ABSTRACT
Circular RNAs (circRNAs) are regulatory endogenous RNAs in human diseases by sponging microRNAs (miRNAs) to affect the gene expression. However, little research focused on the circRNA/miRNA/mRNA axis in diabetic cataract. This study was performed for the exploration of circRNA phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (circPAG1) in diabetic cataract. Human lens epithelial cells were treated with high glucose. The quantitative real-time polymerase chain reaction was used for the expression detection of circPAG1, microRNA-211-5p (miR-211-5p), and E2F transcription factor 3 (E2F3). Cell viability and proliferation were detected using Cell Counting Kit-8 assay and EdU assay. Cell apoptosis was analyzed by flow cytometry. The protein levels were measured by Western blot. Oxidative stress was assessed by malondialdehyde, reactive oxygen species, and superoxide dismutase via the corresponding detection kits. The target interaction was validated using the dual-luciferase reporter assay and RNA immunoprecipitation assay. The expression of circPAG1 was downregulated in diabetic cataract patients. The upregulation of circPAG1 could attenuate the high glucose-induced inhibition of cell viability and proliferation but promotion of cell apoptosis and oxidative stress. CircPAG1 served as a miR-211-5p sponge, and the protective role of circPAG1 was partly achieved by sponging miR-211-5p. MiR-211-5p targeted E2F3 and circPAG1 upregulated the E2F3 level by absorbing miR-211-5p. Inhibition of miR-211-5p repressed the high glucose-mediated cell dysfunction by increasing the expression of E2F3. This study clarified that circPAG1 protected human lens epithelial cells from the high glucose-induced cell damages by the mediation of miR-211-5p/E2F3 axis.
Introduction
Diabetes mellitus (DM) is a frequent chronic systemic disease that can lead to the ocular structural changes, and diabetic cataract (DC) is one of the most common ocular complications resulting from DM [Citation1,Citation2]. High glucose (HG) plays a crucial role in the developing process of DC, in which cell apoptosis and oxidative stress result in lens opacity [Citation3]. In addition, the other complications following cataract extraction in diabetic patients remain to be addressed [Citation4]. Understanding the pathological mechanism of DC and discovering therapeutic targets may be beneficial to the treatment of DC.
Circular RNAs (circRNAs) are single-strand RNA molecules characterized by the closed-loop structures from the backsplicing of mRNAs [Citation5]. CircRNAs have been associated with ophthalmologic diseases such as DC, diabetic retinopathy, retinoblastoma, and age-related macular degeneration [Citation6]. Fan et al. have found that circKMT2E was involved in the pathogenesis of DC [Citation7]. Meanwhile, circRNA phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (circPAG1) was identified to be downregulated in lens tissues from DC patients [Citation7]. It has not been reported about the biological role of circPAG1 in DC.
CircRNAs are known to participate in disease progression by serving as the special sponges for microRNAs (miRNAs) to evoke the gene regulation [Citation8]. MicroRNA-211 (miR-211) inhibited the antioxidative capacities of lens epithelial cells in age-related cataract and promoted lens cell apoptosis in DC [Citation9,Citation10]. Gao et al. have stated that miR-378a-5p and miR-630 repressed lens epithelial cell apoptosis in cataract by regulating the expression of E2F transcription factor 3 (E2F3) [Citation11]. The involvement of miR-211-5p and E2F3 in DC is still unclear.
The present study explored the biological roles of circPAG1, miR-211-5p, and E2F3 in DC. In addition, we researched the sponge potential of circPAG1 for miR-211-5p and the regulatory effect of circPAG1 on E2F3 in DC.
Materials and methods
Human lens samples
This research was ratified and supervised by the Ethics Review Committee of Kunming Children’s Hospital. Before the collection of samples, the written informed consent letters have been signed by overall participants. The lens anterior capsule samples were obtained from DC patients (n = 33) at Kunming Children’s Hospital and the normal controls (n = 33) were collected from the eye bank of Kunming Children’s Hospital. These samples were immediately preserved in enough liquid nitrogen until the extraction of RNA or protein.
Cell culture and treatment
Human lens epithelial cell lines SRA01/04 and B-3 were bought from BioVector NTCC Inc. (Beijing, China). SRA01/04 and B-3 cells were cultured in a complete medium that was comprised of Dulbecco’s modified Eagle’s medium (DMEM; Hyclone, Logan, UT, USA), 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA), and 1% Penicillin–Streptomycin (10,000 U/mL; Gibco) in the 5% CO2 incubator at 37°C. The culture medium of SRA01/04 and B-3 cells was added with glucose (Gibco) by a low dose of 5.5 mM (normal glucose, NG) and a high dose of 25 mM (high glucose, HG) for 48 h [Citation3].
Transient transfection
The pCE-RB-Mam (circ-NC) expression vector (RIBOBIO, Guangzhou, China) was inserted with the sequence of circPAG1 to generate the overexpression vector pCE-RB-Mam-circPAG1 (circPAG1). RNAs were also provided by RIBOBIO, including miRNA mimics (miR-211-5p, NC), miRNA inhibitors (anti-miR-211-5p, anti-NC), and small interfering (si) RNAs (si-circPAG1/si-NC; si-E2F3/scramble). SRA01/04 and B-3 cells were, respectively, inoculated into the 96-well plates, and cell transfection was performed in 60%-confluent cells as per the manual book of the Lipofectamine™ 3000 Kit (Invitrogen, Carlsbad, CA, USA). Cell medium was removed at 6 h post-transfection, and then cells were incubated with DMEM medium containing 25 mM glucose (HG) for 48 h.
The quantitative real-time polymerase chain reaction (qRT-PCR) assay
The qRT-PCR was conducted via SYBR® Green Realtime PCR Master Mix (Toyobo, Kita-Ku, Osaka, Japan) after the RNA extraction by Total RNA Extractor (Sangon, Shanghai, China) and the reverse transcription by ReverTra Ace® qPCR RT Kit (Toyoko). The used primers for qRT-PCR in the current study were displayed as follows: circPAG1, 5ʹ-CAAAGGGAATACATTGCCAGA-3ʹ (forward, F) and 5ʹ-TGTGGCCAACTGCTGAAA-3ʹ (reverse, R); PAG1, 5ʹ-CAGACAAGGAGATGTTCAGCCG-3ʹ (F) and 5ʹ-CTGGACTTCCTCGTAATGCTGC-3ʹ (R); miR-211-5p, 5ʹ-GCCGAGTTCCCTTTGTCATC-3ʹ (F) and 5ʹ-CAGTGCAGGGTCCGAGGTAT-3ʹ (R); E2F3, 5ʹ-AGCGGTCATCAGTACCTCTCAG-3ʹ (F) and 5ʹ-TGGTGAGCAGACCAAGAGACGT-3ʹ (R); 18S rRNA, 5ʹ-GCAATTATTCCCCATGAACG-3ʹ (F) and 5ʹ-GGGACTTAATCAACGCAAGC-3ʹ (R); U6, 5ʹ-CTCGCTTCGGCAGCACA-3ʹ (F) and 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ (R). The comparative cycle threshold (2−∆∆Ct) method was used for the relative expression analysis of each molecule [Citation12]. 18S rRNA was applied to normalize the levels of circRNA and mRNA levels, while a small nuclear RNA U6 acted as the internal reference for miRNA. Meanwhile, the localization of circPAG1 was detected via qRT-PCR assay after the isolation of nuclear and cytoplasmic RNA by PARIS™ Kit (Invitrogen). In addition, the stability of circPAG1 and PAG1 was compared by the qRT-PCR detection following the treatment of Ribonuclease R (RNase R; Epicenter Technologies, Madison, WI, USA) in total RNA or Actinomycin D (Selleck, Houston, TX, USA) in cells.
Cell Counting Kit-8 (CCK-8) assay
SRA01/04 and B-3 cells in the 96-well plates were incubated with 10 μL/well CCK-8 solution (Sangon) at 37°C. 1 h later, the measurement of absorbance was immediately carried out at 450 nm under the microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
EdU assay
After transfection in SRA01/04 and B-3 cells for 48 h, cell proliferative ability was examined by EdU Cell Proliferation Kit (Sangon) according to the manufacturer’s instruction book. Cell pictures were acquired by the fluorescence microscope (Olympus, Tokyo, Japan). EdU/Hoechst-merged cells were indicated as EdU-positive cells.
Flow cytometry
Cells at early apoptosis and late apoptosis were labeled by Annexin V-fluorescein isothiocyanate (Annexin V-FITC) and Propidium Iodide (PI). The detection of cell apoptosis in SRA01/04 and B-3 cells was performed via Annexin V-FITC/PI double staining apoptosis detection kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) as per the user guide. Cells were observed and analyzed through the flow cytometer (BD Biosciences, San Diego, CA, USA). The apoptotic rate was defined as the percentage of Annexin V+/PI- and Annexin V+/PI+-stained cells in total cells.
Western blot
E2F3 protein expression and the apoptotic protein levels were examined using the Western blot Detection Kit (Nanjing Jiancheng Bioengineering Institute) referring to the producer’s direction. The primary antibodies against B-cell lymphoma-2 (Bcl-2; ab32124, 1:1000), Bcl-2-associated X (Bax; ab32503, 1:1000), total-caspase 3 (t-caspase 3; ab32351, 1:1000), Cleaved-caspase 3 (C-caspase 3; ab32042, 1:1000), E2F3 (ab152156, 1:1000), glyceraldehyde-phosphate dehydrogenase (GAPDH; ab9485, 1:1000), and Goat Anti-Rabbit IgG H&L (HRP) secondary antibody (ab205718, 1:3000) were provided by Abcam (Cambridge, UK). The protein expression analysis was performed through the ImageJ software (NIH, Bethesda, MD, USA) with GAPDH as the normalized gene.
Oxidative stress assay
Oxidative stress was assessed by detecting the oxidative indicators. The determination of malondialdehyde (MDA) using the Malondialdehyde (MDA) assay kit, reactive oxygen species (ROS) using Reactive Oxygen Species Assay Kit and superoxide dismutase (SOD) by Superoxide Dismutase (SOD) assay kit were, respectively, implemented following the operating manuals supplied by Nanjing Jiancheng Bioengineering Institute.
Dual-luciferase reporter assay
The targets of circPAG1 and miR-211-5p were predicted through the online Starbase software (http://starbase.sysu.edu.cn). The sequences of wild-type (wt) and mutant (mut) circPAG1 and E2F3 3ʹuntranslated region (3ʹUTR) were respectively cloned into the pmirGLO plasmid (Promega, Madison, WI, USA) to construct the novel luciferase plasmids (circPAG1-wt, circPAG1-mut, E2F3-wt 3ʹUTR, E2F3-mut 3ʹUTR). Each luciferase plasmid and NC or miR-211-5p were co-transfected into SRA01/04 and B-3 cells. 48 h later, cells were harvested for the detection of relative luciferase activity using the Luc-Pair™ Duo-Luciferase HS Assay Kit (GeneCopoeia, Rockville, MA, USA).
RNA immunoprecipitation (RIP) assay
RNA Immunoprecipitation Kit (GENESEED, Guangzhou, China) was performed to explore whether miR-211-5p combined with circPAG1 or E2F3. In brief, cells were incubated with the antibody-coated magnetic beads overnight. RNA complexes were isolated from the magnetic beads, and then the expression detection was conducted using qRT-PCR. Anti-immunoglobulin G (Anti-IgG) was used as the control group for Anti-Argonaute-2 (Anti-Ago2).
Statistical analysis
The data were collected from three independent experiments, and they were expressed as the mean ± standard deviation (SD). The relationship analysis was performed using Pearson’s correlation coefficient, and the statistical analysis was carried out by SPSS 22.0 (SPSS Inc., Chicago, IL, USA). As for the statistical difference, we used Student’s t-test and one-way analysis of variance (ANOVA) followed by Tukey’s test to compare the difference between two groups and multiple groups. It was considered as a significant difference when P< 0.05.
Results
CircPAG1 was signally downregulated in DC patients
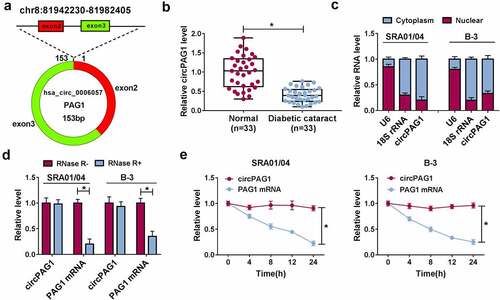
CircPAG1 (hsa_circ_0006057) is derived from exon2 and exon3 of PAG1 gene, and its back-spliced length is 153 bp (). The qRT-PCR analysis demonstrated that circPAG1 level was lower in DC lens samples than that in normal lens samples (). Through the expression detection of U6, 18S rRNA and circPAG1 in the cytoplasm and nucleus, circPAG1 was affirmed to be localized in the cytoplasm of SRA01/04 and B-3 cells (). PAG1 mRNA level was reduced in RNase R+ group compared with RNase R-group, while circPAG1 exhibited high resistance to RNase R (). CircPAG1 expression was also unaffected after the treatment of Actinomycin D, but PAG1 mRNA expression was rapidly decreased (). Therefore, circPAG1 was dysregulated in DC patients and it was stably expressed in human lens epithelial cells.
Figure 1. CircPAG1 was signally downregulated in DC patients. (a) The genic information of circPAG1. (b) The circPAG1 level was assayed using qRT-PCR in normal and DC lens samples. (c) The localization analysis was performed by the qRT-PCR detection in cytoplasm and nucleus of SRA01/04 and B-3 cells. (d-e) The stability of circPAG1 was assessed by the qRT-PCR analysis after RNase R digestion in RNA (d) and Actinomycin D treatment in cells (e). *P< 0.05.
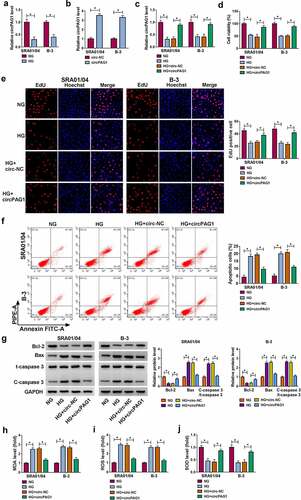
Overexpression of circPAG1 relieved the HG-induced apoptotic and oxidative damages
SRA01/04 and B-3 cells were treated with NG or HG for 48 h. HG was shown to inhibit the expression of circPAG1 relative to NG group (). The stimulative effect of circPAG1 transfection on the circPAG1 expression was great in SRA01/04 and B-3 cells (). Meanwhile, circPAG1 transfection could eliminate the HG-mediated downregulation of circPAG1 level (). CCK-8 assay and EdU assay have showed that HG treatment repressed cell viability () and cell proliferation () in SRA01/04 and B-3 cells, then the introduction of circPAG1 partly mitigated these effects. After the analysis of flow cytometry, we found that the promotion of cell apoptosis by HG was similarly abolished by transfection of circPAG1 (). Furthermore, Western blot was used to detect the protein levels of apoptotic markers. The results manifested that the overexpression of circPAG1 neutralized the HG-triggered Bcl-2 expression inhibition and Bax or C-caspase 3/t-caspase 3 expression upregulation (). HG treatment increased the MDA () and ROS () levels but reduced the SOD level () in SRA01/04 and B-3 cells, whereas the oxidative stress caused by HG was alleviated by upregulation of circPAG1. Altogether, circPAG1 overexpression lightened the HG-induced cell damages in lens epithelial cells.
Figure 2. Overexpression of circPAG1 relieved the HG-induced apoptotic and oxidative damages. (a) The expression level of circPAG1 was quantified by qRT-PCR in NG or HG-treated SRA01/04 and B-3 cells. (b-c) The overexpression efficiency of circPAG1 was evaluated by qRT-PCR in SRA01/04 and B-3 cells (b), as well as in HG-treated cells (c). (d-e) The examination of cell viability (d) and cell proliferation (e) was performed by CCK-8 assay and EdU assay in NG, HG, HG+circ-NC and HG+circPAG1 groups. (f-g) The measurement of cell apoptosis was performed by flow cytometry (f) and Western blot (g). (h-j) The assessment of oxidative stress was performed by the detection of MDA (h), ROS (i) and SOD (j) via the matched kits. *P< 0.05.
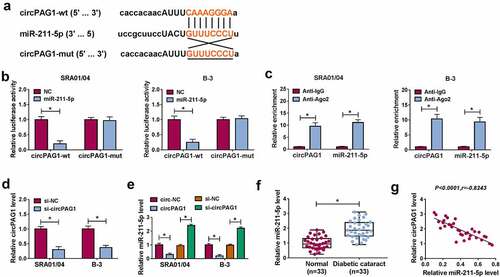
CircPAG1 could interact with miR-211-5p
The target prediction by Starbase revealed that circPAG1 sequence contained the binding sites for miR-211-5p (). The interaction between circPAG1 and miR-211-5p was affirmed using the dual-luciferase reporter assay. The luciferase activity was markedly suppressed by the co-transfection of circPAG1-wt and miR-211-5p contrasted to the co-transfection of circPAG1-wt and NC, while no difference was noticed in circPAG1-mut plasmid after the transfection of NC or miR-211-5p (). In addition, RIP assay showed the enrichment of circPAG1 and miR-211-5p by Ago2 protein in comparison with IgG protein (). The qRT-PCR indicated that si-circPAG1 transfection knocked down the expression of circPAG1 in SRA01/04 and B-3 cells (). The overexpression of circPAG1 evoked the expression downregulation of miR-211-5p, but knockdown of circPAG1 induced an opposite influence on the miR-211-5p level (). Relative to the normal lens samples, miR-211-5p was highly expressed in DC lens samples (). After the linear analysis by Pearson’s correlation coefficient, we observed a negative relationship (P< 0.0001, r= −0.8243) between the expression levels of miR-211-5p and circPAG1 in DC lens samples (). These data could identify that circPAG1 directly targeted miR-211-5p.
Figure 3. CircPAG1 could interact with miR-211-5p. (a) The online starbase software predicted the binding sites between circPAG1 and miR-211-5p. (b-c) Dual-luciferase reporter assay (b) and RIP assay (c) were used to analyze the combination between circPAG1 and miR-211-5p in SRA01/04 and B-3 cells. (d) The qRT-PCR was used to detect the circPAG1 expression after the respective transfection of si-NC and si-circPAG1. (e) The qRT-PCR was used to assess the effect of circPAG1 upregulation or downregulation on the miR-211-5p level. (f) The qRT-PCR was used for the expression quantification of miR-211-5p in normal and DC lens samples. (g) Pearson’s correlation coefficient was used for the linear analysis between circPAG1 and miR-211-5p in DC samples. *P< 0.05.
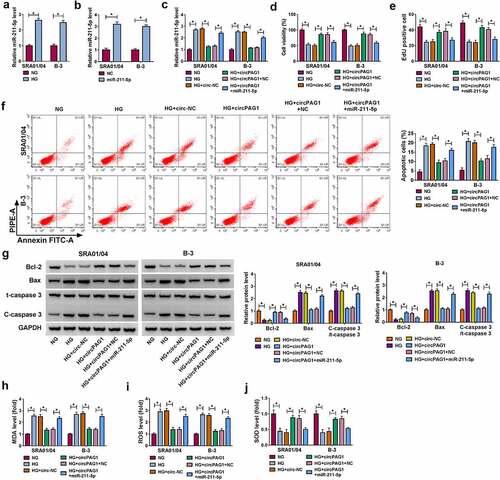
CircPAG1 inhibited the destructive effects of HG on human lens epithelial cells by sponging miR-211-5p
The expression of miR-211-5p was higher in HG-treated SRA01/04 and B-3 cells than that in NG-treated cells (). The miR-211-5p mimic markedly promoted the miR-211-5p level in SRA01/04 and B-3 cells (), and it could abrogate the inhibitory regulation of circPAG1 on the miR-211-5p expression in HG-treated cells (). The overexpression of miR-211-5p overtly abolished the influences of circPAG1 on cell viability repression (), cell proliferation inhibition () and cell apoptosis enhancement () in HG-treated SRA01/04 and B-3 cells. Also, the circPAG1-mediated suppression of oxidative stress under the HG treatment was attenuated by the transfection of miR-211-5p (). CircPAG1 could act as a sponge of miR-211-5p to inhibit the progression of DC in vitro.
Figure 4. CircPAG1 inhibited the destructive effects of HG on human lens epithelial cells by sponging miR-211-5p. (a-b) The miR-211-5p expression analysis was conducted using qRT-PCR after NG or HG treatment (a) and NC or miR-211-5p transfection (b) in SRA01/04 and B-3 cells. (c) The level of miR-211-5p was detected using qRT-PCR in NG, HG, HG+circ-NC, HG+circPAG1, HG+circPAG1+ NC and HG+circPAG1+ miR-211-5p groups. (d-e) CCK-8 assay and EdU assay were respectively applied to determine cell viability (d) and proliferation (e). (f-g) Flow cytometry (f) and Western blot (g) were applied to assess cell apoptosis. (h-j) MDA (h), ROS (i) and SOD (j) levels by the corresponding kits were applied to evaluate oxidative stress. *P< 0.05.
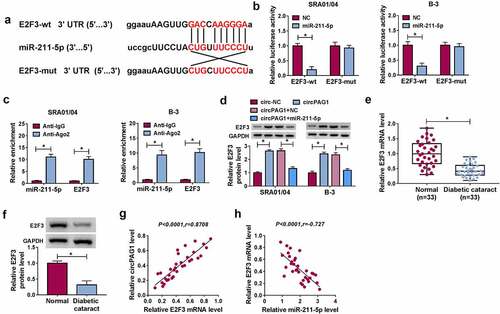
CircPAG1 upregulated the expression of E2F3 by sponging miR-211-5p
The miR-211-5p binding sites were found in the 3ʹUTR sequence of E2F3 as shown in . Dual-luciferase reporter assay indicated that miR-211-5p overexpression exerted an inhibitory effect on the luciferase signal of E2F3-wt group, but it did not affect the luciferase signal of E2F3-mut group (). The binding potential between miR-211-5p and E2F3 was also validated by RIP assay (). Moreover, E2F3 protein expression was elevated after the overexpression of circPAG1, while miR-211-5p inhibited the positive effect of circPAG1 on E2F3 expression (). The mRNA and protein levels of E2F3 were decreased in DC lens samples by comparison with the normal lens samples (). There was a positive correlation (P< 0.0001, r= 0.8708) between E2F3 and circPAG1 () but a negative association (P< 0.0001, r= −0.727) between E2F3 and miR-211-5p () in DC samples. The above findings suggested that circPAG1 regulated the E2F3 expression by sponging miR-211-5p.
Figure 5. CircPAG1 upregulated the expression of E2F3 by sponging miR-211-5p. (a) The binding sites between miR-211-5p and E2F3 3ʹUTR were shown by starbase. (b-c) The binding analysis between E2F3 and miR-211-5p was carried out by dual-luciferase reporter assay (b) and RIP assay (c). (d) Western blot was performed for detecting the protein expression of E2F3 after SRA01/04 and B-3 cells were transfected with circ-NC, circPAG1, circPAG1+ NC or circPAG1+ miR-211-5p. (e-f) The qRT-PCR and Western blot were conducted for the expression detection of E2F3 in normal and DC lens samples. (g-h) The linear relationship between E2F3 and circPAG1 (g) or miR-211-5p (h) was analyzed using Pearson’s correlation coefficient. *P< 0.05.
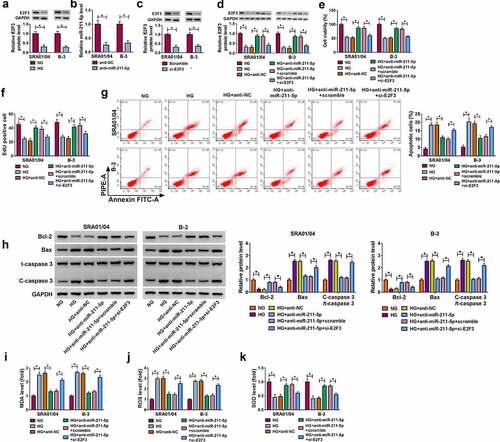
MiR-211-5p/E2F3 axis contributed to the HG-regulated cell dysfunction in human lens epithelial cells
Western blot assay exhibited that E2F3 protein level was downregulated in HG group in contrast to the NG group (). The inhibitory effects of anti-miR-211-5p on the miR-211-5p expression () and si-E2F3 on the E2F3 expression () were verified to be significant in SRA01/04 and B-3 cells by qRT-PCR and Western blot. In HG-treated SRA01/04 and B-3 cells, si-E2F3 reverted the protein expression upregulation of E2F3 induced by anti-miR-211-5p (). The expression inhibition of miR-211-5p promoted cell viability () and cell proliferation () in HG-treated SRA01/04 and B-3 cells, which were overtly countervailed by the knockdown of E2F3. The experimental results of flow cytometry () and Western blot () manifested that the transfection of si-E2F3 prevented the alleviation of anti-miR-211-5p to the HG-promoted cell apoptosis. As for oxidative stress, the miR-211-5p inhibitor rescued the influences of HG on the levels of MDA (), ROS () and SOD () by promoting the expression of E2F3. Taken together, miR-211-5p could aggravate the HG-induced cell dysfunction by targeting E2F3.
Figure 6. MiR-211-5p/E2F3 axis contributed to the HG-regulated cell dysfunction in human lens epithelial cells. (a) E2F3 protein analysis was performed by Western blot after SRA01/04 and B-3 cells were treated with NG or HG. (b-c) The inhibitory efficiency of anti-miR-211-5p or si-E2F3 was evaluated using qRT-PCR or Western blot. (d) The protein level of E2F3 was assayed by Western blot in NG, HG, HG+anti-NC, HG+anti-miR-211-5p, HG+anti-miR-211-5p+scramble and HG+anti-miR-211-5p+si-E2F3 groups. (e-f) Cell viability (e) and proliferation (f) were respectively examined via CCK-8 assay and EdU assay. (g-h) Cell apoptosis was measured via flow cytometry (g) and Western blot (h). (i-k) Oxidative stress was analyzed using the levels of MDA (i), ROS (j) and SOD (k) levels by the detection kits. *P< 0.05.
Discussion
It is necessary to investigate the molecular evolution of DC and develop novel biological targets for DC therapy. Herein, our experimental results have shown that circPAG1 alleviates the HG-induced lens epithelial cell dysfunction by depending upon the regulation of miR-211-5p/E2F3 axis.
CircPAG1 is an exonic circRNA derived from PAG1 gene. The exonic circRNAs are mainly localized in the cytoplasm of eukaryotes [Citation13]. The localization analysis indicated that circPAG1 was enriched in the cytoplasm of human lens epithelial cells. CircRNAs are highly stable due to the closed-loop structures [Citation14]. Our stability assays revealed that circPAG1 was resistant to RNase R and Actinomycin D in comparison with the linear PAG1. The issued studies have exhibited the pivotal roles of circRNAs in cataract. For example, circHIPK3 accelerated cell apoptosis and reduced cell viability in age-related cataract by regulating the miR-193a-mediated CRYAA level [Citation15]. CircMRE11A_013 aggravated lens cell senescence by binding to UBXN1 to enhance the activity of ATM [Citation16]. Liang et al. declared that circZNF292 repressed the apoptotic and oxidative damages by sponging miR-23b-3p in age-related cataract [Citation17]. In this study, circPAG1 was found to be downregulated in DC lens samples and HG-treated lens epithelial cells. After circPAG1 was overexpressed, cell viability and proliferation were promoted, while cell apoptosis and oxidative stress were inhibited in HG-treated cells. Thus, circPAG1 was considered as an inhibitory regulator in the cataractogenesis of DC.
Then, miR-211-5p was identified as a miRNA target for circPAG1, and all those effects of circPAG1 on the biological processes of HG-treated lens cells were suppressed by miR-211-5p overexpression. Many reports indicated that circRNAs functioned as pathogenic regulators by the sponge function of miRNAs. Qiu et al. found that circADAMTS13 restrained cell proliferation in hepatocellular carcinoma via inhibiting the miR-484 activity [Citation18]. Feng et al. asserted that circDLGAP4 protected against Parkinson’s disease by sponging miR-134-5p [Citation19]. The regulation of circKMT2E in DC was also attributed to the miR-204-5p sponge function [Citation7]. Consistently, our findings demonstrated that circPAG1 might inhibit the progression of DC by acting as a natural miR-211-5p sponge.
Furthermore, we found the repressive expression regulation of miR-211-5p on E2F3 by directly interacting with the 3ʹUTR of E2F3. The functional assays have manifested that the knockdown of E2F3 lightens the protective effects of miR-211-5p inhibitor on the HG-treated lens cells. A handful of miRNAs have also been involved in the cataract by targeting the downstream genes. For instance, miR-199a-5p and miR-30a affected the process of epithelial-to-mesenchymal transition in DC by targeting SP1 and SNAI1, respectively, [Citation20,Citation21]. Lens cell proliferation and apoptosis in DC were regulated by miR-214-3p/MMP2 axis [Citation22]. In the current study, it was obvious that miR-211-5p facilitated the DC progression by targeting E2F3.
Moreover, our expression analysis has confirmed that circPAG1 can elevate the expression of E2F3 by sponging miR-211-5p. Increasing circRNA/miRNA/mRNA axes have been reported in different diabetic complications. For instance, the silence of circDMNT3B exacerbated the diabetic retinal vascular dysfunction via regulating the BAMBI expression by targeting miR-20b-5p [Citation23]. CircRNA_0084043 enhanced the HG-induced damages in diabetic retinopathy via the miR-140-3p-mediated TGFA expression change [Citation24]. Circ_010383 promoted renal fibrosis in diabetic nephropathy by working as a miR-135a sponge to induce the inhibition of TRPC1 expression [Citation25]. Overall, the protective role of circPAG1 for lens epithelial cells against DC was also partly attributed to the regulation of miR-211-5p/E2F3 axis.
Conclusion
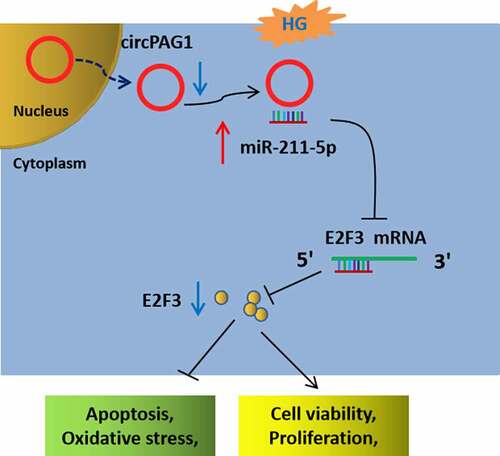
Our results indicated that circPAG1 downregulation reduced the transcription and translation of E2F3 by releasing miR-211-5p in the cytoplasm, thus promoting the HG-induced inhibitory effects on cell viability and proliferation, but also the stimulative effects on cell apoptosis and oxidative stress in human lens epithelial cells (). This study confirmed the involvement of circPAG1/miR-211-5p/E2F3 axis in HG-induced cell damages in DC. More importantly, circPAG1 could be used as a therapeutic biomarker for DC.
Ethical approval
The research related to human use has been complied with all the relevant national regulations, institutional policies and in accordance with the tenets of the Helsinki Declaration, and has been approved the Ethics Review Committee of Kunming Children’s Hospital
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kiziltoprak H, Tekin K, Inanc M, et al. Cataract in diabetes mellitus. World J Diabetes. 2019;10(3):140–153.
- Peterson SR, Silva PA, Murtha TJ, et al. Cataract surgery in patients with diabetes: management strategies. Semin Ophthalmol. 2018;33(1):75–82.
- Gong W, Zhu G, Li J, et al. LncRNA MALAT1 promotes the apoptosis and oxidative stress of human lens epithelial cells via p38MAPK pathway in diabetic cataract. Diabetes Res Clin Pract. 2018;144:314–321.
- Haddad NM, Sun JK, Abujaber S, et al. Cataract surgery and its complications in diabetic patients. Semin Ophthalmol. 2014;29(5–6):329–337.
- Eger N, Schoppe L, Schuster S, et al. Circular RNA Splicing. Adv Exp Med Biol. 2018;1087:41–52.
- Zhang C, Hu J, Yu Y. CircRNA is a rising star in researches of ocular diseases. Front Cell Dev Biol. 2020;8:850.
- Fan C, Liu X, Li W, et al. Circular RNA circ KMT2E is up-regulated in diabetic cataract lenses and is associated with miR-204-5p sponge function. Gene. 2019;710:170–177.
- Tang Q, Hann SS. Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Ther. 2020;13:2067–2092.
- Lu B, Christensen IT, Ma LW, et al. miR-211 regulates the antioxidant function of lens epithelial cells affected by age-related cataracts. Int J Ophthalmol. 2018;11(3):349–353.
- Zeng K, Feng QG, Lin BT, et al. Effects of microRNA-211 on proliferation and apoptosis of lens epithelial cells by targeting SIRT1 gene in diabetic cataract mice. Biosci Rep. 2017;37(4). DOI:10.1042/BSR20170695.
- Gao W, Zhou X, Lin R. miR-378a-5p and miR-630 induce lens epithelial cell apoptosis in cataract via suppression of E2F3. Braz J Med Biol Res. 2020;53(5):e9608.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408.
- Zhou WY, Cai ZR, Liu J, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172.
- Cai H, Li Y, Niringiyumukiza JD, et al. Circular RNA involvement in aging: an emerging player with great potential. Mech Ageing Dev. 2019;178:16–24.
- Liu X, Liu B, Zhou M, et al. Circular RNA HIPK3 regulates human lens epithelial cells proliferation and apoptosis by targeting the miR-193a/CRYAA axis. Biochem Biophys Res Commun. 2018;503(4):2277–2285.
- Liu J, Zhang J, and Zhang G, et al. CircMRE11A_013 binds to UBXN1 and integrates ATM activation enhancing lens epithelial cells senescence in age-related cataract. Aging (Albany NY). 2021;13(4):5383-5402.
- Liang S, Dou S, Li W, et al. Profiling of circular RNAs in age-related cataract reveals circZNF292 as an antioxidant by sponging miR-23b-3p. Aging (Albany NY). 2020;12(17):17271–17287.
- Qiu L, Huang Y, Li Z, et al. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol Oncol. 2019;13(2):441–455.
- Feng Z, Zhang L, Wang S, et al. Circular RNA circDLGAP4 exerts neuroprotective effects via modulating miR-134-5p/CREB pathway in Parkinson’s disease. Biochem Biophys Res Commun. 2020;522(2):388–394.
- Liu X, Gong Q, Yang L, et al. microRNA-199a-5p regulates epithelial-to-mesenchymal transition in diabetic cataract by targeting SP1 gene. Mol Med. 2020;26(1):122.
- Zhang L, Wang Y, Li W, et al. MicroRNA-30a regulation of epithelial-mesenchymal transition in diabetic cataracts through targeting SNAI1. Sci Rep. 2017;7(1):1117.
- Yang J, Zhao S, Tian F. SP1-mediated lncRNA PVT1 modulates the proliferation and apoptosis of lens epithelial cells in diabetic cataract via miR-214-3p/MMP2 axis. J Cell Mol Med. 2020;24(1):554–561.
- Zhu K, Hu X, Chen H, et al. Downregulation of circRNA DMNT3B contributes to diabetic retinal vascular dysfunction through targeting miR-20b-5p and BAMBI. EBioMedicine. 2019;49:341–353.
- Li Y, Cheng T, Wan C, et al. circRNA_0084043 contributes to the progression of diabetic retinopathy via sponging miR-140-3p and inducing TGFA gene expression in retinal pigment epithelial cells. Gene. 2020;747:144653.
- Peng F, Gong W, Li S, et al. circRNA_010383 acts as a sponge for miR-135a, and its downregulated expression contributes to renal fibrosis in diabetic nephropathy. Diabetes. 2021;70(2):603–615.