?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Objective: To explore the relationship between fut3 gene polymorphism and colonic polyps. Methods: Two hundred patients with colonic polyps and 200 healthy people in our hospital in recent 3 years were taken as the research objects, as the disease group and the control group, respectively. The disease group was divided into cancerous colonic polyps group (n = 50) and non-cancerous colonic polyps group (n = 150). The peripheral blood nucleated cells of the subjects were collected and isolated. The fut3 gene polymorphism was obtained by sequencing and analyzed combined with the expression of fut3 gene and the level of tumor markers. Results: The frequency of allele C at rs2561796 locus in the disease group was significantly higher than that in the control group (P < 0.05). The frequency of Ag genotype at rs441158 locus in the disease group was significantly higher than that in the control group, and the frequency of Ca genotype at rs2561796 locus was significantly lower than that in the control group (P < 0.05). In the disease group, the frequency of AA + Ag in the dominant model at rs441158 was significantly higher than that in the control group, and the frequency of Ca + AA in the invisible model at rs2561796 was significantly higher than that in the control group (P < 0.05). The frequency of CGC haplotype in the disease group was higher than that in the control group (P < 0.05). The linkage disequilibrium of rs441158 and rs2561796 loci of fut3 gene was high (d‘ = 0.423). The genotype of rs372725 of fut3 gene was correlated with the expression of fut3 gene (P < 0.05). The expression of fut3 gene in patients with CC genotype was significantly higher than that in patients with other genotypes (P < 0.05). Conclusion: fut3 gene polymorphism is associated with the susceptibility and carcinogenesis of colonic polyps.
KEYWORDS:
1. Background
A colon polyp is a protrusion lesion on the colonic mucosal surface into the lumen of the colon, which is prevalent in the middle-aged and elderly [Citation1,Citation2]. As the society develops and people’s diet habit and work-rest time change, such a disease is also increasingly common in youngsters [Citation3]. The major symptom of colon polyps is bloody stool, which is also the most common and primary symptom, and changes in bowel habits and abnormalities in stool shape are also the indicative factors for colon polyps [Citation4]. Compared with inflammatory polyps, colon polyps are at a much higher risk of carcinogenesis due to atypical hyperplasia [Citation5]. Therefore, searching for and early intervening with the susceptible factors for colon polyps are of great significance for the prevention of conversion of colon polyps to colon cancer.
Gene polymorphisms have been confirmed to be the important influencing factor for the susceptibility to many diseases [Citation6,Citation7]. Moreover, gene polymorphisms mainly affect the development or progression of diseases through altering the base sequences or genotypes of the same allele [Citation8]. There has been evidence that colon polyps are correlated with the polymorphisms of multiple genes such as MTHFR [Citation9] and CYP24A1 [Citation10]. Of them, fucosyltransferase 3 (FUT3) gene encodes fucosyltransferases, and its expression is associated with the human tissue blood group antigens in intestinal mucosal secretion. Moreover, its polymorphisms may affect the expression of Lewis antigens to further influence the susceptibility to colon polyps and disease progression.
Therefore, the paper analyzed and statistically processed the polymorphisms at FUT3 gene loci rs372725, rs441158 and rs2561796 in patients with colon polyps and healthy people and then explored the correlations of the susceptibility to colon polyps, disease progression and carcinogenic tendency with FUT3 gene polymorphisms based on the haplotype and linkage disequilibrium analysis results as well as the expression of this gene and levels of serum tumor markers in patients.
2. Methods
2.1 General information
A total of 200 patients definitely diagnosed with colon polyps in Yantai Traditional Chinese Medicine Hospital in the past 3 years and another 200 healthy people from the Medical Center were selected as the subjects of study and were assigned into disease group and control group, respectively. The disease group was divided into cancerous colonic polyps group (n = 50) and non-cancerous colonic polyps group (n = 150) according to pathological type. The general data of subjects, including name, sex, age, height and weight, and their clinical information, such as family history, marriage and childbirth history, drug allergy history and medical history, were collected from disease group and control group. The statistics revealed that there were no statistically significant differences in the general data, such as age, sex and BMI (P > 0.05).
The patients in disease group were diagnosed with colon polyp based on the following criteria: 1) the symptom of hematochezia appeared previously or recently, or there was a history of bloody, purulent and mucoid stool, 2) polyps were found in a rectal examination, 3) single or more polyps were colonoscopically detected, 4) irregular filling defects were indicated in a barium meal enema, and 5) polyps removed in the colonoscopy were confirmed via pathological biopsy.
2.2 Study methods
2.2.1 Collection and processing of samples
Prior to treatment or drug administration, about 8 mL of peripheral blood was drawn by nurses on duty from the elbow veins of patients in disease group and control group, and within 1.5 h, the blood was centrifuged on a centrifugal machine at 3,000 rpm for 5 min. The nucleated cell layer was separated and transferred into a 1.5 mL centrifugal tube for the extraction of genomic deoxyribonucleic acids (DNAs).
2.2.2 Genomic DNA extraction
The genomic DNAs were extracted from the peripheral blood using the Genomic DNA Extraction Kit (Thermo Fisher Scientific) strictly in accordance with the standard operations as detailed below. First, 200 μL of protease solution was added into a centrifugal tube based on the volume of samples and with the sample of the peripheral blood nucleated cell layer and 1 mL of buffer (GE), mixed using a vortex oscillator for 1 min and let stand at 65°C for 10 min. The resulting sample was added with 2 mL of absolute ethanol, mixed evenly and transferred into an adsorption column. Subsequently, the adsorption column was added with 1.5 mL of buffer and centrifuged at 3,000 rpm for 1 min, which was repeated once. Finally, 200 μL of elution buffer was added to the adsorption column to obtain the solution, namely the genomic DNAs from the peripheral blood in disease group and control group. The DNAs with the purity of 1.8–2.0 determined using a spectrophotometer were qualified samples for subsequent experiments.
2.2.3 Polymerase chain reaction (PCR) amplification and analysis of FUT3 gene polymorphisms
The polymorphic regions at FUT3 gene loci rs372725, rs441158 and rs2561796 were amplified using a PCR instrument. The PCR was performed in the total system (25 μL) composed of 1 μL each of primers, 0.5 μL of DNA templates, 12.5 μL of Taq polymerases and 10 μL of dH2O, under the following conditions: 95°C for 5 min, (95°C for 35 s, 54°C for 45 s and 72°C for 30s)×45 cycles and 72°C for 5 min. The following primers of FUT3 gene polymorphic loci were used: rs372725 Forward: (5ʹ→3ʹ)’CTTCCTGCTAGTCTGTGTCCT’, Reverse: (5ʹ→3ʹ) “ATTGGGGTAGACAGTCCAGGT”, rs441158: Forward: (5ʹ→3ʹ) “TGGACTGTCTACCCCAATGG” Reverse: (5ʹ→3ʹ)’CAGGGTGATGCGGAATACCG’, rs2561796: Forward: (5ʹ→3ʹ) “GCAGCTTCACGACTGGATGT”, Reverse: (5ʹ→3ʹ) “CTCTCTGCGGATCTGTTCCC”. Finally, the PCR products were sent to Anhui Biotechnology Co., Ltd. for sequencing, and after analysis, the polymorphic distributions at the 3 FUT3 gene loci were obtained.
2.2.4 Determination of FUT3 gene expression level
The expression level of FUT3 gene was determined via real-time fluorescence quantitative PCR, and each primer was designed using Primer Premier 5.0 and synthesized by Shanghai Bioengineering Co., Ltd. [FUT3 gene: Forward: (5ʹ→3ʹ) “TACCTCATCCATTGCAGACATCT”, Reverse: (5ʹ→3ʹ) “CTCCTGGGGTGATTGTCCAAG”]. The total PCR system (25 μL) was composed of 1 μL each of primers, 0.5 μL of cDNA templates, 12.5 μL of SYBR premix Taq and 10 μL of ddH2O, and the PCR was completed at 94°C for 2 min, (95°C for 35 s, 56°C for 40 s and 72°C for 35s)×45 cycles and 72°C for 5 min.
2.2.5 Detection of tumor markers
The colon cancer-associated tumor markers serum carcinoembryonic antigen (CEA), carbohydrate antigen 19–9 (CA19-9) and CA50 in the peripheral blood of colon polyp patients were determined in the Biochemistry Room of the Laboratory Department in Yantai Traditional Chinese Medicine Hospital. About 5 mL of peripheral blood was collected by the nurses on duty using pro-coagulation tubes and centrifuged at 3,000 rpm for 5 min. Subsequently, the upper-layer serum was tested using an automatic biochemical analysis machine.
2.2.6 Statistical analysis
IBM SPSS 23.0 software was employed for statistical analysis. All measurement data are in accordance with normal distribution, expressed by (), single-factor analysis of variance is used for comparison between multiple groups and SNK-q test is used for pairwise comparison; count data are expressed by percentage, and comparison between groups is by χ2 test. Haplotype analysis was completed at the SHEsis website. P < 0.05 was considered to be statistically significant.
3. Results
3.1 Allele distributions at FUT3 gene loci rs372725, rs441158 and rs2561796
The distribution of alleles at rs372725, rs441158 and rs2561796 of FUT3 gene is shown in . The frequency of allele C at rs2561796 in the disease group was significantly higher than that in the control group, and the difference was statistically significant (P < 0.05).
Table 1. FUT3 gene rs372725, rs441158 and rs2561796 allele distribution
3.2 Genotype distributions at FUT3 gene loci rs372725, rs441158 and rs2561796
shows the genotype distribution of rs372725, rs441158 and rs2561796 of FUT3 gene in the disease group and control group. The AG genotype frequency at rs441158 in the disease group was significantly higher than that in the control group, and the CA genotype frequency at rs2561796 in the disease group was significantly lower than that in the control group. The difference was statistically significant (P < 0.05).
Table 2. FUT3 gene rs372725, rs441158 and rs2561796 genotype distribution
3.3 Analysis results of polymorphisms at FUT3 gene loci rs372725, rs441158 and rs2561796
The polymorphism analysis of FUT3 gene rs372725, rs441158 and rs2561796 is shown in . The frequency of AA+AG in the dominant model at rs441158 in the disease group was significantly higher than that in the control group, and the frequency of CA+AA in the invisible model at rs2561796 was significantly higher than that in the control group. The difference was statistically significant (P < 0.05).
Table 3. Polymorphism analysis of FUT3 gene rs372725, rs441158 and rs2561796
3.4 Haplotype distributions at FUT3 gene loci rs372725, rs441158 and rs2561796
The haplotype distribution at rs372725, rs441158 and rs2561796 of FUT3 gene is shown in , and the linkage disequilibrium analysis is shown in . The frequency of CGC haplotype in the disease group was higher than that in the control group, and the difference was statistically significant (P < 0.05). The linkage disequilibrium at rs441158 and rs2561796 of FUT3 gene is relatively high (d’ = 0.423).
Table 5. Haplotype distribution at rs372725, rs441158 and rs2561796 loci of FUT3 gene
Table 5. Linkage disequilibrium analysis of FUT3 gene rs372725, rs441158 and rs2561796
Table 6. The relationship between the genotypes of FUT3 gene rs372725, rs441158 and rs2561796 and tumor markers in patients with colonic polyps
3.5 Correlations of genotypes at FUT3 gene loci rs372725, rs441158 and rs2561796 with FUT3 gene expression level
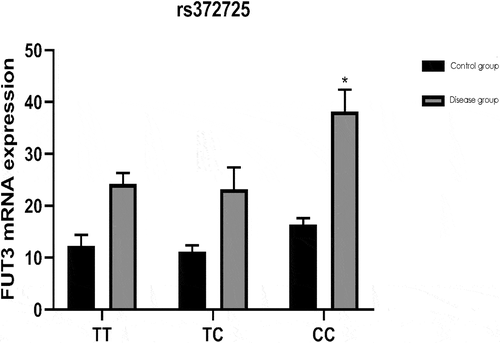
The relationship between the genotypes of FUT3 gene rs372725, rs441158 and rs2561796 loci and gene expression is shown in . The genotype at the rs372725 locus of FUT3 gene is correlated with FUT3 gene expression (P < 0.05). Among them, the expression of FUT3 gene in patients with colon polyps of CC genotype is significantly higher than that of patients with other genotypes, and the difference is statistically significant (P < 0.05).
2.6 Associations of genotypes at FUT3 gene loci rs372725, rs441158 and rs2561796 in colon polyp patients with the carcinogenesis of colon polyps
The genotypes and serum tumor marker levels of FUT3 gene rs372725, rs441158 and rs2561796 in patients with colon polyps are shown in . Among them, the serum CA19-9 level of patients with genotype AA was significantly higher than that of patients with CC and CA genotypes; The serum CEA level of patients with CA genotype was significantly higher than that of patients with AA and CC genotypes, and the difference was statistically significant (P < 0.05).
3.7 Genotype distribution of rs372725, rs441158 and rs2561796 loci of FUT3 gene
shows the genotype distribution of FUT3 gene rs372725, rs441158 and rs2561796 in the non-cancerous colon polyp group and cancer colon polyp group. The AG genotype frequency at rs441158 in the cancerous colon polyp group was significantly higher than that in the non-cancerous colon polyp group, and the CA genotype frequency at rs2561796 was significantly lower than that in the non-cancerous colon polyp group. The difference was statistically significant (P < 0.05).
Table 7. FUT3 gene rs372725, rs441158 and rs2561796 genotype distribution
4. Discussion
Colon polyps are a kind of intestinal disease that occurs in the middle-aged and elderly people [Citation11] and can be divided into inflammatory polyps, tumor polyps, immune polyps and other types [Citation12]. Most inflammatory polyps have less risk of malignant transformation, less threat to the patient’s life and better prognosis. Adenomatous polyps and familial colonic polyposis are often accompanied by cell dysplasia, the number of heterogeneous cells increases and the chance of canceration is greatly increased [Citation13,Citation14]. Compared with colon polyps that are not cancerous, canceration can seriously affect the quality of life and life cycle of patients, which not only brings greater suffering to patients, but also brings a heavy economic burden to the family and society. Therefore, studying the susceptibility factors of colon polyps and the related factors of cancer is of great significance to improve the survival rate of patients, early intervention and early treatment.
Previous studies have shown that Lewis antigens are closely associated with many malignant tumors, and FUT gene-encoded proteins are the key enzymes in their synthesis [Citation15]. Studies have proven that FUT3 gene can affect the proliferation and migration abilities of pancreatic cancer cells [Citation16,Citation17] and the invasion ability of gastric cancer cells as well [Citation18]. Additionally, gene polymorphism may be one of the mechanisms by which FUT3 gene changes the susceptibility of people to diseases. FUT3 gene polymorphisms have been confirmed to be correlated with both the invasiveness of breast cancer cells [Citation19] and the development of ulcerative colitis [Citation20]. The development, progression and change of colon polyps closely associated with colon cancer are likely to be influenced by FUT3 gene polymorphisms to a certain degree. In the present study, the FUT3 gene polymorphisms were analyzed in healthy subjects and colon polyp patients. It was found that there was a difference in the distribution of alleles at rs2561796 between disease group and control group (p = 0.012) and that the frequency of allele C in disease group was obviously higher than that in control group. Moreover, the distributions of genotypes at gene loci rs441158 (p = 0.000) and rs2561796 (p = 0.000) were different between disease group and control group, and the disease group exhibited a higher frequency of genotype AG at rs441158 and a lower frequency of genotype CA at rs2561796 than the control group. The above results suggest that FUT3 gene polymorphisms can affect the occurrence of colon polyps indeed and serve as an important susceptible factor for this disease.
Through the combination of distributions of different genotypes at the same and differentFUT3 gene loci, it was discovered that the distributions of dominant model at rs441158 (P = 0.043) and recessive model at rs2561796 (P = 0.024) in disease group were different from those in control group and that the frequencies of dominant model AA+AG at rs441158 and recessive model CA+AA at rs2561796 were higher in the disease group. The distribution of the haplotype CGC of FUT3 gene was different between disease group and control group (p = 0.032), and its frequency in the disease group was higher than that in control group. FUT3 gene loci rs441158 and rs2561796 were in a higher degree of linkage disequilibrium (d’ = 0.423). Thus, FUT3 gene polymorphisms may affect colon polyps not only via the single genotype at the single gene locus.
In addition, the relationships of genotypes at FUT3 gene loci rs372725, rs441158 and rs2561796 with FUT3 gene expression level were analyzed in this study. According to the results, the genotypes at FUT3 gene locus rs372725 were correlated with the expression level of FUT3 gene (P < 0.05), and genotype CC patients with colon polyps had an evidently higher expression level of FUT3 gene than those with other genotypes, implying that FUT3 gene polymorphisms can change gene expression level and control protein synthesis to affect Lewis antigens and the carcinogenesis of colon polyps. The mRNA expression level of FUT3 was increased in people with genotype CC at FUT3 gene locus rs372725, probably accelerating the carcinogenesis of colon polyps.
Therefore, we studied the relationship between FUT3 gene polymorphism and the content of tumor markers related to colon cancer. It was found that the genotype of FUT3 gene rs441158 locus in patients with colon polyps was related to the serum CA19-9 content (P = 0.003), and the serum CA19-9 content of patients with genotype AA was higher. The genotype of rs2561796 locus is related to the serum CEA content (P = 0.034), and the serum CEA content of patients with the genotype CA is higher. The above results indicate that FUT3 gene polymorphism can indeed affect the content of tumor markers related to colon cancer and then affect the cancerous process of colon polyps. People with specific genotypes, such as FUT3 gene rs441158 locus AA genotype and rs2561796 locus CA genotype, have significantly higher levels of tumor markers, which may have a faster canceration process than people with other genotypes. The AG genotype frequency at rs441158 in the cancerous colon polyp group was significantly higher than that in the non-cancerous colon polyp group, and the CA genotype frequency at rs2561796 was significantly lower in the non-cancerous colon polyp group (P < 0.05). It also further shows that the polymorphism of FUT3 gene is related to colon polyp carcinogenesis.
In summary, in clinical work, it is possible to detect the polymorphisms at rs441158 and rs2561796 of the FUT3 gene to predict the cancerous process and take countermeasures in advance to improve the survival prognosis of patients. However, due to the short study time and the small number of samples in this experiment, there may be some chance in the experimental results. In the future, the experimental time and the number of samples will be expanded to explore the relationship between FUT3 gene polymorphism and colon polyps.
5. Conclusion
Finally, the correlations of FUT3 gene polymorphisms with the content of colon cancer-associated tumor markers were explored in the present study. It was found that in the patients with colon polyps, the genotypes at FUT3 gene locus rs441158 were associated with the content of serum CA19-9 (p = 0.003) and that its content was higher in genotype AA patients. Besides, the genotypes at FUT3 gene locus rs2561796 were correlated with the content of serum CEA, and genotype CA patients had higher content of serum CEA. According to the above findings, it is confirmed that FUT3 gene polymorphisms can influence the content of colon cancer-associated tumor markers, thereby affecting the carcinogenesis of colon polyps. The carcinogenesis in colon polyp patients with specific genotypes, such as genotype AA at FUT3 gene locus rs441158 and genotype CA at rs2561796, who have obviously higher content of tumor markers, may be faster than that in patients with other genotypes. As such, the polymorphisms at FUT3 gene loci rs441158 and rs2561796 can be clinically detected to predict the carcinogenesis of colon polyps, and countermeasures need to be taken in advance to improve the survival prognosis of patients.
Authors’ contributions
XH wrote the manuscript. XH and FC worked on collection and processing of samples. WJ and NL helped with determination of FUT3 gene expression level. XH and YW detected tumor markers. All authors read and approved the final manuscript.
Availability of Data and Materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study was approved by the ethics committee of Yantai Traditional Chinese Medicine Hospital, and written informed consents were signed by the patients.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Worland T, Cronin O, Harrison B, et al. Clinical and financial impacts of introducing an endoscopic mucosal resection service for treatment of patients with large colonic polyps into a regional tertiary hospital. Endosc Int Open. 2019;7(11):E1386–E1392.
- Wehbeh A, Gerson MC, Rex DK. Enterococcus faecalis endocarditis after endoscopic mucosal resection of a large sessile colonic polyp. ACG Case Rep J. 2019;6:1–3.
- Tanaka Y, Inoue T, Kakimoto K, et al. Evaluation of the impact of linked color imaging for improving the visibility of colonic polyp. Oncol Lett. 2019;18(5):5555–5560.
- Konishi K, Igarashi H, Maeda S, et al. Distribution of regulatory T cells in inflammatory colorectal polyps of miniature dachshunds. Vet Immunol Immunopathol. 2019;218:109938.
- Lowe D, Saleem S, Arif MO, et al. Role of endoscopic resection versus surgical resection in management of malignant colon polyps: a national cancer database analysis. J Gastrointest Surg. 2020;24(1):177–187.
- Pereira-Martins DA, Coelho-Silva JL, Domingos IF, et al. Association between ANXA2*5681 polymorphism (rs7170178) and osteonecrosis in haemoglobin SS-genotyped patients. Br J Haematol. 2020;188(3):e8–e11.
- Pejanovic-Skobic N, Markovic I, Bozina N, et al. Lack of association of SCN2A rs17183814 polymorphism with the efficacy of lamotrigine monotherapy in patients with focal epilepsy from herzegovina area, bosnia and herzegovina. Epilepsy Res. 2019;158:106221.
- Yokoyama N, Kawasaki A, Matsushita T, et al. Association of NCF1 polymorphism with systemic lupus erythematosus and systemic sclerosis but not with ANCA-associated vasculitis in a Japanese population. Sci Rep. 2019;9(1):16366.
- Sun M, Zhong J, Zhang L, et al. Genetic impact of methylenetetrahydrofolate reductase (MTHFR) polymorphism on the susceptibility to colorectal polyps: a meta-analysis. BMC Med Genet. 2019;20(1):94.
- Chen XQ, Mao JY, Li WB, et al. Association between CYP24A1 polymorphisms and the risk of colonic polyps and colon cancer in a Chinese population. World J Gastroenterol. 2017;23(28):5179–5186.
- Yadav S, Loftus EV Jr, Harmsen WS, et al. Outcome of endoscopic resection of colonic polyps larger than 10 mm in patients with inflammatory bowel disease. Endosc Int Open. 2019;7(8):E994–E1001.
- Kono M, Takeuchi Y, Higashino K, et al. Circumferential ileocecal valve removal for a colonic polyp using underwater endoscopic mucosal resection. Endoscopy. 2020;52(1):E7–E8.
- Miwa T, Ibuka T, Ozawa N, et al. Idiopathic ileocolonic varices coexisting with a colon polyp treated successfully by endoscopy: a case report and literature review. Intern Med. 2019;58(23):3401–3407.
- de Neree Tot Babberich MPM, Bronzwaer MES, Andriessen JO, et al. Outcomes of surgical resections for benign colon polyps: a systematic review. Endoscopy. 2019;51(10):961–972.
- Do Nascimento JC, Ferreira SA, Vasconcelos JL, et al. Fut3 role in breast invasive ductal carcinoma: investigating its gene promoter and protein expression. Exp Mol Pathol. 2015;99(3):409–415.
- Gao HF, Wang QY, Zhang K, et al. Overexpressed N-fucosylation on the cell surface driven by FUT3, 5, and 6 promotes cell motilities in metastatic pancreatic cancer cell lines. Biochem Biophys Res Commun. 2019;511(2):482–489.
- Zhan L, Chen L, Chen Z. Knockdown of FUT3 disrupts the proliferation, migration, tumorigenesis and TGF-β induced EMT in pancreatic cancer cells. Oncol Lett. 2018;16(1):924–930.
- Cai YJ, Zheng XF, Lu CH, et al. Effect of FUT3 gene silencing with miRNA on proliferation, invasion and migration abilities of human KATO-III gastric cancer cell line. Cell Mol Biol (Noisy-le-grand). 2016;62(7):15–20.
- Do Nascimento JCF, de Oliveira Vasconcelos A, Seabra MABL, et al. The challenge of determining the impact of FUT3 tumor-associated polymorphism rs2306969 (−6951 C> T) in invasive breast cancer cells. Mol Biol Rep. 2019;46(3):3531–3536.
- Hu D, Zhang D, Zheng S, et al. Association of ulcerative colitis with FUT2 and FUT3 polymorphisms in patients from Southeast China. PLoS One. 2016;11(1):e0146557.