_circ_0001756 sequence (). RNA pull-down assay further confirmed that IGF2BP2 protein could be pulled down by biotinylated has_circ_0001756-WT (P < 0.0001) but not biotinylated has_circ_0001756-MUT (P = 0.91) or biotinylated NC (). Furthermore, transfection of pLO-circ_0001756 significantly increased IGF2BP2 expression (P < 0.0001), while transfection of si-hsa_circ_0001756 decreased IGF2BP2 expression in SKOV3 cells (P < 0.0001) (). All results indicated that hsa_circ_0001756 could bind with IGF2BP2 and positively regulated IGF2BP2 expression in OC cells.
Figure 3. Hsa_circ_0001756 binds with IGF2BP2 in OC cells. (a) The potential IGF2BP2 target motif in hsa_circ_0001756 sequence was predicted by starBase. Red fonts represent the wild-type (WT) binding motifs; blue fonts represent the mutant-type (MUT) binding motifs. (b) RNA pull-down assay was used to verify the binding of hsa_circ_0001756 and IGF2BP2. (c) pLO-circ_0001756 (30 nM), si-hsa_circ_0001756 (50 nM) and their negative controls were transfected into SKOV3 cells, respectively. The expression of IGF2BP2 protein in SKOV3 cells was measured by using Western blotting after transfection for 48 h. Data were presented as mean ± SD. **P < 0.01.
3.4 Has_circ_0001756 regulates OC cell proliferation and invasion and EMT by promoting IGF2BP2 expression
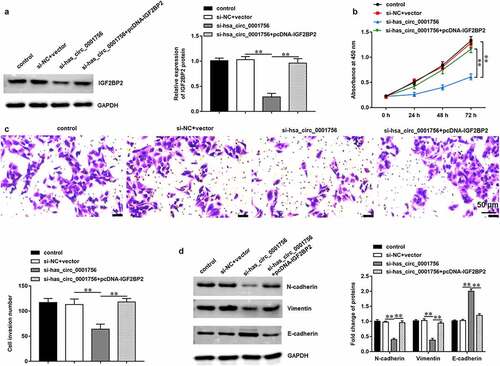
To explore whether hsa_circ_0001756 exerted its functions in OC progression by regulating IGF2BP2, SKOV3 cells were transfected with si-hsa_circ_0001756 alone or together with pcDNA-IGF2BP2. We found that transfection of si-hsa_circ_0001756 decreased IGF2BP2 expression (P < 0.0001), while transfection of pcDNA-IGF2BP2 increased IGF2BP2 expression (P < 0.0001) (). Moreover, hsa_circ_0001756 knockdown inhibited proliferation (P < 0.0001) () and invasion (P = 0.0006) () of SKOV3 cells, while IGF2BP2 overexpression reversed these effects (P < 0.0001; P = 0.0003). Besides, has_circ_0001756 knockdown increased E-cadherin expression (P = 0.0004) and decreased the expression of N-cadherin (P < 0.0001) and Vimentin (P < 0.0001) in SKOV3 cells, while IGF2BP2 overexpression reversed this expression pattern (P = 0.0005; P = 0.0002; P = 0.0002) ().
Figure 4. IGF2BP2 overexpression reversed the effects of hsa_circ_0001756 knockdown on OC cell proliferation, invasion and EMT. SKOV3 cells were transfected with si-hsa_circ_0001756 (50 nM) alone or together with pcDNA-IGF2BP2 (30 nM). (a) The expression of IGF2BP2 protein in SKOV3 cells was measured by using Western blotting assay after transfection for 48 h. (b) SKOV3 cell proliferation was detected by using CCK-8 assay. (c) SKOV3 cell invasion was measured by using Transwell assay. (d) The expression of EMT marker proteins (E-cadherin, N-cadherin and Vimentin) was detected by using Western blotting assay. Data were presented as mean ± SD. **P < 0.01.
3.5 IGF2BP2 interacts with RAB5A protein
The potential interacting proteins of IGF2BP2 were predicted with the Genemania tool (http://genemania.org/), which showed that there was a potential interaction between IGF2BP2 protein and RAB5A protein (). Subsequently, Co-IP assay verified that both IGF2BP2 and RAB5A proteins could be detected by immunoprecipitation with IGF2BP2 antibody but not with IgG in SKOV3 and A2780 cells (). Moreover, we found that IGF2BP2 overexpression significantly increased RAB5A expression (P < 0.0001; P = 0.0007), while IGF2BP2 knockdown decreased RAB5A expression in SKOV3 (P < 0.0001) () and A2780 cells (P = 0.0003) (). In addition, RNA pull-down assay further confirmed that both IGF2BP2 and RAB5A proteins could be pulled down by biotinylated has_circ_0001756-WT but not biotinylated NC in SKOV3 (P < 0.0001) () and A2780 cells (P < 0.0001) ().
Figure 5. IGF2BP2 interacts with RAB5A and promoting IGF2BP2 expression in SKOV3 cells. (a) The online Genemania tool (http://genemania.org/) was used to predict the potential interacting proteins of IGF2BP2. (b, c) The interaction between IGF2BP2 protein and RAB5A protein in SKOV3 and A2780 cells was verified by using Co-IP assay. (d, e) pcDNA-IGF2BP2 (30 nM), si-IGF2BP2 (50 nM) and their negative controls were transfected into SKOV3 cells, respectively. Western blotting was used to measure RAB5A protein expression in SKOV3 and A2780 cells. (f, g) RNA pull-down assay was performed to verify the interactions among hsa_circ_0001756, IGF2BP2 and RAB5A in SKOV3 and A2780 cells. Data were presented as mean ± SD. **P < 0.01.
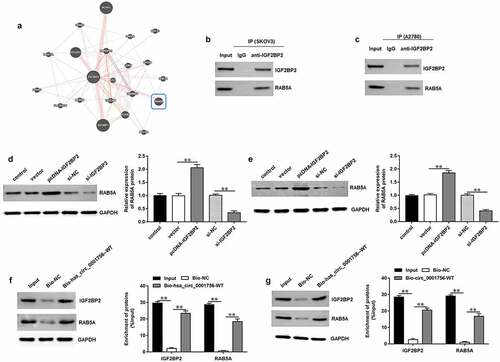
3.6 IGF2BP2 regulates OC cell proliferation, invasion, and EMT by promoting RAB5A expression and activating EGFR/MAPK signaling pathway
To further investigate the effects of IGF2BP2 and RAB5A on OC progression, SKOV3 cells were transfected with si-IGF2BP2 alone or together with pcDNA-RAB5A. Western blotting results showed that IGF2BP2 knockdown inhibited RAB5A expression (P < 0.0001), while transfection of pcDNA-RAB5A increased RAB5A expression (P = 0.0001) (). Furthermore, IGF2BP2 knockdown inhibited proliferation (P < 0.0001) () and invasion (P < 0.0001) () of SKOV3 cells, while RAB5A overexpression reversed these effects (P < 0.0001; P < 0.0001). In addition, IGF2BP2 knockdown decreased the expression of N-cadherin and Vimentin (P < 0.0001; P = 0.0002) and increased E-cadherin expression (P < 0.0001) in SKOV3 cells, while RAB5A overexpression reversed this expression pattern (P = 0.0002; P = 0.0003; P < 0.0001) (). Besides, we further found that IGF2BP2 knockdown inhibited p-EGFR and p-38 expression (P < 0.0001; P < 0.0001), and RAB5A overexpression increased p-EGFR and p-p38 expression in SKOV3 cells (P = 0.0002; P = 0.0007) (), indicating that the IGF2BP2/RAB5A axis may modulate OC progression by regulating EGFR/MAPK signaling pathway.
Figure 6. RAB5A overexpression reversed the effects of IGF2BP2 knockdown on OC cell proliferation, invasion and EMT. SKOV3 cells were transfected with si-IGF2BP2 (50 nM) alone or together with pcDNA-RAB5A (30 nM). (a) The expression of RAB5A protein in SKOV3 cells was detected by using Western blotting after transfection for 48 h. (b) CCK-8 assay was used to detect SKOV3 cell proliferation. (c) Transwell assay was conducted to evaluate SKOV3 cell invasion. (d) Western blotting assay was performed to detect the expression of EMT marker proteins (E-cadherin, N-cadherin and Vimentin). (e) The expression of p-EGFR/EGFR and p-p38/p38 proteins in SKOV3 cells was detected by Western blotting assay. Data were presented as mean ± SD. **P < 0.01.
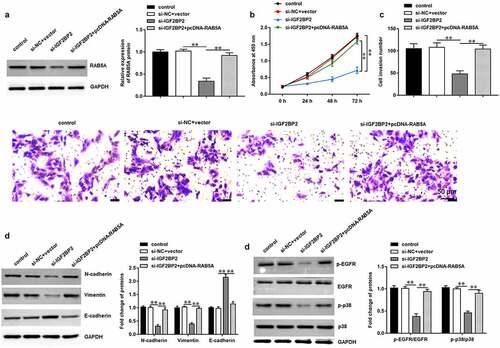
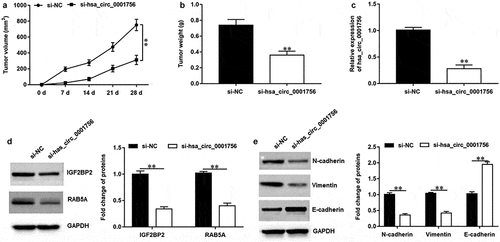
3.7 Hsa_circ_0001756 knockdown suppresses OC progression in vivo
The role of hsa_circ_0001756 in OC progression was further investigated in xenograft tumor mice in vivo. SKOV3 cells transfected with si-hsa_circ_0001756 or si-NC were injected into nude mice through tail vein. It was shown that the tumor volumes (P < 0.0001) and tumor weights (P = 0.0012) were lower in si-hsa_circ_0001756 group compared with that in si-NC group (). Moreover, we confirmed the decreased hsa_circ_0001756 (P < 0.0001) (), IGF2BP2 and RAB5A expression (P < 0.0001; P = 0.0003) () in tumor tissues of si-hsa_circ_0001756 group. Besides, our results showed that E-cadherin expression was increased (P < 0.0001), and the expression of N-cadherin and Vimentin was decreased (P = 0.0003; P = 0.0004) in si-hsa_circ_0001756 group (). Thus, we demonstrated that hsa_circ_0001756 knockdown inhibited OC progression in vivo.
Figure 7. Hsa_circ_0001756 knockdown inhibited the tumor growth of OC in vivo. Mice were randomly divided into two groups including si-NC group and si-hsa_circ_0001756 group (n = 8 per group). SKOV3 cells (2 × 106) transfected with si-NC or si-hsa_circ_0001756 were injected into nude mice through tail vein. (a) Hsa_circ_0001756 knockdown suppressed the tumor volume in vivo. (b) Hsa_circ_0001756 knockdown decreased the tumor weights of nude mice. (c) The expression of hsa_circ_0001756 in tumor tissues was detected by using RT-qPCR. (d) The protein expression of IGF2BP2 and RAB5A in tumor tissues was determined by using Western blotting assay. (e) The expression of E-cadherin, N-cadherin and Vimentin in tumor tissues was determined by using Western blotting assay. Data were presented as mean ± SD. **P < 0.01.
4. Discussion
In recent years, circRNAs, as prognostic biomarkers and therapeutic targets for human cancers, has attracted rising attention [Citation20,Citation21]. A profiling and bioinformatics analysis demonstrated that chr7:139415731–139416814- (hsa_circ_0001756) was upregulated in serum samples of OC patients and might act as a viable novel OC biomarker [Citation13]. However, the underlying role of hsa_circ_0001756 in OC progression has not been explored. In the present study, we found that hsa_circ_0001756 was upregulated in OC tissues and OC cell lines. Moreover, knockdown of hsa_circ_0001756 significantly inhibited OC cells proliferation, invasion and EMT in vitro and reduced tumor growth in vivo. More notably, circRNAs have been reported to play important roles in human physiological and pathological processes by acting as miRNA sponges or protein sponges [Citation22–24]. Our study found that hsa_circ_0001756 could bind with IGF2BP2 and positively regulated its expression in OC cells, thereby modulating the proliferation, invasion, and EMT in OC cells.
Previous studies have suggested that IGF2BP2 plays oncogenic roles in various human cancers [Citation25,Citation26]. More importantly, the mRNA expression of IGF2BP2 was found to be significantly upregulated in epithelial ovarian tumors compared to normal ovarian samples, indicating that IGF2BP2 might play an important role in ovarian cancer progression [Citation16]. Moreover, it was reported that IGF2BP2 expression was higher both in ovarian and endometrial high-grade serous carcinomas (HGSC) than that in endometrioid carcinomas, and knockdown of IGF2BP2 significantly decreased the proliferation of ovarian HGSC cells [Citation27]. IGF2BP2 is identified as an RBP, and it was reported that some noncoding RNAs participated in cancer progression by binding with IGF2BP2 and regulating IGF2BP2 expression. For example, LncRNA Long Intergenic Noncoding RNA for IGF2BP2 Stability (LINRIS) promoted the aerobic glycolysis in colorectal cancer by binding with IGF2BP2 and stabilizing IGF2BP2 expression [Citation28]. LncRNA 91 H promoted IGF2 expression and promotes tumorigenesis in colorectal cancer by interacting with IGF2BP2 [Citation29]. Based on previous studies, we further found that IGF2BP2 knockdown inhibited OC cell proliferation, invasion and EMT. In addition, IGF2BP2 was an RBP of hsa_circ_0001756 and was positively regulated by hsa_circ_0001756 in OC cells.
RAB5A is a member of Rab subfamily, which has been identified as an oncogene in various human cancers. And upregulation of RAB5A is associated with tumor metastasis. For instance, higher expression of RAB5A was observed in CRC tissues, and clinical data analysis showed that RAB5A is significantly correlated with CRC metastasis and prognosis [Citation30]. RAB5A was overexpressed in oral cancer tissue samples and promoted the malignant phenotype through EMT and the ERK/MMP‑2 signaling pathway [Citation31]. Significantly, it was found that RAB5A promoted OC cell proliferation, which may be associated with the APPL1-related EGF signaling pathway [Citation19]. Similarly, our study found that RAB5A overexpression promoted proliferation, invasion, and EMT in OC cells. Besides, RAB5A was previously reported to be upregulated in hepatocellular carcinoma and promoted HCC cell metastasis and proliferation, which is associated with EGF-EGFR signaling [Citation32]. Remarkably, EGF-EGFR signaling plays critical roles in tumor metastasis. The abnormal activation of EGFR enhanced cancer cell motility, promoted EMT, and upregulated MMPs for invasion and migration through activating several signaling cascades, mostly including MAPK and PI3K/Akt signaling pathways [Citation33,Citation34]. In this study, we further demonstrated that the IGFABP2/RAB5A axis might modulate OC progression by regulating EGFR phosphorylation and further activating the MAPK signaling pathway in OC cells.
5. Conclusion
Our findings suggested that hsa_circ_0001756 was upregulated in OC tissues and cell lines. Knockdown of hsa_circ_0001756 significantly inhibited OC cell proliferation, invasion, EMT in vitro and reduced xenograft tumor growth in vivo. Mechanistically, hsa_circ_0001756 probably modulated OC progression through regulating the IGF2BP2/RAB5A axis and the EGFR/MAPK signaling pathway. Our study may provide a novel regulatory mechanism and potential therapeutic target for the treatment of OC.
Availability of materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30.
- Elzek MA, Rodland KD. Proteomics of ovarian cancer: functional insights and clinical applications. Cancer Metastasis Rev. 2015;34(1):83–96.
- Boussios S, Zarkavelis G, Seraj E, et al. Non-epithelial ovarian cancer: elucidating uncommon gynaecological malignancies. Anticancer Res. 2016;36:5031–5042.
- Boussios S, Karihtala P, Moschetta M, et al. Veliparib in ovarian cancer: a new synthetically lethal therapeutic approach. Invest New Drugs. 2020;38(1):181–193.
- Revythis A, Shah S, Kutka M, et al. Unraveling the wide spectrum of melanoma biomarkers. Diagnostics (Basel). 2021;11(8):1341.
- Ning L, Long B, Zhang W, et al. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int J Oncol. 2018;53(6):2637–2646.
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
- Belousova EA, Filipenko ML, Kushlinskii NE. Circular RNA: new regulatory molecules. Bull Exp Biol Med. 2018;164(6):803–815.
- Zhan W, Liao X, Chen Z, et al. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. J Cell Physiol. 2020;235(2):1733–1745.
- Sun Y, Jin JG, Mi W-Y, et al. Long noncoding RNA UCA1 targets miR-122 to promote proliferation, migration, and invasion of glioma cells. Oncol Res. 2018;26(1):103–110.
- Zhang M, Xia B, Xu Y, et al. Circular RNA (hsa_circ_0051240) promotes cell proliferation, migration and invasion in ovarian cancer through miR-637/KLK4 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):1224–1233.
- Guan X, Zong ZH, Liu Y, et al. circPUM1 promotes tumorigenesis and progression of ovarian cancer by sponging miR-615-5p and miR-6753-5p. Mol Ther Nucleic Acids. 2019;18:882–892.
- Wang J, Wu A, Yang B, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in ovarian cancer. Gene. 2020;724:144150.
- Tang Q, Hann SS. Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Ther. 2020;13:2067–2092.
- Dahlem C, Barghash A, Puchas P, et al. The insulin-like growth factor 2 mRNA binding protein IMP2/IGF2BP2 is overexpressed and correlates with poor survival in pancreatic cancer. Int J Mol Sci. 2019;20(13):3204.
- Gu L, Shigemasa K, Ohama K. Increased expression of IGF II mRNA-binding protein 1 mRNA is associated with an advanced clinical stage and poor prognosis in patients with ovarian cancer. Int J Oncol. 2004;24(3):671–678.
- Yang X, Liu Z, Li Y, et al. Rab5a promotes the migration and invasion of hepatocellular carcinoma by up-regulating Cdc42. Int J Clin Exp Pathol. 2018;11(1):224–231.
- Yang PS, Yin PH, Tseng LM, et al. Rab5A is associated with axillary lymph node metastasis in breast cancer patients. Cancer Sci. 2011;102(12):2172–2178.
- Zhao Z, Liu XF, Wu HC, et al. Rab5a overexpression promoting ovarian cancer cell proliferation may be associated with APPL1-related epidermal growth factor signaling pathway. Cancer Sci. 2010;101(6):1454–1462.
- Wei J, Wei W, Xu H, et al. Circular RNA hsa_circRNA_102958 may serve as a diagnostic marker for gastric cancer. Cancer Biomark. 2020;27(2):139–145.
- Gao Y, Zhang C, Liu Y, et al. Circular RNA profiling reveals circRNA1656 as a novel biomarker in high grade serous ovarian cancer. Biosci Trends. 2019;13(2):204–211.
- Shi Y, Fang N, Li Y, et al. Circular RNA LPAR3 sponges microRNA-198 to facilitate esophageal cancer migration, invasion, and metastasis. Cancer Sci. 2020;111(8):2824–2836.
- Shi N, Shan B, Gu B, et al. Circular RNA circ-PRKCI functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-3680-3p in esophageal squamous cell carcinoma. J Cell Biochem. 2019;120(6):10021–10030.
- Liu Z, Wang Q, Wang X, et al. Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov. 2020;6(1):72.
- Barghash A, Golob-Schwarzl N, Helms V, et al. Elevated expression of the IGF2 mRNA binding protein 2 (IGF2BP2/IMP2) is linked to short survival and metastasis in esophageal adenocarcinoma. Oncotarget. 2016;7(31):49743–49750.
- Kessler SM, Lederer E, Laggai S, et al. IMP2/IGF2BP2 expression, but not IMP1 and IMP3, predicts poor outcome in patients and high tumor growth rate in xenograft models of gallbladder cancer. Oncotarget. 2017;8(52):89736–89745.
- Hiramatsu K, Yoshino K, Serada S, et al. Similar protein expression profiles of ovarian and endometrial high-grade serous carcinomas. Br J Cancer. 2016;114(5):554–561.
- Wang Y, Lu JH, Wu QN, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18(1):174.
- Gao T, Liu X, He B, et al. Long non-coding RNA 91H regulates IGF2 expression by interacting with IGF2BP2 and promotes tumorigenesis in colorectal cancer. Artif Cells Nanomed Biotechnol. 2020;48(1):664–671.
- Yu MH, Luo Y, Qin SL, et al. Increased expression of Rab5A predicts metastasis and poor prognosis in colorectal cancer patients. Int J Clin Exp Pathol. 2015;8:6974–6980.
- Zhang D, Lu C, Ai H. Rab5a is overexpressed in oral cancer and promotes invasion through ERK/MMP signaling. Mol Med Rep. 2017;16(4):4569–4576.
- Fukui K, Tamura S, Wada A, et al. Expression of Rab5a in hepatocellular carcinoma: possible involvement in epidermal growth factor signaling. Hepatol Res. 2007;37(11):957–965.
- Kajanne R, Miettinen P, Mehlem A, et al. EGF-R regulates MMP function in fibroblasts through MAPK and AP-1 pathways. J Cell Physiol. 2007;212(2):489–497.
- Bhat FA, Sharmila G, Balakrishnan S, et al. Quercetin reverses EGF-induced epithelial to mesenchymal transition and invasiveness in prostate cancer (PC-3) cell line via EGFR/PI3K/Akt pathway. J Nutr Biochem. 2014;25(11):1132–1139.
