ABSTRACT
The number and quality of oocytes in the ovarian reserve are related to fertility and reproductive lifespan in mammals. Some transcription factors have been demonstrated to determine oogenesis. The insulinoma-associated 2 (Insm2) gene is a member of the Snail transcriptional repressor superfamily. Recent studies have demonstrated Insm2 plays an essential role for insulin secretion and glucose intolerance in mice, but the functions of Insm2 in reproduction remain elusive. Here, by examination of Insm2 knockout mice, we found Insm2 was essential for female fertility. Loss of Insm2 resulted in female infertility with major defects in primordial follicle pool, ovarian folliculogenesis and ovulation. Transcriptomic profiling of ovaries suggests that loss of Insm2 caused defects in oocyte meiosis and steroid synthesis. Both oocyte- and granulosa cell-expressed genes were dysregulated, including Foxo1 and other known genes involved in primary ovarian insufficiency. Together, these studies show that Insm2 is required for oocyte development and their communication with ovarian somatic cells.
KEYWORDS:
Introduction
In mammals, oogenesis is an extended multistage process that begins in the embryo but arrests at metaphase and will not continue without fertilization. Females are born with a finite, non-renewing pool of oocyte enclosed by somatic cells, called primordial follicles. The limited supply of non-growing follicles is often referred to as the ovarian reserve. The size of the primordial follicle pool determines the reproductive potential of mammalian females. Premature ovarian insufficiency (POI) represents a condition characterized by the absence of normal ovarian function due to an incipient (by 3–10 y) ovarian aging [Citation1]. The underlying cause of POI is early oocyte depletion leading to infertility. Many cases of POI are idiopathic, and POI is highly heterogeneous in genetic etiology. Previous studies have identified POI candidate genes, including breast cancer type susceptibility protein 2 (Brca2) [Citation2], deleted in azoospermia-like (Dazl) [Citation3], insulin-like growth factor 1 (Igf-1) [Citation4], forkhead box O1 (Foxo1) [Citation5], methylsterol monooxygenase 1 (Msmo1) [Citation6], and squalene epoxidase (Sqle) [Citation6].
Early folliculogenesis begins with the activation of the follicle and ends with the formation of the follicular antrum [Citation7]. In this process, the early follicles go through four stages, namely primary, early secondary, middle secondary, and late secondary [Citation8]. Many biological events such as zona pellucida formation, oocyte enlargement, follicular theca formation, follicular antrum formation, and granulosa cell proliferation and differentiation occur in early folliculogenesis. The ovarian follicles are regulated by oocyte-somatic cell interaction. When the bidirectional communication is disrupted, follicle assembly can be impaired, leading to early follicle reserve loss or inability to acquire meiotic competence [Citation9]. Steroid synthesis is important for the synchronization of follicle growth and oocyte development [Citation10]. The ovary contains multiple distinctive steroid-producing cells, including stromal cells, theca cells, granulosa cells [Citation11]. The high apoptosis rate of granulosa cells is significantly related to decreased pregnancy outcome from in vitro fertilization (IVF) [Citation12].
Insulinoma-associated-2 (IA-2 or INSM2) is a zinc-finger transcription factor that was identified and isolated from a human insulinoma subtraction library [Citation13]. INSM2 is an intronless gene encoding a 566-amino acid protein that contains five zinc-finger DNA-binding motifs and several potential functional domains. The complementary DNA (cDNA) of INSM2 is expressed in normal human brain, pituitary, pancreas, and brain tumor cell lines, but not in a variety of other normal or tumor tissues [Citation13]. In mice, northern blot analysis showed that Insm2 mRNA was expressed in insulinoma cell lines and enriched normal islets. Cai et al. detected Insm2 transcript in expressed in adult mouse tissues using northern blot [Citation14] They also demonstrated that Insm2 is the direct target of neurogenin 3 (Ngn3) and neurogenic differentiation 1 (NeuroD1) in pancreatic islets [Citation14]. However, besides the above results in pancreatic islets, the biological role of Insm2 in other tissues is largely unknown.
To determine the reproductive function of Insm2, homozygous deletion mice (Insm2 −/−) were generated using CRISPR-Cas9 technique [Citation15]. We found Insm2 is not essential for male fertility in mice. However, loss of Insm2 causes female infertility in mice. Insm2 −/− female mice have severe defects in the ovarian reserve. Furthermore, RNA-Seq analysis revealed that oocyte meiosis and steroid synthesis pathways were disrupted in Insm2 −/− ovary. We found a higher level of apoptosis companying by downregulation of Foxo1 and Nsdhl (NADPH steroid dehydrogenase-like) in mutant ovary granulosa cells, suggesting that Insm2 may control follicular development through steroid synthesis.
Materials and methods
Declarations and ethics statements
All the animal procedures were approved by the Institutional Animal Care and Use Committee of Tongji Medical College, Huazhong University of Science and Technology. All experiments with mice were conducted ethically according to the Guide for the Care and Use of Laboratory Animal guidelines.
Mice
Insm2 deficient mice were generated as previously described [Citation15]. Mice were all housed in pathogen-free facility of Huazhong University of Science and Technology. Homozygous Insm2 −/− knockout mice were obtained from Insm2 +/− mice in C57BL/6 J background. Primers for genotyping (forward: 5’-CTGGTGAAGCGAACCAAGCG-3’, reverse: 5’-TCCTGCGGAGGTGCGAGGCA-3’) were located on flanking sides of the deleted region of Insm2. See main text for detailed study design.
Fertility assessment
Eight-wk-old Insm2 −/− and WT mice were mated for 2–4 months (2 females: 1 male). The number of offspring from each pregnant female was recorded after birth.
Histology analysis
Genotyped mice were euthanized for isolation of ovary tissues, which were then treated in 4% formalin buffer for paraffin embedding. Five micrometer thick sections on slides were stained with hematoxylin and eosin (H&E) for further analysis. Early growing follicles at primary (type 3a, b), secondary (type 4, 5a, 5b), small antral follicle stages of development, and atretic follicles were counted in all sections based on the well-accepted standards established by the Pedersen and Peters criterion [Citation16]. Briefly, type 3a, b follicles were characterized by a monolayer of cubic follicle cells surrounding an oocyte; type 4 follicles, two layers of follicle cells surrounding an oocyte; type 5a follicles have three layers of follicle cells; and type 5b follicles were characterized by many layers of follicle cells surrounding a fully grown oocyte, but the follicular antrum has not yet formed [Citation17]. Five sections of ovary were randomly chosen for counting the follicles, and the numbers were normalized by the total ovarian area in the section. The ovarian area was measured using a FluoView 1000 microscope (Olympus, Japan).
Immunohistochemistry and immunofluorescence
Sections of mouse ovaries were dewaxed, hydrated, incubated in citrate buffer (pH 6.0) and then heated in a microwave for antigen retrieval. Briefly, the section was incubated with the primary antibody: anti-NSDH1 (1:100, Proteintech), anti-FOXO1 (1:100, Cell Signaling Technology), or anti-γ-H2AX (1:100, Solarbio Life Science) at 4°C overnight. Following three 5 min washes in PBS. Immunohistochemistry was performed with goat anti-rabbit IgG conjugated HRP. Positive signals were developed with DAB solution (ZLI-9018; ZSGB-BIO, China). Counterstaining was conducted with hematoxylin. The slides were observed under a light microscope. Immunofluorescence was performed using secondary antibodies were conjugated with Alexa Fluor 594 or 488. Invitrogen ProLong Gold Slowfade media containing 4’,6-diamidino-2- phenylindole (DAPI) was used to mount the slides. Laser confocal scanning images were captured using FluoView 1000 microscope (Olympus, Japan).
Oocyte count from natural- and superovulation
For natural ovulation, estrous cycling of 6-wk-old Insm2 −/− and WT mice was determined by vaginal smears for 2 wk after which animals were killed at estrus. For superovulation, 6-wk-old Insm2 −/− and WT mice were given a single intraperitoneal injection of 5 U of pregnant mare serum gonadotropin (Sigma, St. Louis, MO) followed 48 h later by 5 U of human chorionic gonadotropin (Sigma). After an additional 18 h, the oocyte-cumulus mass was released from the oviduct ampulla with a syringe and cultured in M2 medium. After treating with 0.1% hyaluronidase (Sigma) to make granulosa cells shed from the oocytes, oocytes were collected and counted.
RNA isolation and RNA sequencing
Total RNA was isolated with TRIzol reagents (Invitrogen) from the ovaries of 6-wk-old (a transition from juveniles into sexually mature) female mice at estrus. The RNA concentration was verified using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). One microgram of total RNA was used from each sample to prepare the mRNA libraries using TruSeq Stranded mRNA Library Preparation Kit Set A (Cat. No. RS-122-2101, Illumina) according to the manufacturers’ instructions. All libraries were sequenced using the Illumina HiSeq 4000 platform. The FASTX-Toolkit was used to remove adaptor sequences and low-quality reads from the sequencing data. To identify all the transcripts, we used Tophat2 and Cuffiinks to assemble the sequencing reads based on the UCSC MM10 mouse genome. The differential expression analysis was performed by Cuffdiff. The differential expressed genes were set with the threshold of P < 0.05 and fold change ≥2.
Quantitative RT-PCR
Reverse transcriptional reactions contained 500 ng of purified total RNA using a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa) to remove the DNA contamination. RT-qPCR was performed with SYBR green master mix (TaKaRa) on Stepone Real-Time PCR system according to manufactures’ instructions: 95°C for 10 min, 40 cycles of 95°C for 1 min and 60°C for 30 s. Insm2 (forward: 5′-gctccggcagctcctacc-3′; reverse: 5′-ggctcctccggtgaggatt-3′). The relative gene expression was quantified using the comparative cycle threshold method, with the Gapdh expression used for normalization [Citation18]. The primer sequences for other genes and retrotransposons and were listed in Table S1.
Molecular function and pathway analysis
To further understand the biological relevance of the hub genes and their regulators in oocyte development and cellular metabolism, we performed functional enrichment analysis using ClueGO [Citation19]. ClueGO facilitates the visualization of functionally related genes displayed as a clustered network and chart. The statistical test used for the enrichment was based on right-sided hypergeometric option with a Benjamini–Hochberg correction and kappa score of 0.3.
Detection of apoptosis
The extent of cell death in the ovary was established by TUNEL analysis using an TUNEL apoptosis assay kit (C1098, Beyotime). The staining was performed according to the protocol provided by the manufacturer.
Statistical analysis
All data are presented as mean ± SD unless otherwise noted in the figure legends. Statistical differences between datasets were assessed by one-way ANOVA or Student’s t-test. P-values are denoted in figures by *P < 0.05 and **P < 0.01.
Results
Insm2 deficiency did not affect testis histology and male fertility
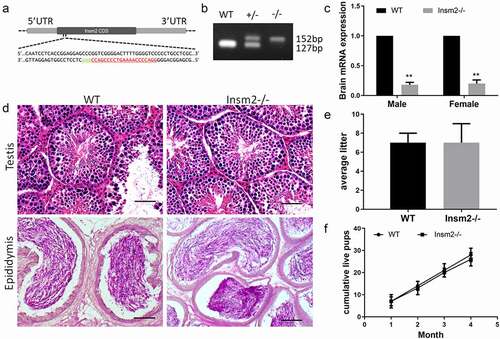
To identify the biological roles of Insm2 in reproduction, Insm2 knockout mice by CRISPR/Cas9 technology were used. The deletion (25 bp) induces a frameshift and a premature stop codon, producing global disruption of the Insm2 gene (Insm2 −/−) ()). Insm2 deficient mice were identified by PCR amplification ()). Both published sequencing data [Citation20] and our RT-PCR result showed Insm2 transcriptional activity is extremely low in the adult ovary (Figure S1(a,b)), so we examined Insm2 mRNA level in knockout mice through brain tissues. Quantitative qRT-PCR analysis showed that the level of Insm2 mRNA was dramatically reduced in knockout mice when compared with the wild-type (WT) mice ()). A very small amount of unstable Insm2 mRNAs probably exist in the mutant mice due to the frameshifted deletion. Then, we examined mouse testes histology by H&E staining, and did not find a visible morphological difference when Insm2 was deficient ()). Furthermore, breeding tests demonstrated no significant difference in the average litter size ()) or the cumulative live pups between Insm2-deficient and control mice ()). The above results indicate that Insm2 is dispensable for male fertility.
Figure 1. Insm2 deletion has no defects in the testis histology and the male infertility. (a) Schematic of the sgRNA targeting site of Insm2 allele. The sgRNA targeting sequence is labeled in red. (b) Representative PCR genotyping results showing the WT and knockout (−/−) alleles can be detected at 152 and 127 bp bands, respectively. (c) Quantitative RT-PCR analysis indicates that Insm2 mRNA is markedly decreased in Insm2−/− mouse brain as compared to normal controls. (d) H&E staining of the testes and epididymis from WT and Insm2−/− mice. Scale bars = 20 μm. (e) Aggregate pups per litter among pairings that produced a litter. (f) Continuous breeding assessment shows cumulative number of litters per male. Three samples per genotype were analyzed in each experiment. **P < 0.01.
Insm2 deficiency induced infertility in female mice
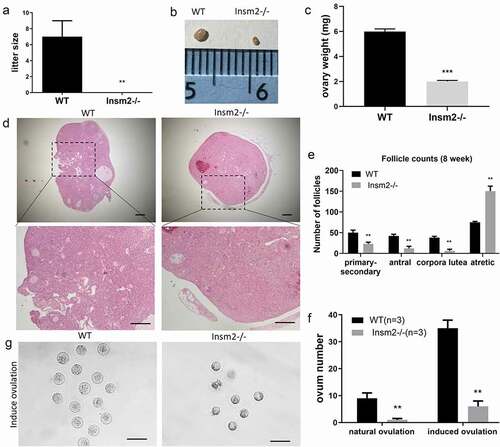
To determine whether Insm2 is required for female fecundity, we performed the breeding tests by crossing WT males with adult Insm2 −/− females and its littermate controls in one cage till 4 months old. The results showed none of the gene mutant females were pregnant ()). To investigate the effect of Insm2 deficiency on the ovarian structure of the mice, ovary size and weight were examined. Compared with the ovaries of the WT mice, the ovary size ()) and weight ()) of Insm2 −/− mice was significantly decreased. The H&E staining results by serial sections showed that the total ovarian follicles, including primary follicles, antral follicles and corpora lutea were significantly decreased in Insm2 −/− mice ()). These results indicated that Insm2 deficiency led to decreased ovarian reserve. Under natural ovulation, we only collected about 2 oocytes in average from every three Insm2 −/− female mice. To examine whether Insm2 −/− female mice could ovulate more oocytes, we tried to collect oocytes from the oviducts after superovulation. About 36 oocytes in average were obtained from every three WT females (12 oocytes per mice), while only about 6 oocytes were recovered in every three Insm2 −/− females (2 oocytes per mice) ()). In addition, we noticed that Insm2 has a relatively stronger transcriptional level in infant ovary compared with adult ovary (Figure S1C), suggesting Insm2 appear to be much more active at the early stage of ovary development.
Figure 2. Insm2 deletion results in ovarian reserve and follicular development defects in mice. (a) Average litter sizes of WT and Insm2−/− female mice during fertility assay. (b) Gross morphology of the ovary from 8-wk-old WT and Insm2−/− mice. (c) The weight of ovaries. (d) Representative images of ovarian sections from 8-wk-old WT and Insm2−/− mice. Scale bars = 25 μm. (e) NumbeFrs represent total counts of every fifth section from serially sectioned ovaries. (f) Number of ovulated oocytes from mated females. Superovulation was induced with 5 IU (international units) each of PMSG and hCG at 48-hour intervals. (g) Morphology of MII oocytes derived from WT and Insm2−/− females after superovulation. Scale bars = 100 μm. Three samples per genotype were analyzed in each experiment. **P < 0.01.
The transcriptome of Insm2 null ovary was dramatically perturbed
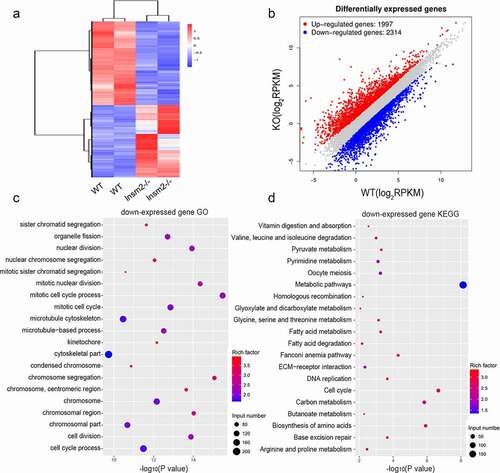
To understand the molecular changes occurring during folliculogenesis in the ovaries of Insm2 −/− mice, we further analyzed transcriptional profile through RNA sequencing. The transcriptome results showed that 4311 genes were significantly changed in 6-wk-old mice Insm2 −/− ovary, which indicated that Insm2 is crucial for the gene expression during folliculogenesis. Based on the differentially expressed genes, trees with clear distinctions between WT and Insm2 −/− ovary were generated by cluster analysis ()). Specifically, Insm2 −/− ovary showed 2314 down-regulated genes and 1997 upregulated genes ()). Then, we conducted Gene Ontology (GO) and KEGG analysis on the up and down regulated genes. GO analysis showed that down-regulated genes are significantly enriched in sister chromatid segregation, organelle fission, nuclear division, mitotic cell cycle, cell division and cell cycle process ()). KEGG analysis revealed that down-regulated genes are mostly involved in oocyte meiosis, cell cycle, and cellular metabolism of vitamin/amino acid/glucose/fatty acid ()). The up-regulated genes were significantly enriched in the different GO terms, such as regulation of immune response and cell activation (Figure S2(a)). KEGG analysis showed that the upregulated genes were mainly involved in immunity, including Fc gamma R-mediated phagocytosis, B cell receptor signaling pathway and NF-kappa B signaling pathway (Figure S2(b)). RNA-Seq analyses revelated that global gene expression changes in Insm2 −/− ovary, in which dysregulation of several factors may contribute to diminished ovarian reserve.
Figure 3. Loss of Insm2 alters the ovarian transcriptome. (a) Hierarchical clustering of significantly different genes in an RNA-seq analysis of 8-wk-old Insm2−/− ovaries compared with control ovaries (P < 0.05 and fold change ±2). Two independent pools of two ovaries per genotype were analyzed. Downregulated genes are shown in blue and upregulated genes are indicated in red. (b) Graph plot showing dysregulated genes in Insm2−/− ovary. Significantly regulated genes have a P-value of < 0.05 and fold change of > 2. The blue dots represent significantly decreased transcripts; the red dots represent the transcripts for which expression levels were significantly increased. (c) GO analysis of differentially down-expressed genes ordered by −log10 (P-value). (d) KEGG analysis of differentially down-expressed genes ordered by −log10 (P-value). Two samples per genotype were analyzed in each experiment.
Insm2 deficiency impaired oocyte meiosis and steroidogenesis
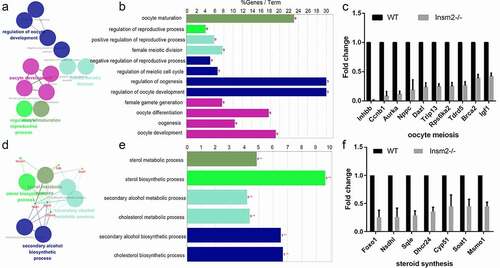
To gain the biological insight on the differentially down-expressed genes, we performed enrichment analysis using ClueGO software. Overall, oocyte meiosis and steroid synthesis were significantly enriched, which could be respectively categorized into 12 and 6 terms of GO. For oocyte meiosis, the main GO categories were as follows: regulation of oocyte development, female meiotic division, female meiotic division, oocyte development, oocyte maturation, etc. ()). We found the oocyte development-related genes: Aurka, Ccnb1, Dazl, Igf1, Inhbb, Nppc, Rps6ka2, Tdrd5, Trip13, and Brca2 were significantly decreased compared with the WT group () and Figure S1(d)). For steroid synthesis, the main GO categories were as follows: sterol metabolic process, sterol biosynthetic process, cholesterol metabolic process, cholesterol biosynthetic process, etc. ()). We found that steroid synthesis genes: Foxo1, Cyp51, Dhcr24, Msmo1, Nsdhl, Soat1, and Sqle were obviously reduced in mice lacking Insm2 gene () and Figure S1(e)). The above results showed that Insm2 may regulate ovarian folliculogenesis through sterol synthetic signaling.
Figure 4. ClueGO analysis of down-regulated genes in oocyte meiosis and steroid synthesis. (a) and (d) Functionally grouped network with terms as nodes linked based on their kappa score level (≥0.3), where only the label of the most significant term per group is shown. The node size represents the term enrichment significance. (b) and (e) GO terms specific for down-regulated genes. The bars represent the number of genes associated with the terms. The percentage of genes per term is shown as bar label. (c) and (f) Column graph shows the relative expression levels of oocyte meiosis and steroid synthesis related genes expressed in the ovaries from WT and Insm2−/− mice.
Foxo1 and Nsdhl were down-regulated in Insm2 deficiency ovary
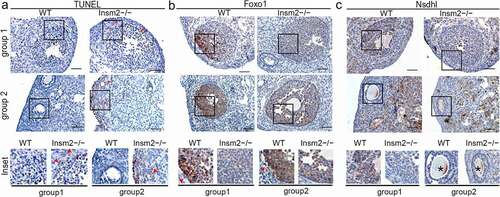
To explore the potential mechanisms behind the infertility induced by Insm2 deficiency, we examined cell apoptosis with TUNEL assay. The apoptosis of the ovarian granulosa cell was significantly increased in the Insm2 −/− ovary compared with the WT ovary. Immunochemistry results suggested that apoptosis in follicular cells occurred in growing follicles (primary and mature) of Insm2 −/− mice ()). Follicular atresia may be subsequently induced and early occurred by granulosa cell apoptosis. The AKT/PI3K pathway represents a potential cross-link by which insulin signaling could affect folliculogenesis and ovulation [Citation21]. Foxo1, one of the main targets of AKT signaling in granulosa cells, is expressed in high amounts in granulosa cells of growing follicles [Citation22]. Compared with controls, Foxo1 exhibited a remarkably decreased expression in Insm2 −/− granulosa cells both in corpus luteum and antral follicles () and Figure S3(a)). Moreover, we found that Nsdhl, an enzyme responsible for the cholesterol biosynthesis, was also downregulated in the granulosa cells of Insm2 −/− follicles () and Figure S3(b)). This repression was consistent with the steroid synthesis changes observed in RNA-seq data.
Figure 5. Insm2 deletion results in increased apoptosis and abnormal expression of Foxo1 and Nsdhl. (a) The images from 8-wk-old WT and Insm2−/− mice show apoptotic signal in the corpus luteum and primary follicle. (b) Foxo1 and (c) Nsdhl expression was downregulated by Insm2 knockout. Red triangles indicate the positive staining. Solid box indicates the location of the higher magnification images in the lower panels. Asterisk indicates nonspecific staining. Scale bars = 50 μm. Three samples per genotype were analyzed in each experiment.
Retrotransposons were activated in Insm2 knockout ovary
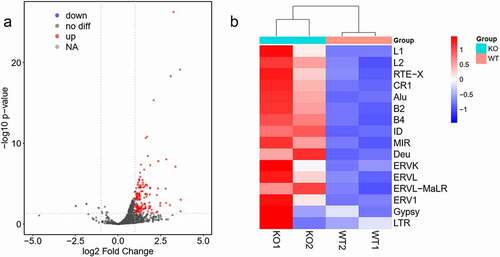
To further understand how Insm2 functions, we compared the expression levels of retrotransposon between Insm2 −/− and control ovaries. Volcano plots show that many retrotransposons were strongly activated in Insm2 −/− ovary ()). The transcript levels of long terminal repeats (such as ERVL and ERVL-MaLR), long interspersed elements (such as LINE1 and LINE2) and short interspersed elements SINE (including Alu, B2, B4, ID, MIR and Deu) were significantly higher in Insm2 −/− ovary than in control () and Figure S1(f)). We propose that the upregulated retrotransposon may invade genome by random insertion and cause DNA double-strand breaks (DSBs), checkpoint activation and cell death in mutant ovary [Citation23]. The immunofluorescent staining demonstrated that the number of γ-H2AX positive granulosa cells in 8-wk-old Insm2 −/− ovaries was significantly higher than those in control ovaries (Figure S3(c)). These findings suggest that, in the absence of Insm2, there was more DNA damage in the granulosa cells of ovaries.
Figure 6. Retrotransposons were activated in Insm2 knockout ovary. (a) Volcano plot representation of up- and down-regulated transposons as measured by RNA-seq between WT and Insm2 knockout mice. Red dots: upregulated transposons; blue dots: downregulated transposons (fold change >2 and P-value <0.05). (b) Heat map with hierarchical clustering of retrotransposons from WT and Insm2 knockout mice ovary RNA-Seq results. Individual names of retrotransposons are shown at the right of the chart. Colors represent the average read count for an element in a given family. Two samples per genotype were analyzed in each experiment.
Discussion
Insm2 expression has been found in human fetal pancreas and mouse embryos, as well as adult pancreatic islets [Citation14]. Although no obvious diabetic symptoms were observed in Insm2 −/− mice, deletion of Insm2 affects pancreatic islet cell functions are attributed to lower serum insulin secretion and lower C-peptide levels [Citation15]. However, fasting blood glucose levels in Insm2 −/− were significantly higher than control mice [Citation15]. Normal follicular growth is the result of complementary action of follicle-stimulating hormone (FSH) and luteinizing hormone (LH). Decreased LH response to gonadotropin hormone-releasing hormone (GnRH) stimuli is closely related to high fasting glucose, severe insulin deficiency or reduced C-peptide levels [Citation24]. In patients with type 1 diabetes, insulin deficiency usually induces hypogonadism and hyperandrogenism, leading to the fertility problems [Citation25].
Insm2 mRNA has been found to be expressed in mouse testis [Citation14]. Here, our fertility assay and histology analysis suggested that Insm2 deficiency was not essential for male mice fertility. Conversely, Insm2 deficiency causes infertility in female mice. Compared with WT mice ovary, Insm2 −/− female mice had smaller ovaries and fewer numbers of follicles and corpora lutea. Superovulation treatment of Insm2 −/− mice only induced fewer numbers of oocytes. To identify potential factors that contribute to impaired folliculogenesis and ovulation, we performed RNA-seq analysis and made a comprehensive transcriptomic analysis on ovaries from mutant and control mice. GO and KEGG analysis was performed on the downregulated and upregulated genes exhibiting greater than twofold difference in expression, respectively. We noticed that the downregulated genes mainly enriched in the biological processes of oocyte meiosis and steroid synthesis. Additionally, the complexity of RNA-Seq results in our study may be attributed to loss of follicles (loss of oocytes, loss of granulosa cells, etc.) in Insm2 −/− ovary. This could be fixed by including histology and morphometrics to define a time period in which the follicle numbers of the mutant mice are the similar. Ovarian follicle status in Insm2 −/− female mice before puberty will be determined in the further study.
It is known that the follicle atresia accounts for the oocyte’s loss during folliculogenesis. Apoptosis plays crucial rules in the follicle atresia [Citation26]. In the TUNEL analysis, we found corpus luteum of Insm2 −/− ovary contained much more apoptotic cells in the comparison with control. The Forkhead box O (FOXO) transcription factors regulate multiple cellular functions, including cell proliferation, cell survival, apoptosis and metabolism [Citation27]. Foxo1 is highly expressed in granulosa cells of ovarian follicles and functions as a critical mediator of normal ovarian follicular development and luteinization. Foxo1 or Foxo3 knockout alone led to no overt phenotype, while simultaneous Foxo1 and Foxo3 knockout led to female infertility and severe follicle defects [Citation28]. Nsdhl is a decarboxylase enzyme involved in cholesterol biosynthesis. Meiosis-activating sterols (MAS) are substrates of Nsdhl in the cholesterol pathway and are important for normal organismal development. Byskov et al. isolated a class of C-29 sterols named follicle fluid meiosis-activating sterol (FF-MAS) and testicular meiosis-activating sterol (T-MAS) [Citation29]. MAS have at high concentrations in the testis and ovary, and as the name suggests, they play an important role in meiosis activation. Deficiency in Nsdhl dramatically impairs cholesterol biosynthesis in both humans and mice [Citation30]. Loss-of-function mutations in Nsdhl cause Congenital Hemidysplasia with Ichthyosiform erythroderma and Limb Defects (CHILD). Occasionally, CHILD syndrome patients have unilateral absence or hypoplasia of the ovary and fallopian tubes [Citation31]. In our results, we found both Foxo1 and Nsdhl were downregulated in the ovary of Insm2 −/− mice, indicating that steroidogenic pathways was altered after loss of Insm2.
Retrotransposons are genetic mobile elements that make up about 40% of the mouse genome. In the long-term evolution of mammals, retrotransposons are conducive to the diversity of the genome. However, in the short term, the activation of retrotransposon can also threaten genome integrity, thus leading to tumorigenesis and infertility. In male germ cells, retrotransposons are repressed by DNA methylation, histone modification and piRNA machinery, thereby maintaining normal spermatogenesis. Mice mutants lacking piRNA pathway show normal female fertility. In Lsh-deficient mice, oocytes had de-repressed retrotransposon by extensive DNA hypomethylation, thus triggering insufficient ovarian follicle formation and severe oocyte loss. In addition, elevated LINE-1 expression leads to DNA damage and meiotic defects in mammalian oocyte. LINE-1 activation correlates with increased fetal oocyte attrition (FOA), reduced ovarian reserves and oocyte aneuploidy. Collectively. Retrotransposons are essential for maintaining the stability of the genome. Excessive retrotransposon activation can lead to meiotic defects, which decreases ovarian reserves and causes oocyte attrition.
INSM2 shares high similarities in its amino acid sequence with the Snail superfamily members, a kind of C2H2 zinc-finger transcription factors, which plays an important role in the development disease [Citation32]. For example, mice lacking both Snai2 and Snai3 have been reported to be infertile in males [Citation33]. Previous study showed that Insm2 mRNA expressed in mouse embryonic development from E6.5 to E18.5, in which is transiently enhanced in E11.5-E13.5, suggesting that a potential role of Insm2 in the development of mouse embryonic development. Insm2 was recently found to be involved in the transcriptional regulation of ultraconserved (uc) RNAs 372, which suppresses the maturation of miR-195/miR-4668 to regulate expression of genes related to lipid synthesis [Citation34]. Moreover, genetic analysis by GWAS in a cohort of Africa Americans with type 2 diabetes revealed a significant association of the disease to an SNP (rs1952392, MAF = 0.0188; P < 0.001) at the proximal promoter region of the human INSM2 gene [Citation35]. These studies indicated INSM2 plays multiple roles in the development of other tissues and organs. This study provides a potential clue of exploring the underlying mechanism in the disease that are affected by INSM2.
In summary, we found Insm2 is required for female fertility, and it acts as a novel positive regulator during folliculogenesis. We also showed that Insm2 is involved in oocyte development, steroid synthesis pathway and retrotransposon repression in mouse ovary. Our results will provide valuable information for clinical treatment of infertility patients with diminished ovarian reserve.
Author contributions
LQZ and ZML conceived and designed the research. ZML and LQZ wrote the paper. ZML, YYL and CFF performed bench experiments.
Supplemental Material
Download MS Word (3.1 MB)Acknowledgments
We thank Dr Tao Cai from National Institute of Dental and Craniofacial Research/National Institutes of Health, Bethesda, for sharing the Insm2 knockout mice.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available from the corresponding author, LQZ, upon reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2092816
Additional information
Funding
References
- Luisi S, Orlandini C, Regini C, et al. Premature ovarian insufficiency: from pathogenesis to clinical management. J Endocrinol Invest. 2015 Jun;38(6):597–603. PubMed PMID: 25596661.
- Sharan SK, Pyle A, Coppola V, et al. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development. 2004 Jan;131(1):131–142. PubMed PMID: 14660434.
- De Vos M, Devroey P, Fauser BC. Primary ovarian insufficiency. Lancet. 2010 Sep 11;376(9744):911–921. PubMed PMID: 20708256.
- Louhio H, Hovatta O, Sjoberg J, et al. The effects of insulin, and insulin-like growth factors I and II on human ovarian follicles in long-term culture. Mol Hum Reprod. 2000 Aug;6(8):694–698. PubMed PMID: 10908278.
- Venturella R, De Vivo V, Carlea A, et al. The genetics of non-syndromic primary ovarian insufficiency: a systematic review. Int J Fertil Steril. 2019 Oct;13(3):161–168. PubMed PMID: 31310068; PubMed Central PMCID: PMCPMC6642427.
- Zhang Y, Ouyang X, You S, et al. Effect of human amniotic epithelial cells on ovarian function, fertility and ovarian reserve in primary ovarian insufficiency rats and analysis of underlying mechanisms by mRNA sequencing. Am J Transl Res. 2020;12(7):3234–3254. PubMed PMID: 32774697; PubMed Central PMCID: PMCPMC7407690.
- Chen Y, Liu Q, Liu R, et al. A prepubertal mice model to study the growth pattern of early ovarian follicles. Int J Mol Sci. 2021 May 12;22(10):5130. PubMed PMID: 34066233; PubMed Central PMCID: PMCPMC8151218.
- Chen Y, Wang X, Yang C, et al. A mouse model reveals the events and underlying regulatory signals during the gonadotrophin-dependent phase of follicle development. Mol Hum Reprod. 2020 Dec 10;26(12):920–937. PubMed PMID: 33063120.
- Rodrigues P, Limback D, McGinnis L, et al. Germ-somatic cell interactions are involved in establishing the follicle reserve in mammals. Front Cell Dev Biol. 2021;9:674137. PubMed PMID: 34195191; PubMed Central PMCID: PMCPMC8236641.
- Azhar S, Tsai L, Medicherla S, et al. Human granulosa cells use high density lipoprotein cholesterol for steroidogenesis. J Clin Endocrinol Metab. 1998;83(3):983–991.
- Gupta MK, Chia S-Y. Ovarian hormones: structure, biosynthesis, function, mechanism of action, and laboratory diagnosis. In: Falcone T, Hurd W, Editors. Clinical reproductive medicine and surgery. New York, NY: Springer; 2013. p. 1–30 https://doi.org/10.1007/978-1-4614-6837-0_1.
- Bencomo E, Perez R, Arteaga MF, et al. Apoptosis of cultured granulosa-lutein cells is reduced by insulin-like growth factor I and may correlate with embryo fragmentation and pregnancy rate. Fertil Steril. 2006 Feb;85(2):474–480. PubMed PMID: 16595230.
- Lan MS, Lu J, Goto Y, et al. Molecular cloning and identification of a receptor-type protein tyrosine phosphatase, IA-2, from human insulinoma. DNA Cell Biol. 1994 May;13(5):505–514. PubMed PMID: 8024693.
- Cai T, Chen X, Wang R, et al. Expression of insulinoma-associated 2 (INSM2) in pancreatic islet cells is regulated by the transcription factors Ngn3 and NeuroD1. Endocrinology. 2011 May;152(5):1961–1969. PubMed PMID: 21343251; PubMed Central PMCID: PMCPMC3075939.
- Wang L, Sun ZS, Xiang B, et al. Targeted deletion of Insm2 in mice result in reduced insulin secretion and glucose intolerance. J Transl Med. 2018 Oct 25;16(1):297. PubMed PMID: 30359270; PubMed Central PMCID: PMCPMC6202866.
- Pedersen T, Peters H. Proposal for a classification of oocytes and follicles in the mouse ovary. J Reprod Fertil. 1968 Dec;17(3):555–557. PubMed PMID: 5715685.
- Yang C, Liu Q, Chen Y, et al. Melatonin delays ovarian aging in mice by slowing down the exhaustion of ovarian reserve. Commun Biol. 2021 May 6;4(1):534. PubMed PMID: 33958705; PubMed Central PMCID: PMCPMC8102596.
- Li Z, Chen S, Yang Y, et al. Novel biomarker ZCCHC13 revealed by integrating DNA methylation and mRNA expression data in non-obstructive azoospermia. Cell Death Discov. 2018 Dec;4(1):36. PubMed PMID: 29531833; PubMed Central PMCID: PMCPMC5841273.
- Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009 Apr 15;25(8):1091–1093. PubMed PMID: 19237447; PubMed Central PMCID: PMCPMC2666812.
- Pan L, Gong W, Zhou Y, et al. A comprehensive transcriptomic analysis of infant and adult mouse ovary. Genomics Proteomics Bioinformatics. 2014 Oct;12(5):239–248. PubMed PMID: 25251848; PubMed Central PMCID: PMCPMC4411413.
- Sekulovski N, Whorton AE, Shi M, et al. Periovulatory insulin signaling is essential for ovulation, granulosa cell differentiation, and female fertility. FASEB J. 2020 Feb;34(2):2376–2391. PubMed PMID: 31908002.
- Liu Z, Rudd MD, Hernandez-Gonzalez I, et al. FSH and FOXO1 regulate genes in the sterol/steroid and lipid biosynthetic pathways in granulosa cells. Mol Endocrinol. 2009 May;23(5):649–661. PubMed PMID: 19196834; PubMed Central PMCID: PMCPMC2675958.
- Mita P, Sun X, Fenyo D, et al. BRCA1 and S phase DNA repair pathways restrict LINE-1 retrotransposition in human cells. Nat Struct Mol Biol. 2020 Feb;27(2):179–191. PubMed PMID: 32042152; PubMed Central PMCID: PMCPMC7082080.
- Volpi R, Chiodera P, Gramellini D, et al. Influence of residual insulin secretion and duration of diabetes mellitus on the control of luteinizing hormone secretion in women. Eur J Clin Invest. 1998 Oct;28(10):819–825. PubMed PMID: 9792995.
- Codner E, Merino PM, Tena-Sempere M. Female reproduction and type 1 diabetes: from mechanisms to clinical findings. Hum Reprod Update. 2012 Sep-Oct;18(5):568–585. PubMed PMID: 22709979.
- Yu YS, Sui HS, Han ZB, et al. Apoptosis in granulosa cells during follicular atresia: relationship with steroids and insulin-like growth factors. Cell Res. 2004 Aug;14(4):341–346. PubMed PMID: 15353131.
- Paik JH, Kollipara R, Chu G, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007 Jan 26;128(2):309–323. PubMed PMID: 17254969; PubMed Central PMCID: PMCPMC1855089.
- Liu Z, Castrillon DH, Zhou W, et al. FOXO1/3 depletion in granulosa cells alters follicle growth, death and regulation of pituitary FSH. Mol Endocrinol. 2013 Feb;27(2):238–252. PubMed PMID: 23322722; PubMed Central PMCID: PMCPMC3683807.
- Byskov AG, Andersen CY, Nordholm L, et al. Chemical structure of sterols that activate oocyte meiosis. Nature. 1995 Apr 6;374(6522):559–562. PubMed PMID: 7700384.
- Liu XY, Dangel AW, Kelley RI, et al. The gene mutated in bare patches and striated mice encodes a novel 3beta-hydroxysteroid dehydrogenase. Nat Genet. 1999 Jun;22(2):182–187. PubMed PMID: 10369263.
- Morimoto M, Souich C, Trinh J, et al. Expression profile of NSDHL in human peripheral tissues. J Mol Histol. 2012 Feb;43(1):95–106. PubMed PMID: 22113624.
- Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002 Mar;3(3):155–166. PubMed PMID: 11994736.
- Pioli PD, Dahlem TJ, Weis JJ, et al. Deletion of Snai2 and Snai3 results in impaired physical development compounded by lymphocyte deficiency. PLoS One. 2013;8(7):e69216. PubMed PMID: 23874916; PubMed Central PMCID: PMCPMC3713067.
- Guo J, Fang W, Sun L, et al. Ultraconserved element uc.372 drives hepatic lipid accumulation by suppressing miR-195/miR4668 maturation. Nat Commun. 2018 Feb 9;9(1):612. PubMed PMID: 29426937; PubMed Central PMCID: PMCPMC5807361.
- Liang J, Le TH, Edwards DRV, et al. Single-trait and multi-trait genome-wide association analyses identify novel loci for blood pressure in African-ancestry populations. PLoS Genet. 2017 May;13(5):e1006728. PubMed PMID: 28498854; PubMed Central PMCID: PMCPMC5446189.
