ABSTRACT
Gastric cancer (GC) is one of the most common malignant tumors. Circular RNA (circRNA) has been shown to be involved in the progression of GC. However, the function of circ_0008035 in GC has not been studied. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect the expression of circ_0008035, microRNA-1256 (miR-1256) and carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6). 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay, 5-ethynyl-2’-deoxyuridine (EdU) assay, flow cytometry, and transwell assay were used to detect cell function. Western blot examined the protein levels of Ki67, Bax, MMP-2, and CEACAM6. The relationship between miR-1256 and circ_0008035 or CEACAM6 was verified by dual-luciferase reporter assays and RNA pull down. The xenotransplantation model was established in BALB/c nude mice to study the role of circ_0008035 in vivo. Circ_0008035 and CEACAM6 were significantly high-expressed in GC tissues and cells. Silencing of circ_0008035 reduced GC cell proliferation, migration, and invasion while enhancing apoptosis. MiR-1256 was a target of circ_0008035. The inhibition effect of circ_0008035 knockdown on the malignant behavior of GC cells could be reversed by miR-1256 inhibitor. In addition, CEACAM6 was a target of miR-1256. Overexpression of CEACAM6 partially restored the inhibitory effect of miR-1256 on cell progression. Animal experiments confirmed the anti-tumor effect of circ_0008035 knockdown in vivo. Collectively, circ_0008035 regulated the expression of CEACAM6 by sponging miR-1256, thereby promoting the development of GC. Our data provided a novel targeted therapy for GC.
Introduction
Gastric cancer (GC), as one of the most common gastrointestinal malignancies worldwide, ranks the second in the mortality rate of cancer-related diseases [Citation1–3]. There are many factors that induce GC, mainly due to helicobacter pylori infection. Meanwhile, stress and dietary habits also make the incidence of GC to become younger [Citation4–6]. Although some progress has been made in the diagnosis and treatment of GC, the 5-year survival rate of patients with advanced GC is low [Citation7]. Therefore, it is very necessary to find new and effective therapeutic targets of GC.
Circular RNAs (circRNAs) are non-coding RNAs with covalently closed continuous cycles, lacking 3’ and 5’ termini [Citation8,Citation9]. CircRNAs are involved in various cancer diseases, including GC. In addition, circRNAs sponge microRNAs (miRNAs) and affect gene expression at the post-transcriptional level [Citation10,Citation11]. For instance, Yue et al. established that circ_0004104 worked as a ceRNA for miR-539-3p to contribute to the development of GC [Citation12]. Chen et al. uncovered that circ_ASAP2 exerted a strong oncogenic activity in GC via inducing CDK6 through miR-770-5p competition [Citation13]. Similarly, circ_0008035 enhanced the proliferation of GC cells [Citation14]. Therefore, we believed that circ_0008035 might be a key circRNA regulating GC progression, and further studies on its role and new mechanisms in GC progression were worth continuing.
MicroRNAs (miRNAs), which negatively regulate target genes by interacting with the 3ʹuntranslated region (3ʹUTR), are also 22-nucleotide non-coding RNAs [Citation15,Citation16]. MiRNAs can be involved in the growth process of various cancer cells [Citation17,Citation18]. Recently, a large number of literatures have reported the regulatory impact of miRNA in the occurrence and progression of GC. For example, Zheng et al. showed that miR-146b-5p as an anticancer factor regulated the progression of GC [Citation19]. Similarly, it had been found that miR-96-5p might promote the proliferation of GC cells [Citation20]. In a preliminary survey for targeted miRNAs of circ_0008035 using target prediction program circinteractome, we observed a putative miR-1256 binding region within circ_0008035. In previous studies, miR-1256 plays a pro-cancer or anti-cancer role in a variety of cancers, such as non-small cell lung cancer (NSCLC), papillary thyroid cancer, and GC [Citation21–23]. MiRNAs perform their functions by binding to the targeted mRNA 3ʹuntranslated region (3ʹUTR) [Citation24]. It is well known that carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) is an oncogene [Citation25]. Studies have shown that CEACAM6 is taken part in various cancer progress and mainly regulates cell proliferation, such as breast cancer, gallbladder cancer, and GC [Citation26–28].
Here, we investigated the impact of circ_0008035 in GC by silencing circ_0008035. In addition, the effect of circ_0008035/miR-1256/CEACAM6 axis on GC proliferation, metastasis, and apoptosis was explored. The results showed that circ_0008035 could promote the malignant behavior of GC, confirming that it might be a new target for the treatment of GC.
Materials and methods
Clinical tissue samples
The study was audited by the Ethics Committee of the First Affiliated Hospital and College of Clinical Medicine of Henan University of Science and Technology. 30 cases of GC tumor tissue samples and paracancerous tissue samples were gathered from 30 GC patients at the First Affiliated Hospital and College of Clinical Medicine of Henan University of Science and Technology. All patients have written informed consent. Subsequently, the samples were frozen and preserved at −80°C for use. All GC patients signed written informed consent.
Cell lines and cell culture
Human gastric mucosal epithelial cells (GES-1) and GC cell lines (AGS and N87) were purchased from American-type culture specimens (ATCC; Manassas, VA, USA). All of these cells were cultured in Dulbecco’s modified eagle medium (DMEM; Thermo Fisher, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) and 1% streptomycin and penicillin (Invitrogen, Carlsbad, CA, USA) at 37°C in 5% CO2 environment.
Cell Transfection
Short hairpin RNA (shRNA) targeting circ_0008035 (sh-circ_0008035#1 and sh-circ_0008035#2) and its corresponding control group (sh-NC), miR-1256 inhibitor (anti-miR-1256) and its matched (anti-NC), miR-1256 overexpression (miR-1256) and its matched (miR-NC), CEACAM6 overexpression vector (CEACAM6) and matched control (pcDNA) were synthesized by Ribobio Co., Ltd. (Guangzhou, China). There were transfected into AGS and N87 cells using Lipofectamine 2000 (Thermo Fisher).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Trizol Reagent (Thermo Fisher) was used to separate total RNA. The quality of RNA was detected by NanoDrop ND-1000 spectrophotometer (Thermo Fisher). The total RNA was reverse transcribed using Reverse Transcription Kit (Thermo Fisher). Then, qRT-PCR was performed using SYBR Green qRT-PCR Mix (Takara, Shiga, Japan). The amplification process was as follows: 95°C for 30 sec; followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec; and 1 cycle of 95°C for 5 sec, 60°C for 60 sec; lastly, 1 cycle of 50°C for 30 sec. GAPDH or U6 was used as internal reference, and the relative expression was calculated by 2−ΔΔCT method. Primer sequences are listed in .
Table 1. Primer sequences for qRT-PCR
Cellular distribution analysis
RNA was extracted from the cytoplasmic and nuclear sections using Paris kits (Thermo Fisher). Using U6 or GAPDH as internal reference, qRT-PCR was used to detect the circ_0008035 level in each part of cytoplasm or nuclear.
CircRNA validation
Oligo (dT)18 primers and Random primers were added to verify whether circ_0008035 was a circRNA. QRT-PCR was used to measure the corresponding expression level.
3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay
AGS and N87 cells were incubated in 96-well plates for 24, 48, or 72 h. 10 μL MTT (Sigma, St. Louis, MO, USA) was added to each well for 4 h, and 100 μL dimethyl sulfoxide (DMSO; Sigma) was added to dissolve crystal violet precipitates. Finally, the optical density (OD) value of cells was recorded at 570 nm using a microplate reader.
5-ethynyl-2’-deoxyuridine (EdU) assay
The transfected AGS and N87 cells (2 × 104/mL) were seeded in 200 μL medium supplemented with 10% FBS, and then treated with 50 μM the Cell-Light EdU DNA Cell Proliferation Kit (Ribobio) for 24 h and 37°C for 2 h. The images were obtained using a fluorescence microscope (Leica, Wetzlar, Germany) after staining with a mixture of Apollo staining solution and DAPI.
Flow cytometry assay
Flow cytometry (BD Biosciences, San Jose, CA, USA) was used to detect cell apoptosis and cell cycle. For detecting cell apoptosis, the transfected cells were incubated with Annexin-FITC and Propidium iodide (PI; BD Biosciences), and then cell apoptosis was analyzed. For detecting cell cycle, the transfected cells were incubated with PI/TritonX-100 staining solution (containing 0.2 mg RNase A, 20 μg of PI, and 0.1% TritonX-100) at 4°C for 30 min, and then cell cycle was analyzed.
Transwell assay
Transwell chambers coated with or without Matrigel (Corning, New York, Madison, USA) were used for the ability of GC cells invasion or migration, respectively. 5 × 104 cells were added into the upper of Transwell chambers. Meanwhile, the Transwell lower chambers were added with fresh culture medium containing 10% FBS. The amounts of invaded and migration cells were counted under a high-powered microscope.
Western blot analysis
The total proteins of GC tissues or cells were separated from using Radioimmunoprecipitation analysis buffer (RIPA; Thermo Fisher). Protein was loaded into 12% SDS-PAGE, and then transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA) by electrophoresis. After blocking the membrane with 5% skim milk, anti-β-actin (1:1,000, ab8226, Abcam, Cambridge, MA, USA), anti-Bax (1:200, ab32503, Abcam), anti-matrix metallopeptidase 2 (MMP-2; 1:1,000, ab92536, Abcam), anti-Ki67 (1:1,000, ab92742, Abcam), and anti-CEACAM6 (1:1,000, ab78029, Abcam) were used to incubate with the membrane, respectively. After incubated with secondary antibody, protein signal on the membrane was visualized using BeyoECL Plus kit (Beyotime, Shanghai, China).
Dual-luciferase reporter assay
The wild type (WT) or mutant type (MUT) sequences of circ_0008035 or CEACAM6 3ʹUTR containing presumed miR-1256 interacting sites were cloned into pGL3-basic vectors (Realgene, Nanjing, China), respectively. AGS and N87 cells in 48-well plates (8 × 103 cells/well) were transfected with luciferase reporter plasmid in combination with miR-1256 mimic or miR-NC by Lipofectamine 2000 (Thermo Fisher). The relative luciferase activities were measured by Dual-Luciferase Reporter Assay Kit (GeneCopoeia, Rockville, MD, USA).
Immunohistochemistry (IHC)
Resected tissues were fixed with 4% buffered paraformaldehyde, dehydrated and embedded in paraffin. Paraffin sections (5 μM) were dewaxed and rehydrated for antigen stripping. Then anti-Ki-67 (1:200, ab92742; Abcam) and anti-CEACAM6 (1:100, ab78029, Abcam) was placed at 4°C overnight, and matched with the secondary antibody. Sections were stained with diaminobenzidine (DAB) kit (Sigma) according to protocol. The positive staining was observed with a light microscope.
RNA Pull-down assay
Biotinylated-miR-NC (Bio-miR-NC) and Bio-miR-1256 probes were provided by GENESEED (Guangzhou, China). GC cells were transfected with Bio-miR-NC and Bio-miR-1256 for 48 h. Cells were lyzed using Pierce™ Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher) and then incubated with streptavidin-coated magnetic beads. RNA samples were purified from beads and used for qRT-PCR analysis.
Xenograft models
All animal experiments were approved by the Animal Ethics Committee of the First Affiliated Hospital of Henan University of Science and Technology and the School of Clinical Medicine. Twelve 8-week-old female BALB/c nude mice were purchased from Vital River (Beijing, China) and randomly divided into two groups. The mice were raised in SPF animal laboratory. AGS cells transfected with sh-NC or sh-circ_0008035 were injected subcutaneously into the mice. Tumor volume was measured once a week after the intervention (volume formula: volume = length×width2/2). Mice were executed by neck break on day 35 after the intervention, and the tumors were removed and weighed. The expression of circ_0008035 and miR-1256 were analyzed by qRT-PCR, and the protein expression of CEACAM6 was analyzed by Western blot. IHC detected the change of Ki67, Bax and MMP-2 positive cells in mice tumor tissues.
Statistical analysis
GraphPad Prism 7 (GraphPad, San Diego, CA, USA) was utilized for data analysis. Difference comparison between groups was determined using Student’s t-test or ANOVA. Pearson’s correlation analysis was applied to measure the correlation between two groups. Each experiment was carried out at least three times and the final data were shown as the mean ± standard deviation (SD). Using P < 0.05 meant significant difference.
Results
Circ_0008035 was highly expressed in GC tissues and cells
Analysis of the GSE78092 database showed that circ_0008035 was highly expressed in GC tissues (). Here, qRT-PCR showed that circ_0008035 was significantly upregulated in GC tissues and cells (). In addition, Oligo (dT)18 primers were only able to pair with mRNA, while they had no amplification effect on circ_0008035 (). Finally, we isolated RNAs from the cytoplasm and nucleus of AGS and N87 cells and found that circ_0008035 was mainly distributed in the cytoplasm relative to the nucleus (). In a word, circ_0008035 was up-regulated circRNA in GC.
Figure 1. Circ_0008035 was highly expressed in GC tissues and cells. (a) CircRNA expression profile of GSE78092. (b) Hsa_circ_0008035 in GSE78092 database. (c, d) The expression of circ_0008035 was detected by qRT-PCR. (e, f) Oligo (dT)18 primers was used to verify circRNAs. (g, h) The distribution of circ_0008035 in nucleus or cytoplasm was determined by qRT-PCR.
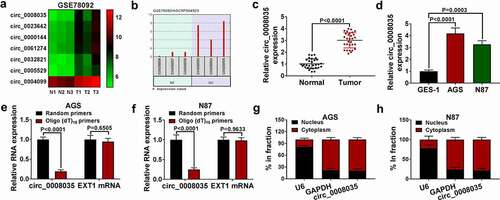
Circ_0008035 silencing inhibited the malignant behavior of GC cells
Next, we verified the role of circ_0008035 in GC cells by knocking down circ_0008035. The transfected efficiency of circ_0008035 was detected by qRT-PCR, and the results showed that the knockdown efficiency of sh-circ_0008035#1 was better (). Functionally, sh-circ_0008035#1 notably decreased the cell viability and proliferation of AGS and N87 cells (). The cell cycles in AGS and N87 cells were arrested by silencing circ_0008035 (). Flow cytometry showed that sh-circ_0008035#1 significantly promoted the apoptosis of AGS and N87 cells (). Mechanically, knockdown of circ_0008035 significantly reduced the number of migration and invasion in AGS and N87 cells (). Finally, Western blot showed that sh-circ_0008035#1 remarkably decreased the levels of Ki67 and MMP-2, and increased the expression of Bax (). Taken together, knockdown of circ_0008035 inhibited the cell growth and metastasis of GC cells.
Figure 2. Circ_0008035 silencing inhibited the malignant behavior of GC cells. (a, b) The expression of circ_0008035 and EXT1 mRNA was detected by qRT-PCR. (c, d) MTT was used to detect cell viability. (e) Cell proliferation was measured by EdU assay. (f, g) The cell cycle was detected by flow cytometry. (h) The cell apoptosis was detected by flow cytometry. (i, j) Transwell was used to detect the cell migration and invasion. (k, l) Western blot was used to detect the expression of relative proteins.
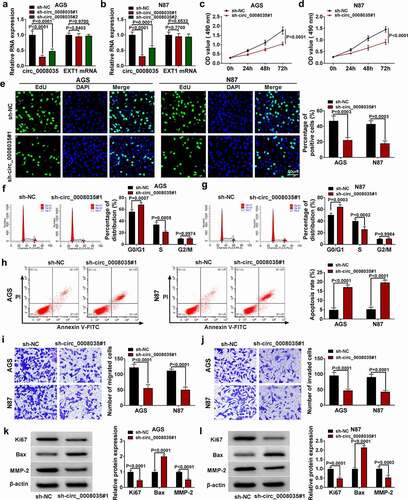
MiR-1256 was a target of circ_0008035
As shown in , we predicted that circ_0008035 could sponge eight miRNAs, qRT-PCR detected that miR-1256, miR-647, and miR-508-3p were significantly down-regulated in GC tumor tissues. Then, we used the probe to filter and found that miR-1256 was the most suitable one (). Circinteractome (https://circinteractome.nia.nih.gov/mirna_target_sites.html) predicted the binding sites between miR-1256 and circ_0008035 (). The transfected efficiency of miR-1256 mimic is shown in . Dual-luciferase reporter assay results showed that the combined transfection of miR-1256 mimic and circ_0008035 WT vector significantly inhibited luciferase activity, while the combined transfection of miR-1256 mimic and circ_0008035 MUT vector showed no significant change (). And RNA pull down experiment showed that Bio-miR-1256 enriched circ_0008035 in large amount (). The expression of miR-1256 was significantly decreased in GC tissues (). Pearson’s correlation analysis showed that miR-1256 expression was negatively correlated with circ_0008035 expression in GC tumor tissues (). Similarly, qRT-PCR detection showed that the expression of miR-1256 was significantly decreased in AGS and N87 cells (). Lastly, knockdown of circ_0008035 significantly up-regulated the expression of miR-1256, while co-transfection of miR-1256 inhibitor reversed the effect of sh-circ_0008035#1 (). In shorts, miR-1256 could be sponged by circ_0008035.
Figure 3. MiR-1256 was a target of circ_0008035. (a, b) The relative miRNA expression was detected by qRT-PCR. (c) The binding sites between circ_0008035 and miR-1256. (d) QRT-PCR was used to detect the expression of miR-1256. (e, f) Dual-luciferase reporter assay was performed to confirm the association between circ_0008035 and miR-1256. (g) RNA pull down was performed to measure the interaction between circ_0008035 and miR-1256. (h) QRT-PCR was used to detect the expression of miR-1256 in GC tissues. (i) Pearson’s correlation analysis. (j, k) QRT-PCR was used to detect the expression of miR-1256 in GC cells.
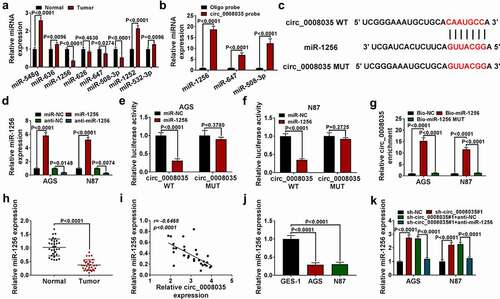
The effect of sh-circ_0008035#1 on cell behavior was eliminated by miR-1256 inhibitor in GC cells
Functionally, sh-circ_0008035#1 notably decreased the cell viability and proliferation of AGS and N87 cells, while miR-1256 inhibitor partially reversed it (). And anti-miR-1256 recovered the cell cycle of AGS and N87 cells, which were blocked due to silencing of circ_0008035 (). Conversely, anti-miR-1256 reversed the cell apoptosis promoted by sh-circ_0008035#1 (). Mechanically, silencing circ_0008035 significantly reduced the migration and invasion of AGS and N87 cells, while they were partially reversed by anti-miR-1256 (). Finally, Western blot showed that sh-circ_0008035#1 remarkably decreased the levels of Ki67 and MMP-2, and increased the expression of Bax, while these effects could be partially reversed by down-regulation of miR-1256 ( and 4 J). In general, miR-1256 inhibitor restored cell malignant behavior reduced by sh-circ_0008035#1.
Figure 4. The effect of sh-circ_0008035#1 on cell behavior was eliminated by miR-1256 inhibitor in GC cells. (a, b) MTT was used to detect cell viability. (c) Cell proliferation was measured by EdU assay. (d, e) The cell cycle was detected by flow cytometry. (f) The cell apoptosis was detected by flow cytometry. (g, h) Transwell was performed to measure the cell migration and invasion. (i, j) Western blot was utilized to measure the expression of relative proteins.
CEACAM6 was a target of miR-1256
As shown in , Starbase predicted the binding sites of miR-1256 and CEACAM6. Dual-luciferase reporter assay results showed that the combined transfection of miR-1256 mimic and CEACAM6 3ʹUTR WT vector significantly inhibited luciferase activity (). RNA pull down experiment showed that Bio-miR-1256 enriched CEACAM6 in large amount (). Functionally, qRT-PCR results showed that CEACAM6 expression was increased in GC tumor tissues compared with normal tissues (). And Pearson’s correlation analysis showed that CEACAM6 expression was negatively correlated with miR-1256 expression, and positively correlated with circ_0008035 expression in GC tumor tissues (). Similarly, analysis in GEPIA database showed that CEACAM6 was highly expressed in GC tissues (). Mechanically, Western blot analysis showed that CEACAM6 protein was significantly increased in GC tissues and cells (). Then, overexpression of miR-1256 decreased the level of CEACAM6 (). Finally, the miR-1256 inhibitor significantly restored the expression of CEACAM6 protein reduced by knockdown of circ_0008035 in AGS and N87 ().
Figure 5. CEACAM6 was the direct target of miR-1256. (a) The complementary sequences between miR-1256 and CEACAM6 were shown. (b, c) Dual-luciferase reporter assay was performed to confirm the association between miR-1256 and CEACAM6. (d) RNA pull down was performed to measure the interaction between CEACAM6 and miR-1256. (e) QRT-PCR was used to detect the expression of CEACAM6 in GC tissues (n = 30). (f, g) Pearson’s correlation analysis. (h) The expression of CEACAM6 detected by qRT-PCR in GEPIA database. (i, j) Western blot was performed to measure the protein expression of CEACAM6 in GC tissues (n = 3) and cells. (k-m) Western blot was used to detect the protein expression of CEACAM6.
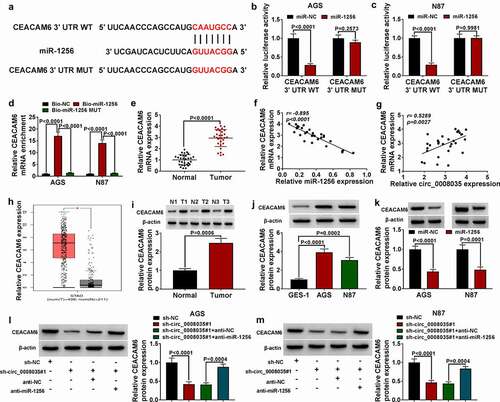
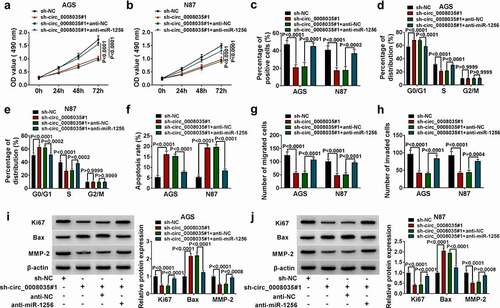
The effect of miR-1256 on cell behavior was eliminated by overexpression of CEACAM6 in GC cells
Western blot tested the overexpression efficiency of CEACAM6 overexpression vector in AGS and N87 cells (). Functionally, CEACAM6 restored the viability and proliferation of AGS and N87 cells reduced by overexpression of miR-1256 (). The overexpression of CEACAM6 recovered the cell cycle of AGS and N87 cells, which were blocked due to miR-1256 (). Conversely, CEACAM6 reversed the cell apoptosis promoted by miR-1256 (). Mechanically, overexpression of miR-1256 significantly reduced the migration and invasion in AGS and N87 cells, while all of them were partially reversed by overexpression of CEACAM6 (). Finally, Western blot showed that miR-1256 remarkably decreased the levels of Ki67 and MMP-2, and increased the expression of Bax, while all of them partially were reversed by overexpression of CEACAM6 (). Taken together, CEACAM6 restored cell malignant behavior reduced by overexpression of miR-1256.
Figure 6. The effect of miR-1256 on cell behavior was eliminated by CEACAM6 in GC cells. (a) The protein expression of CEACAM6 was detected by Western blot. (b, c) MTT was used to detect cell viability. (d) Cell proliferation was measured by EdU assay. (e, f) The cell cycle was detected by flow cytometry. (g) The cell apoptosis was detected by flow cytometry. (h, i) Transwell was used to detect the cell migration and invasion. (j) Western blot was performed to measure the expression of relative proteins.
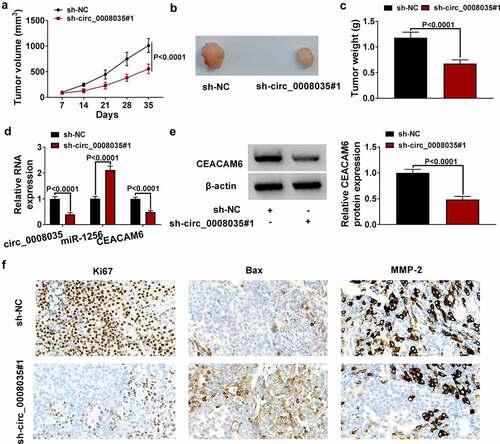
Circ_0008035 knockdown inhibited tumor growth in vivo
In order to better explore the influence of circ_0008035 in GC, we constructed a GC xenotransplantation model. By recording and observing tumor volume () and tumor weight (), we found that knockdown of circ_0008035 significantly inhibited tumor growth. Then, qRT-PCR showed that the expression of circ_0008035 and CEACAM6 was reduced, while the expression of miR-1256 was enhanced in the tumor tissues of sh-circ_0008035 group (). Similarly, Western blot detected that the expression of CEACAM6 was diminished by sh-circ_0008035#1 (). Finally, IHC results showed that knockdown of circ_0008035 significantly reduced the positive cells of Ki67 and MMP-2 while increasing the positive cells of Bax (). In conclusion, down-regulation of circ_0008035 inhibited tumor growth of GC in vivo.
Figure 7. Circ_0008035 knockdown inhibited tumor growth in vivo. (a-c) Tumor volume and weight after circ_0008035 knockdown in vivo. (d) Relative expression levels of circ_0008035, miR-1256 and CEACAM6 in xenografts were detected by qRT-PCR. (e) The protein expression of CEACAM6 was analyzed by Western blot. (f) The positive cells of Ki67, Bax and MMP-2 in mice tumor tissues were analyzed by IHC.
Discussion
Numerous studies have demonstrated that the crosstalk of ceRNA via shared miRNAs is involved in the pathogenesis of human diseases including cancer [Citation29]. Here we show, for the first time, circ_0008035 acts as a novel regulator of GC cell functional characteristics by regulating the miR-1256/CEACAM6 axis.
The mechanism of circRNAs in GC progression has become a new research hotspot. For example, down-regulation of circ_HN1 suppressed the cell malignant behavior of GC cells [Citation30]. And circ_001653 acted as a cancer-promoting gene in GC [Citation31]. Consistent with these findings, we screened circ_0008035 by analyzing the expression profile of GC using GEO database (GSE78092). And circ_0008035 was abnormally up-regulated in GC tissues and cell lines. Functionally, silencing circ_0008035 restricted the cell malignant behavior in vitro. Furthermore, animal studies revealed that circ_0008035 knockdown repressed the growth of GC tumors in vivo.
Then, we predicted that miR-1256 was a target of circ_0008035 by circinteractome. Numerous studies have documented the anti-tumor activity of miR-1256 in human cancers. For instance, studies had shown that miR-1256 suppressed the proliferation of NSCLC cells [Citation21,Citation32]. Similarly, some studies had shown that miR-1256 was down-regulated in GC, which was consistent with our results [Citation23]. Circ-DCAF6, a highly expressed circRNA in GC, had been reported to promote GC progression depending on the modulation of miR-1256 [Citation33]. Consistent with this finding, our data showed that miR-1256 inhibitor reversed the cell growth and metastasis of GC cells inhibited by knockdown of circ_0008035. We also identified that CEACAM6 was a direct and functional target of miR-1256. CEACAM6 has also been established as a functionally downstream effector of miR-128 in the context of repressing cervical cancer cell growth [Citation34]. The study of Zhang et al. had shown that CEACAM6 was highly expressed in GC, and this result was consistent with ours [Citation25]. More importantly, we pointed out that circ_0008035 regulated CEACAM6 expression through sponging miR-1256.
Collectively, we established that circ_0008035 could serve as a novel regulator of GC. We uncovered a novel circ_0008035/miR-1256/CEACAM6 ceRNA network in GC development. Since downregulation of circ_0008035 suppressed tumor growth, we confirmed that targeted inhibition of circ_0008035 might be a promising method for the development of new anti-tumor therapies in GC.
Highlight
Silencing of circ_0008035 inhibited the progress of GC.
MiR-1256 could be sponged by circ_0008035.
CEACAM6 was the target of miR-1256.
Acknowledgments
None
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
- Correa P. Gastric cancer: overview. Gastroenterol Clin North Am. 2013;42(2):211–217.
- Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12(3):354–362.
- Tsugane S, Sasazuki S. Diet and the risk of gastric cancer: review of epidemiological evidence. Gastric Cancer. 2007;10(2):75–83.
- Vainio H, Heseltine E, Wilbourn J. Priorities for future IARC monographs on the evaluation of carcinogenic risks to humans. Environ Health Perspect. 1994;102(6–7):590–591.
- Ge S, Feng X, Shen L, et al. Association between habitual dietary salt intake and risk of gastric cancer: a systematic review of observational studies. Gastroenterol Res Pract. 2012;2012:808120.
- Qi X, et al. Management of advanced gastric cancer: an overview of major findings from meta-analysis. Oncotarget. 2016;7:78180–78205.
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388.
- Liu J, Liu T, Wang X, et al. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16(1):58.
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777.
- Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612.
- Yue F, Peng K, Zhang L, et al. Circ_0004104 accelerates the progression of gastric cancer by regulating the miR-539-3p/RNF2 axis. Dig Dis Sci. 2021;66(12):4290–4301.
- Chen B, Ji F, Wen X, et al. Circular RNA circ_ASAP2 promotes cell viability, migration, and invasion of gastric cancer cells by regulating the miR-770-5p/CDK6 axis. Int J Clin Exp Pathol. 2020;13(11):2806–2819.
- Huang S, et al. A novel circular RNA hsa_circ_0008035 contributes to gastric cancer tumorigenesis through targeting the miR-375/YBX1 axis. Am J Transl Res. 2019;11(4):2455–2462.
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610.
- Melo SA, Esteller M. Disruption of microRNA nuclear transport in human cancer. Semin Cancer Biol. 2014;27:46–51.
- Zhen Q, et al. LncRNA DANCR promotes lung cancer by sequestering miR-216a. Cancer Control. 2018;25(1):1073274818769849.
- Zhao W, Geng D, Li S, et al. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/ HMGA2 axis in breast cancer. Cancer Med. 2018;7(3):842–855.
- Ding JN, Zang YF, Ding YL. MiRNA-146b-5p inhibits the malignant progression of gastric cancer by targeting TRAF6. Eur Rev Med Pharmacol Sci. 2020;24(17):8837–8844.
- He X, Zou K. MiRNA-96-5p contributed to the proliferation of gastric cancer cells by targeting FOXO3. J Biochem. 2020;167(1):101–108.
- Liu W, et al. MiR-1256 suppresses proliferation and migration of non-small cell lung cancer via regulating TCTN1. Oncol Lett. 2018;16(2):1708–1714.
- Wu C, et al. MiR-1256 inhibits cell proliferation and cell cycle progression in papillary thyroid cancer by targeting 5-hydroxy tryptamine receptor 3A. Hum Cell. 2020;33(3):630–640.
- Cai J, et al. circHECTD1 facilitates glutaminolysis to promote gastric cancer progression by targeting miR-1256 and activating beta-catenin/c-Myc signaling. Cell Death Dis. 2019;10(8):576.
- Muniategui A, et al. Quantification of miRNA-mRNA interactions. PLoS One. 2012;7(2):e30766.
- Zhang Y, et al. CEACAM6 promotes tumor migration, invasion, and metastasis in gastric cancer. Acta Biochim Biophys Sin (Shanghai). 2014;46(4):283–290.
- Balk-Moller E, et al. A marker of endocrine receptor-positive cells, CEACAM6, is shared by two major classes of breast cancer: luminal and HER2-enriched. Am J Pathol. 2014;184(4):1198–1208.
- Tian C, Zhang B, Ge C. Effect of CEACAM6 silencing on the biological behavior of human gallbladder cancer cells. Oncol Lett. 2020;20(3):2677–2688.
- Zang M, et al. CEACAM6 promotes tumor angiogenesis and vasculogenic mimicry in gastric cancer via FAK signaling. Biochim Biophys Acta. 2015;1852(5):1020–1028.
- Karreth FA, P P. Pandolfi, ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121.
- Wang D, et al. Circular RNA circ_HN1 facilitates gastric cancer progression through modulation of the miR-302b-3p/ROCK2 axis. Mol Cell Biochem. 2021;476(1):199–212.
- Zhou W, et al. hsa_circ_001653 up-regulates NR6A1 expression and elicits gastric cancer progression by binding to microRNA-377. Exp Physiol. 2020;105(12):2141–2153.
- Chang H, Qu J, Wang J, et al. Circular RNA circ_0026134 regulates non-small cell lung cancer cell proliferation and invasion via sponging miR-1256 and miR-1287. Biomed Pharmacother. 2019;112:108743.
- Wu L, Liu D, Yang Y. Enhanced expression of circular RNA circ-DCAF6 predicts adverse prognosis and promotes cell progression via sponging miR-1231 and miR-1256 in gastric cancer. Exp Mol Pathol. 2019;110:104273.
- Chuang PC, Lu CW, Tsai CC, et al. MicroRNA-128 confers anti-endothelial adhesion and anti-migration properties to counteract highly metastatic cervical cancer cells’ migration in a parallel-plate flow chamber. Int J Mol Sci. 2020;22(1):215.
