ABSTRACT
Increasing evidence reveals that circular RNAs (circRNAs) regulate multiple biological functions in glioma. Previously, several reports have illustrated that circFAM53B contributes to cancer development. However, the functions and mechanisms of circFAM53B in glioma remain elusive. Here, we gauged the circFAM53B profile in glioma tissues and cell lines and conducted gain-of-function assays of circFAM53B to verify circFAM53B’s influence on the proliferation and metastasis of glioma cells (including A172 and LN18). As a result, circFAM53B was up-regulated in glioma tissues (vs. the matched non-tumor tissues). Higher levels of circFAM53B predicted poorer survival of glioma patients. Functionally, circFAM53B up-regulation accelerated cell proliferation, colony formation, invasion and epithelial-mesenchymal transition (EMT), and heightened Bax/Bcl2 ratio. By contrast, circFAM53B down-regulation repressed glioma development in vitro. Mechanistically, bioinformatics analysis suggested that circFAM53B served as a competitive endogenous RNA (ceRNA) by sponging miR-532-3p, which targeted proto-oncogene (MET) and receptor tyrosine kinase (c-MET). miR-532-3p up-regulation delayed glioma development and inactivated the PI3K/AKT axis. Moreover, the treatment of the c-MET inhibitor SGX523, the PI3K inhibitor LY294002, and the Akt inhibitor MK-2206 reduced circFAM53B-mediated oncogenic effects. Conclusively, circFAM53B aggravated glioma progression by up-regulating the c-MET/PI3K/AKT pathway and down-regulating miR-532-3p. Thus, the circFAM53B/miR-532-3p/c-MET/PI3K/AKT axis is a potential treatment target for glioma.
1. Introduction
Glioma is the most frequent tumor in the central nervous system (CNS) characterized by high morbidity and mortality and a low five-year survival rate [Citation1,Citation2]. Surgery is the main therapeutic therapy for glioma. However, glioma patients’ median survival rate remains low even with advanced surgery, radiotherapy, chemotherapy and interventional therapy due to its strong invasiveness, high recurrence, and drug resistance [Citation3,Citation4]. Hence, it is urgent to probe the pathological mechanism of glioma and to search for new treatment options.
Circular RNAs (circRNAs) are closed-loop noncoding RNAs formed by reverse splicing without 3′-end and 5′-end [Citation5]. Recently, increasing studies have uncovered that circRNAs are dysregulated in multiple cancers and exert vital roles in tumorigenesis and tumor progression [Citation6,Citation7]. Moreover, circRNAs also affect glioma progression and act as therapeutic targets [Citation8]. CircFAM53B (ID: hsa_circ_0020318), a novel circRNA located on chr10:126,305,648–126,395,456, was first identified by Salzman J et al. [Citation9]. The latest research revealed that circFAM53B is overexpressed in ovarian cancer (OC) and is related to clinical staging and poor prognosis of OC patients. Moreover, circFAM53B accelerates OC cell proliferation, migration, and invasion through the miR‑646/VAMP2 and miR‑647/MDM2 pathways [Citation10]. Nonetheless, the biological functions of circFAM53B in glioma remain unknown.
MicroRNAs (miRNAs) are noncoding RNAs with a length of 19–22 bases and are widely expressed in cells. By targeting the 3ʹ-untranslated region (3ʹUTR) of mRNAs, miRNAs modulate the protein-coding gene expression at the post-transcriptional level [Citation11,Citation12]. What’s more, miRNAs contribute to tumorigenesis and tumor development, including cell proliferation, survival, tumor angiogenesis, invasion, and metastasis [Citation13,Citation14]. For example, down-regulated miR-145 leads to increased expression of metastatic protein ROCK1, thus facilitating glioma invasion [Citation15]. Additionally, miR-483-3p facilitates neuroblastoma cell proliferation and migration as an oncogene [Citation16]. In parallel, miR-532-3p mediates various cancer as a tumor suppressor [Citation17,Citation18], while its role in glioma needs further studies.
At present, we discover that circFAM53B is up-regulated in glioma tissues and is correlated with glioma patients’ poorer survival rate. Through gain-of-function experiments, we observed that up-regulation of circFAM53B aggravated glioma. Meanwhile, miR-532-3p was an underlying target of circFAM53B. Furthermore, bioinformatics analysis substantiated that MET was targeted by miR-532-3p. Thus, we posit that there is a circFAM53B -miR-532-3p-c-MET axis in glioma.
2. Materials and methods
2.1. Collection of clinical samples
Forty paired cancerous and adjacent non-cancerous tissues in primary glioma patients were collected from the First People’s Hospital of Suzhou. None of the patients were treated with preoperative chemotherapy or radiotherapy. The specimens were diagnosed independently by two pathologists. The Research Ethics Committee of the First People’s Hospital of Suzhou granted this study (approval number: SZF2018-223), and all involved patients signed informed consent.
2.2. Cell culture and transfection
Human glioma cells (U251, A172, TJ861, TJ905, and LN18) and healthy glial cells HEB were ordered from the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in a DMEM-F12 medium comprising 10% FBS and 1% penicillin/streptomycin (Beijing Solarbio Science&Technology Co., Ltd, Beijing, China) at 37°C with 5% CO2. The DMEM-F12 and FBS were acquired from Thermo Fisher Scientific (Shanghai, China). We substituted the medium every 2–3 days. The MET inhibitor SGX523 (4 nM, Cat. No. S1112, Selleck), the PI3K inhibitor LY294002 (1 µM, Cat. No. S1105, Selleck), and the AKT inhibitor MK-2206 2HCL (5 µM, Cat. No. S1078, Selleck) were administered to glioma cells for 24 hours for inhibiting MET, PI3K, AKT, respectively.
CircFAM53B overexpression plasmids, small interfering RNA targeting circFAM53B (si-circFAM53B) and the blank vectors (vector, or si-NC) were designed and synthesized by Shanghai Genechem Co., Ltd. miR-532-3p mimics and the negative controls (miR-NC) were provided by Shanghai Integrated Biotechnology Co., Ltd. Twenty-four hours before the transfection, A172 and LN18 cells were cultured in 24-well plates (1 × 105 cells/well). Next, the cells were transfected with the above vectors using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Twenty-four hours later, the medium was replaced with a fresh compete medium. Then, we applied quantitative reverse transcription-polymerase chain reaction (qRT-PCR) to assess the profile of circFAM35B or miR-532-3p to evaluate the transfection efficiency.
2.3. qRT-PCR
qRT-PCR was conducted to verify the profiles of circFAM35B, miR-532-3p, and cancer-related genes (MET and FZD6). Glioma cells were harvested, and the total RNA was extracted out of cells with the TRIzol reagent (Invitrogen). The RNA purity and content were calculated by spectrophotometry. The absorbance ratio between 1.8 and 2.0 at 260 nm and 280 nm is considered acceptable. 2 μg RNA was transcribed into cDNA with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA). Next, 1 μL cDNA was taken, and qRT-PCR was conducted using SYBR Green qPCR Master Mix (MedChemExpress, NJ, USA) on the ABI 7500 Fast Real‐Time PCR System (Applied Biosystems, Foster City, CA). Finally, the PCR amplification was evaluated by the melting curve. The endogenous control of miR-532-3p was U6, and GAPDH was the housekeeping gene of other genes. The expression was quantified by the 2−∆∆CT method. All experiments were conducted in triplicate.
2.4. Cell counting kit-8 (CCK8) assay
Each group of A172 and LN18 cells in the exponential growth phase were made into single-cell suspensions. After cell counting, the cells were inoculated into six 96-well plates (1000 cells/well) and cultured for different durations (24, 48, 72, 96 hours). Then, 90 μL culture medium and 10 μL CCK-8 solution (Beyotime Biotechnology, Wuhan, China) were added to the sample, and blank control wells were supplemented with the culture medium only. After incubation for 2 hours, the absorbance (A) value (450 nm) was observed and recorded with a microplate. Cell proliferation in each group was evaluated by subtracting the A value of the corresponding blank group from the A value of cells in each group according to the standard curve. All experiments were made three times.
2.5. Transwell assay
The 24-well Boyden chamber with 8-μm pore size polycarbonate membranes (Corning, NY, USA) was adopted to test cell invasion. Cells were placed in the Matrigel-coated upper chamber (Beyotime Biotechnology, Wuhan, China) at 2 × 104 cells/well, and the lower chamber was supplemented with a 600 μL complete medium (containing 20% FBS). The cells were maintained at 37°C for 24 hours. Afterward, cell suspension in the upper chamber was removed, and the lower chamber cells were fastened with 4% paraformaldehyde (PFA) and stained with 0.1% crystal violet. Finally, the invaded cells were counted. All tests were repeated three times.
2.6. Colony formation assay
A172 and LN18 cells transfected with circFAM35B overexpression plasmids, miR-532-3p mimics, or miR-NC were harvested. Next, phosphate-buffered saline (PBS) was utilized to resuspend the cells. Next, the suspension was inoculated into 6-well plates (150 cells/well). The next week, the plates were put in the incubator (37°C, 5% CO2). Finally, the colonies were dyed with crystal violet (Beyotime Biotechnology, Wuhan, China) and observed with a light microscope (Olympus, Japan).
2.7. Western blot
Total cellular proteins were isolated with the RIPA lysis solution (Beyotime Biotechnology, Wuhan, China). They were then quantified by the BCA method and reserved at −80°C. After denaturing, 20 μg of total proteins were separated by 10% SDS-PAGE and transferred to PVDF membranes (300 mA constant current). After that, the membranes were sealed with TBST solution containing 5% skim milk for 1 hour at room temperature (RT) and maintained overnight at 4°C with primary antibodies of MET (also known as c-MET) (Abcam, ab68141, 1:1000), p-PI3K (Abcam, ab182651, 1:1200), PI3K (Abcam, ab191606, 1:1200), p-AKT (Abcam, ab38449, 1:1000), AKT (Abcam, ab8806, 1:1000), E-cadherin (Abcam, ab40772, 1:1000), Vimentin (Abcam, ab92547, 1:1000), Bax (Abcam, ab32503, 1:1000) and Bcl2 (Abcam, ab182858, 1:1000). Subsequently, TBST was applied to rinse the membranes (4 times, 8 minutes/time). Next, the membranes were kept with the corresponding HRP-conjugated secondary antibodies (WuHan AmyJet Scientific Inc.) (concentration: 1:2000) at RT for 1.5 hours, followed by cleaning with TBST 4 times (8 minutes/time). The Pierce ECL Western blot Substrate kit (Thermo, Shanghai, China) was employed for X-ray development. All tests were conducted in triplicate.
2.8. Dual-luciferase reporter gene experiment
Here, the dual-luciferase reporter assay system (Promega, Madison, WI, USA) was adopted. Objective fragments of wild-type circFAM35B (circFAM35B-wt), wild-type MET (MET-wt), mutant type MET (MET-mut) and mutant circFAM35B (circFAM35B-mut1, circFAM35B-mut2, circFAM35B-mut1+ mut2) were integrated into the pGL3 vector (Promega, Madison, WI, USA) to synthesize corresponding reporter vectors. Then those vectors were co-transfected with miR-532-3p mimics or miR-NC into A172 cells. The luciferase activity was detected 48 hours after the transfection. All experiments were done three times.
2.9. RNA immunoprecipitation (RIP)
RIP was implemented to determine the targeting correlation between miR-532-3p and circFAM35B using the EZ-Magna RIP Kit (Millipore, Billerica, MA, USA). A172 cells were lysed. Next, the lysates were incubated with the RIP buffer, which contained magnetic beads conjugated with the normal mouse IgG (as negative control) or human anti-Ago2 antibody (Millipore). Subsequently, proteinase K was applied to remove the proteins, and the immunoprecipitated RNA was collected. At last, qRT-PCR was conducted to estimate circFAM35B and miR-532-3p expression.
2.10. Xenograft tumor model in nude mice
In-vivo experiments were implemented in BALB/c-nude mice to figure out the impact of circFAM53B on tumor proliferation and metastasis. Twenty-four mice (6-week-old, 22–25 g) were commercially provided by the Laboratory Animal Center of Nanjing Medical University (Nanjing, China). All of them could eat and drink at random and were housed in a 50% humidity environment with 12 hours of light/dark cycles. A172 cells transfected with circFAM35B overexpression plasmids or negative vectors were subcutaneously injected into the right side of the mouse’s back. Tumor volumes were monitored every 5 days. Thirty days later, we killed the mice and stripped the tumors and weighed them. Tumor volume (V) = (width2 × length)/2. The animal experiments were authorized by the Animal Research Ethics Committee of China Pharmaceutical University (approve number: CXY2019-059) following the ARRIVE guidelines and the Basel Declaration. All animals received humane care required by the National Institutes of Health (USA) guidelines.
2.11. Immunohistochemistry (IHC)
IHC was performed to examine EMT markers (E-cadherin and Vimentin) and Ki67 in tumors. The isolated tumor tissues were immobilized by 4% PFA for 24 hours at RT, followed by paraffin-embedding and sectioning (4-μm thick). After that, the sections were adopted for IHC. The antibodies (all from Abcam) applied in this study included E-cadherin (ab40772, 1:200), Vimentin (ab92547, 1:200) and Ki67 (ab15580, 1:200).
2.12. Statistics
SPSS17.0 (SPSS Inc., Chicago, IL, USA) was adopted for data analysis. The measurement data were presented as mean ± standard deviation (x ± s). The pairwise comparison was made by t test, while a one-way analysis of variance was employed to compare multiple means. SNK-q test was adopted for pairwise comparison between the mean values of various data. P< 0.05 represented statistical significance.
3. Results
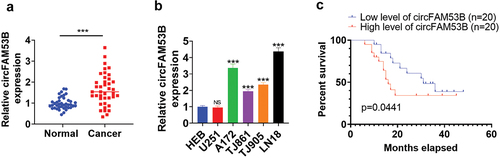
3.1. CircFAM53B was highly expressed in glioma
We determined circFAM53B expression in glioma tissues and cell lines with qRT-PCR to probe circFAM53B’s function in glioma. It turned out that the circFAM53B level was elevated in glioma tissues (vs. normal adjacent tissues) (P< 0.001, ). Four glioma cell lines, including A172, TJ861, TJ905, and LN18, had significantly higher levels of circFAM53B compared to that in HEB cells (P< 0.001, )). Besides, higher circFAM53B levels were linked to poorer survival of glioma patients (p = 0.0441, ). Therefore, we supposed that circFAM53B was an oncogene in glioma.
Figure 1. The circFAM53B level in glioma.
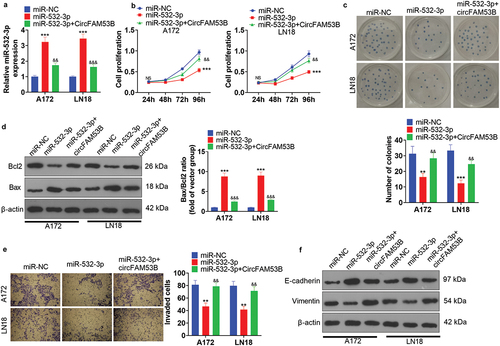
3.2. CircFAM53B accelerated glioma cell proliferation, invasion, EMT and choked apoptosis
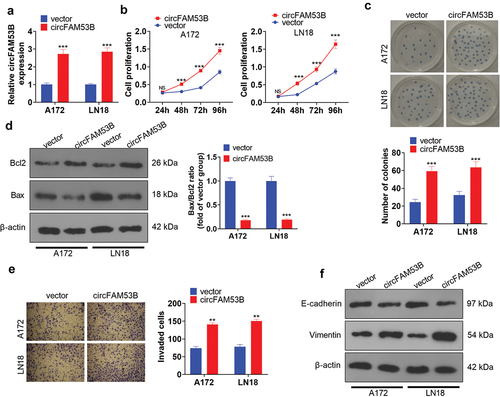
We established circFAM53B overexpression models in A172 and LN18 cells to make certain the functions of circFAM53B in glioma ()). Next, the CCK8 assay, colony formation assay and Western blot were utilized to assess cell proliferation and apoptosis, respectively. The outcomes revealed that circFAM53B up-regulation markedly enhanced the proliferation of A172 and LN18 cells (-c)). Western blot data demonstrated that forced circFAM53B overexpression heightened Bcl2 levels, curbed Bax levels, and declined the Bax/Bcl2 ratio, suggesting that circFAM53B reduced cell apoptosis ()). Furthermore, transwell assay and Western blot were implemented to test cell invasion and EMT markers. Notably, up-regulating circFAM53B intensified invasion, restrained E-cadherin, and facilitated Vimentin expression in glioma cells () and (f)). Overall, circFAM53B was an oncogene in glioma.
Figure 2. Up-regulation of circFAM53B intensified the malignant behaviors of glioma cells.
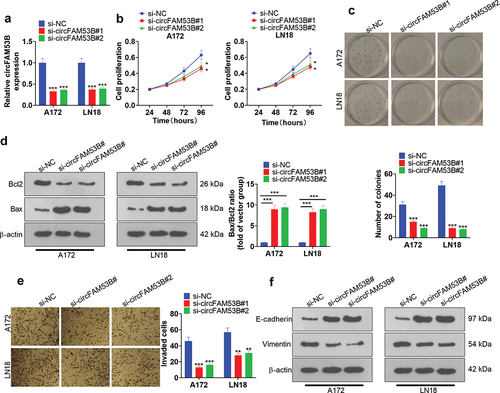
3.3. Effect of knocking down CircFAM53B on cell proliferation, invasion, EMT and apoptosis
We established circFAM53B knockdown models in A172 and LN18 cells to validate the function of circFAM53B in glioma ()). Secondly, the CCK8 assay, colony formation assay and Western blot were employed to assay cell proliferation and apoptosis, respectively. As a result, circFAM53B knockdown dramatically restrained the proliferation and viability of A172 and LN18 cells (-c)). As evidenced by Western blot outcomes, circFAM53B knockdown enhanced the Bax/Bcl2 ratio, suggesting that it intensified apoptosis ()). Besides, we adopted the Transwell assay and Western blot to test cell invasion and EMT. Notably, knocking down circFAM53B impeded cell invasion, elevated E-cadherin expression and reduced Vimentin expression () and (f)). These outcomes revealed that circFAM53B knockdown hampered proliferation, invasion and EMT of glioma cells.
Figure 3. Influence of CircFAM53B knockdown on cell proliferation, invasion, EMT and apoptosis.
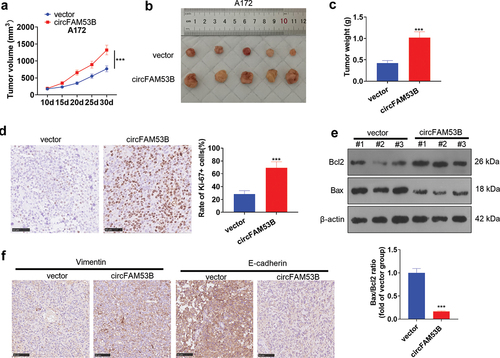
3.4. CircFAM53B triggered A172 cell growth and EMT in vivo
The in-vivo experiment was applied to make certain the impact of circFAM53B on A172 cell proliferation. Our findings displayed that A172 cells up-regulating circFAM53B formed larger tumors (-c)). Then, IHC was conducted to monitor Ki67 (a proliferation marker) in tumor tissues, which disclosed that the circFAM53B group had a higher Ki67-positive rate (vs. the vector group) ()). The Bcl2 and Bax expression in tumor tissues was examined by Western blot, revealing that Bax was down-regulated while Bcl2 was up-regulated in the circFAM53B group versus the vector group ()). Furthermore, IHC illustrated that the E-cadherin expression was declined and the Vimentin profile was obviously heightened in the circFAM53B group versus the vector group ()). These data confirmed that circFAM53B accelerated A172 cell proliferation and metastasis in vivo.
Figure 4. CircFAM53B boosted A172 cell proliferation and EMT in vivo.
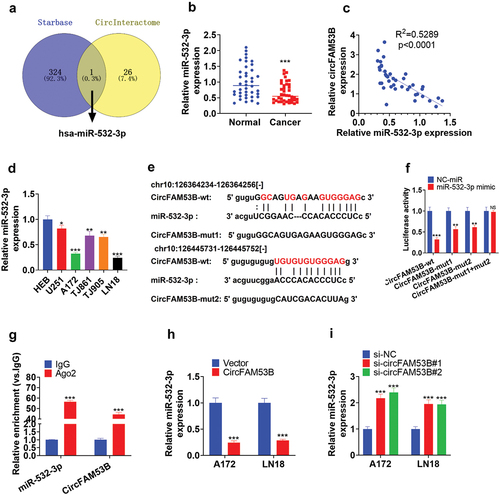
3.5. CircFAM53B targeted and dampened miR-532-3p
We adopted the bioinformatics database Starbase (htttp://starbase.sysu.edu.cn/index.php) and circInteractome (https://circinteractome.nia.nih.gov/) to probe the downstream molecular mechanism of circFAM53B. Meanwhile, Venn’s diagram was utilized, which illustrated that miR-532-3p was targeted by circFAM53B ()). In addition, qRT-PCR manifested that miR-532-3p was notably down-regulated in glioma tissues and cells ()). Besides, the circFAM53B expression was reversely related to miR-532-3p expression (-d)). The binding relationship between circFAM53B and miR-532-3p was shown in ). As testified by the dual-luciferase reporter assay, miR-532-3p mimics markedly hampered the luciferase activity of A172 cells transfected with circFAM53B-wt, while they had little influence on cells transfected with circFAM53B-mut1+ mut2 vectors ()). RIP testified that circFAM53B and miR-532-3p were more enriched in the anti-Ago2 group, suggesting that circFAM53B bound with miR-532-3p through Ago2 ()). In parallel, we modulated the expression of circFAM53B. qRT-PCR revealed that circFAM53B overexpression led to down-regulation of miR-532-3p, while circFAM53B knockdown up-regulated miR-532-3p (-i)). In summary, miR-532-3p was down-regulated in glioma tissues and cells, and it was targeted by circFAM53B.
Figure 5. miR-28-5p was a sponge RNA of circFAM53B.
3.6. The role of circFAM53B /miR-532-3p in glioma
To probe the mutual regulation of circFAM53B and miR-532-3p in glioma development, miR-532-3p mimics were employed to overexpress miR-532-3p, and circFAM53B overexpression plasmids were supplemented to up-regulate circFAM53B. qRT-PCR manifested that miR-532-3p was up-regulated after the miR-532-3p mimic transfection, while circFAM53B up-regulation impeded the miR-532-3p level (vs. the miR-532-3p group) ()). Next, glioma cell proliferation, colony formation, apoptosis, invasion and EMT were monitored. The data revealed that miR-532-3p up-regulation hindered glioma cells’ proliferation, colony formation, invasion and EMT and facilitated apoptosis (-f)). In contrast, overexpression of circFAM53B strengthened the proliferation, colony formation, invasion and EMT of glioma cells (vs. the miR-532-3p group) (-f)). Those outcomes collectively hinted that miR-532-3p suppressed glioma, and circFAM53B might function as an oncogene via dampening miR-532-3p.
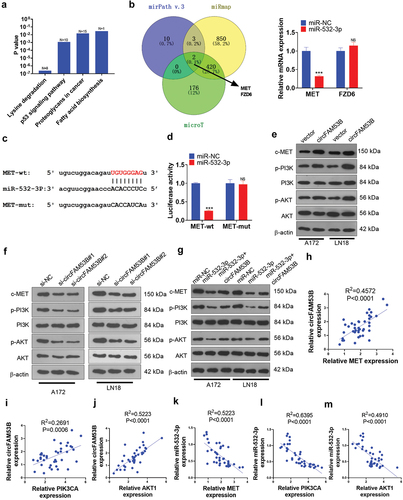
3.7. CircFAM53B/miR-532-3p modulated c-MET/PI3K/AKT
We conducted bioinformatics analysis through miRPath v.3 (http://snf-515788.vm.okeanos.grnet.gr/index.php?r=miRpath) to clarify the potential mechanisms that circFAM53B/miR-532-3p regulated glioma. As a result, we discovered that there were four pathways regulated by miR-532-3p ()). Through miRanda and microT databases, we observed that MET and FXD6 were possible targets of miR-532-3p. Next, we implemented qRT-PCR to verify the two candidate genes in glioma cells. The data substantiated that miR-532-3p overexpression substantially restrained MET expression, while it exerted no effect on FXD6 ()). In this setting, we conducted the dual-luciferase reporter assay and discovered that miR-532-3p targeted MET (). Moreover, we tested the c-MET/PI3K/AKT profile under selectively modulation of circFAM53B and miR-532-3p. As a result, overexpressing circFAM53B elevated c-MET, p-PI3K and p-AKT levels, while knockdown of circFAM53B led to the opposite result (). Similar to the rescue experiment, circFAM53B overexpression in cells up-regulating miR-532-3p reactivated c-MET/PI3K/AKT pathway (compared with the miR-532-3p group, ). Moreover, by examining the correlation between circFAM53B and miR-532-3p and the c-MET/PI3K/AKT pathway, we observed that circFAM53B was positively correlated with the c-MET/PI3K/AKT pathway and miR-532-3p was negatively correlated with the c-MET/PI3K/AKT pathway (-m)). Therefore, we believe that the circFAM53B-miR-532-3p axis affects glioma progression by mediating the c-MET/PI3K/AKT pathway.
Figure 7. CircFAM53B/miR-532-3p modulated the MAPK/ERK pathway.
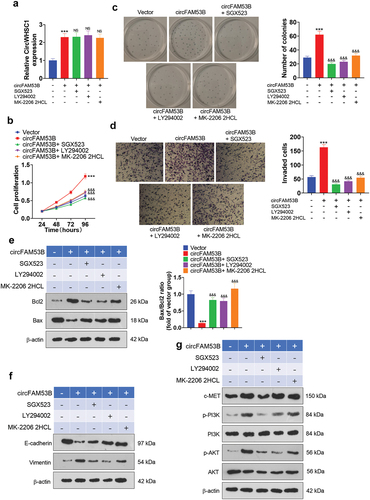
3.8. Inhibition of the c-MET/PI3K/AKT pathway expression reduced the oncogenic effects of circFAM53B
To probe the influence of suppressing the expression of the c-MET/PI3K/AKT pathway on the carcinogenic effect of circFAM53B, we added the MET inhibitor SGX523 (4 nM, Cat. No. S1112, Selleck), the PI3K inhibitor LY294002 (1 µM, Cat. No. S1105, Selleck), and the AKT inhibitor MK-2206 2HCL (5 µM, Cat. No. S1078, Selleck) into A172 cells stably transfected with circFAM53B overexpression plasmids. qRT-PCR results displayed that the application of the above inhibitors had no significant effects on circFAM53B expression (P> 0.05 vs.circFAM53B group, )). Subsequently, we gauged cell proliferation and invasion using the CCK8 assay, colony formation assay and Transwell assay, which illustrated that the proliferative and migrative ability of the cells was notably choked by the addition of these inhibitors (-d)). Furthermore, Western blot was adopted to check the expression of apoptosis-related proteins (Bax and Bcl2) and EMT-related proteins (E-cadherin and Vimentin). As a result, the addition of these inhibitors resulted in up-regulation of Bax and E-cadherin and down-regulation of Bcl2 and Vimentin in the cells, suggesting that these inhibitors intensified apoptosis and restrained EMT (-f)). Meanwhile, administration of SGX523 diminished the expression levels of c-MET, p-PI3K and p-AKT. LY294002 treatment repressed p-PI3K and p-AKT, whereas MK-2206 2HCL only attenuated the p-AKT level ()). Thus, inactivating the c-MET/PI3K/AKT pathway suppressed cell proliferation, invasion and EMT, reversing the oncogenic effect of circFAM53B.
Figure 8. Impacts of inactivating the c-MET/PI3K/AKT pathway expression on the oncogenic effect of circFAM53B.
4. Discussion
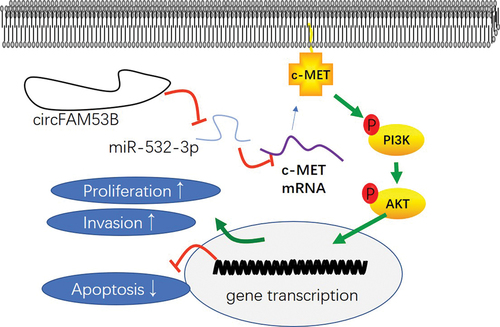
Here, we observed that circFAM53B was notably up-regulated in glioma. Meanwhile, the circFAM53B/miR-532-3-5p/c-MET/PI3K/AKT axis has a vital role in mediating glioma progression ().
Figure 9. The mechanism diagram of circFAM53B in glioma.
Uncontrolled cell proliferation and metastasis remain the obstacles in treating glioma. Emerging evidence has illustrated that circRNAs contribute to the malignant process of glioma by regulating proliferation, apoptosis, migration, invasion, and chemoresistance [Citation19,Citation20]. For instance, circPVT1 is up-regulated in glioma tissues, and it facilitates glioma cell proliferation and metastatic potential by modulating miR-199a-5p [Citation21]. Similarly, overexpressing CircPIP5K1A strengthens glioma cell proliferation, invasion, and EMT and impedes apoptosis both in vivo and in vitro [Citation22]. Among multiple circRNAs, circFAM35B exerts a prominent role in tumorigenesis. For instance, circFAM35B accelerates OC development [Citation10]. Here, our data revealed that circFAM35B was significantly up-regulated in glioma tissues, which predicted unfavorable survival of glioma patients. Furthermore, functional assays verified that down-regulating circFAM35B dampened the malignant behaviors of glioma cells. Hence, circFAM35B was an oncogene in glioma.
Abundant reports have established that miRNAs affect tumorigenesis and tumor development by modulating gene expression [Citation23]. For example, miR-10b is overexpressed in glioma samples and expedites cell invasion by regulating MMP-14 and uPAR via HOXD10 [Citation24]. Meanwhile, overexpressing miR-106b-5p facilitates glioma cell proliferation and abates cell apoptosis [Citation25]. miR-532-3p has higher expression in hepatocellular carcinoma (HCC) tissues and is associated with the advanced stages of HCC. Functionally, miR-532-3p enhances HCC cell migration, invasion and proliferation by targeting receptor protein tyrosine phosphatase T (PTPRT) [Citation26]. In parallel, miR-532-3p reduces the proliferation and EMT of colorectal cancer cells through ETS1 and ETS1/TGM2 [Citation27], suggesting that it is a tumor suppressor. What’s more, dysregulated miR-532-3p is involved in glioma. A recent study indicated that SNHG10 aggravates glioma through miR-532-3p/FBXL19 [Citation28]. Here, we discovered that miR-532-3p was a sponge of circFAM53B, and it was down-regulated in glioma tissues. Besides, up-regulating miR-532-3p led to reduced glioma cell proliferation, invasion and EMT. Therefore, overexpressing circFAM53B functioned as an oncogene by restraining miR-532-3p, a tumor suppressor in glioma.
The MET pathway is a classical signaling pathway controlling diversified essential cell biological processes in tumorigenesis and tumor development, including apoptosis, migration, translation, metabolism, and is of interest as a therapeutic target [Citation29,Citation30]. Besides, MET and its ligand hepatocyte growth factor (HGF) trigger glioma cells’ proliferation, survival, migration, invasion, angiogenesis, stemness, and therapeutic resistance [Citation31]. Thus, modulating the MET pathway activation is potentially relevant for glioma therapy. For example, the pharmacologic (crizotinib) inhibition of c-MET leads to enhanced reprogramming of fatty acid metabolism and production of reactive oxygen species in glioblastoma [Citation32]. Interestingly, miRNAs also target the c-MET pathway. For instance, miRNA-128-3p enhances the curative effect of temozolomide on glioblastoma cells by repressing c-Met and EMT [Citation33]. In parallel, overexpressing miR-23b-3p hampers the proliferative, migrative, and invasive ability of cervical cancer cells [Citation34]. Here, bioinformatics analysis illustrated that c-MET was a promising target of miR-532-3p in glioma. miR-532-3p was confirmed to target the 3ʹ-UTR of MET (also known as c-MET) and abate MET expression. Besides, Western blot manifested that overexpressing circFAM53B enhanced c-MET, p-PI3K, and p-AKT levels. Inhibiting the c-MET/PI3K/AKT pathway using the related inhibitors obviously reduced circFAM53B-mediated oncogenic effects. Thus, circFAM53B/miR-532-3p regulated glioma development by modulating the c-MET/PI3K/AKT pathway.
Overall, our study confirms that circFAM53B facilitates glioma cell proliferation and invasion and regulates glioma evolvement by regulating the miR-532-3p-c-MET/PI3K/AKT axis (). This study has provided further insights into the molecular mechanisms regulating glioma development and progression, which facilitates the diagnosis and treatment of gliomas.
Ethics statement
Our study was approved by the Ethics Review Board of China Pharmaceutical University.
Authors’ contributions
Conceived and designed the experiments: Xiaozhao Deng;
Performed the experiments: Jiaping Pei, Hui Dou;
Statistical analysis: Hui Dou;
Wrote the paper: Jiaping Pei.
All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s)
Data availability statement
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Rasmussen BK, Hansen S, Laursen RJ, et al. Epidemiology of glioma: clinical characteristics, symptoms, and predictors of glioma patients grade I-IV in the Danish neuro-oncology registry. J Neurooncol. 2017;135(3):571–579.
- Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–1850.
- Kilday JP, Bartels UK, Bouffet E. Targeted therapy in pediatric low-grade glioma. Curr Neurol Neurosci Rep. 2014;14(4):441.
- Sturm D, Pfister SM, Jones DTW. Pediatric gliomas: current concepts on diagnosis, biology, and clinical management. J Clin Oncol. 2017;35(21):2370–2377.
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338.
- Long Z, Gong F, Li Y, et al. Circ_0000285 regulates proliferation, migration, invasion and apoptosis of osteosarcoma by miR-409-3p/IGFBP3 axis. Cancer Cell Int. 2020;20:481.
- Chen L, Zhang X, Wang S, et al. Circ_0084927 facilitates cervical cancer development via sponging miR-142-3p and up-regulating ARL2. Cancer Manag Res. 2020;12:9271–9283.
- Jin C, Zhao J, Zhang ZP, et al. CircRNA EPHB4 modulates stem properties and proliferation of gliomas via sponging miR-637 and up-regulating SOX10. Mol Oncol. 2021;15(2):596–622. doi:https://doi.org/10.1002/1878-0261.12830
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013 Dec; 9(12). [published correction appears in. PLoS Genet. 2013;9(9):e1003777. DOI:https://doi.org/10.1371/annotation/f782282b-eefa-4c8d-985c-b1484e845855].
- Sun D, Liu J, Zhou L. up-regulation of circular RNA circ-FAM53B predicts adverse prognosis and accelerates the progression of ovarian cancer via the miR-646/VAMP2 and miR-647/MDM2 signaling pathways. Oncol Rep. 2019;42(6):2728–2737.
- Vaschetto LM. miRNA activation is an endogenous gene expression pathway. RNA Biol. 2018;15(6):826–828.
- Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. 2016;17(10):1712. [Published 2016 Oct 13]
- Van Roosbroeck K, Calin GA. Cancer hallmarks and MicroRNAs: the therapeutic connection. Adv Cancer Res. 2017;135:119–149.
- Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23.
- Wan X, Cheng Q, Peng R, et al. ROCK1, a novel target of miR-145, promotes glioma cell invasion. Mol Med Rep. 2014;9(5):1877–1882.
- Wu K, Wang J, He J, et al. miR-483-3p promotes proliferation and migration of neuroblastoma cells by targeting PUMA. Int J Clin Exp Pathol. 2018;11(2):490–501.
- Liu Y, Li Q, Dai Y, et al. miR-532-3p inhibits proliferation and promotes apoptosis of lymphoma cells by targeting β-catenin. J Cancer. 2020;11(16):4762–4770.
- Jiang W, Zheng L, Yan Q, et al. miR-532-3p inhibits metastasis and proliferation of non-small cell lung cancer by targeting FOXP3. J BUON. 2019;24(6):2287–2293.
- Sun J, Li B, Shu C, et al. Functions and clinical significance of circular RNAs in glioma. Mol Cancer. 2020;19(1):34.
- Cheng J, Meng J, Zhu L, et al. Exosomal noncoding RNAs in Glioma: biological functions and potential clinical applications. Mol Cancer. 2020;19(1):66.
- Chi G, Yang F, Xu D, et al. Silencing hsa_circ_PVT1 (circPVT1) suppresses the growth and metastasis of glioblastoma multiforme cells by up-regulation of miR-199a-5p. Artif Cells Nanomed Biotechnol. 2020;48(1):188–196.
- Zheng K, Xie H, Wu W, et al. CircRNA PIP5K1A promotes the progression of glioma through up-regulation of the TCF12/PI3K/AKT pathway by sponging miR-515-5p. Cancer Cell Int. 2021;21(1):27.
- El-Osaily HH, Ibrahim IH, Essawi ML, et al. Impact of miRNAs expression modulation on the methylation status of breast cancer stem cell-related genes [published online ahead of print, 2021 Jan 12]. Clin Transl Oncol. 2021. DOI:https://doi.org/10.1007/s12094-020-02542-0
- Sun L, Yan W, Wang Y, et al. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res. 2011;1389(none):9–18.
- Liu F, Gong J, Huang W, et al. MicroRNA-106b-5p boosts glioma tumorigensis by targeting multiple tumor suppressor genes. Oncogene. 2014;33(40):4813–4822.
- Wang Y, Yang Z, Wang L, et al. miR-532-3p promotes hepatocellular carcinoma progression by targeting PTPRT. Biomed Pharmacother. 2019;109:991–999.
- Gu C, Cai J, Xu Z, et al. miR-532-3p suppresses colorectal cancer progression by disrupting the ETS1/TGM2 axis-mediated Wnt/β-catenin signaling. Cell Death Dis. 2019;10(10):739.
- Jin L, Huang S, Guan C, et al. ETS1-activated SNHG10 exerts oncogenic functions in glioma via targeting miR-532-3p/FBXL19 axis. Cancer Cell Int. 2020;20(1):589.
- Zambelli A, Biamonti G, Amato A. HGF/c-Met signalling in the tumor microenvironment. Adv Exp Med Biol. 2021;1270:31–44.
- Malik R, Mambetsariev I, Fricke J, et al. MET receptor in oncology: from biomarker to therapeutic target. Adv Cancer Res. 2020;147:259–301.
- Cheng F, Guo D. MET in glioma: signaling pathways and targeted therapies. J Exp Clin Cancer Res. 2019;38(1):270.
- Zhang Y, Nguyen TTT, Shang E, et al. MET inhibition elicits PGC1α-dependent metabolic reprogramming in glioblastoma. Cancer Res. 2020;80(1):30–43.
- Zhao C, Guo R, Guan F, et al. MicroRNA-128-3p enhances the chemosensitivity of temozolomide in glioblastoma by targeting c-Met and EMT. Sci Rep. 2020;10(1):9471.
- Campos-Viguri GE, Peralta-Zaragoza O, Jiménez-Wences H, et al. miR-23b-3p reduces the proliferation, migration and invasion of cervical cancer cell lines via the reduction of c-Met expression. Sci Rep. 2020;10(1):3256.