ABSTRACT
To elaborate on the role of circular RNA 0001955 (circ_0001955) on the proliferation and apoptosis of non-small cell lung cancer (NSCLC) cells and its underlying mechanism. Circ_0001955 expression in NSCLC was screened out through bioinformatics analysis based on GEO database. Circ_0001955, microRNA-769-5p (miR-769-5p), and epidermal growth factor receptor (EGFR) expression in NSCLC tissues and cell lines was examined using quantitative real-time PCR (qRT-PCR) and Western blot. Cell proliferation and apoptosis were examined using the CCK-8 method, BrdU experiment and flow cytometry analysis, respectively. Bioinformatics prediction, dual-luciferase reporter gene experiment and RNA immunoprecipitation (RIP) experiments were applied to validate the targeting relationship between miR-769-5p and circ_0001955 and the 3’ UTR of EGFR. Pearson’s correlation analysis was employed to validate the correlations among them. Circ_0001955 expression was up-regulated in NSCLC tissues and cell lines, and its overexpression was strongly associated with increased tumor TNM stage and lymph node metastasis. Circ_0001955 overexpression enhanced the proliferation and restrained the apoptosis in NSCLC cells, whereas knocking down circ_0001955 exerted the opposite effects. Circ_0001955 directly targeted miR-769-5p and negatively regulated its expression. EGFR, a target gene of miR-769-5p, could be indirectly and positively regulated by circ_0001955. Correlation analysis indicated that circ_0001955 was negatively correlated with miR-769-5p expression, while circ_0001955 was positively correlated with EGFR expression. Circ_0001955 facilitates the proliferation and represses the apoptosis of NSCLC cells by modulating miR-769-5p/EGFR axis.
1. Introduction
Lung cancer (LC) is the most common tumors worldwide [Citation1]. LC is mainly divided into non-small-cell LC (NSCLC) and small-cell LC (SCLC), with NSCLC taking up 80%–85% of all LC cases [Citation2]. Currently, the main treatment options for NSCLC include surgery, radiotherapy, targeted therapy, and immunotherapy [Citation3]. Early-stage NSCLC is still mainly treated surgically, but 30% to 70% of patients are already in advanced stage at the time of initial diagnosis and cannot be cured by radical surgery, thus the prognosis is particularly unfavorable [Citation4,Citation5]. Therefore, it is imperative to probe the mechanism of NSCLC pathogenesis to find valuable tumor markers and new therapeutic targets.
Circular RNAs (circRNA) are non-coding RNAs without a 5’-end cap and a 3’-end poly-A tail, and have a closed-loop structure [Citation6]. CircRNAs are more conserved and stable and less susceptible to degradation than linear RNAs [Citation7]. Reportedly, circRNAs expression is closely associated with disease development, and is likely to serve as biomarkers for diseases such as tumors [Citation8]. For instance, circ_006100 is overexpressed in gastric cancer tissues, and its overexpression is linked to an adverse prognosis of the patients [Citation9]. Down-regulation of circ-ZKSCAN1 expression is observed in bladder cancer, and circ-ZKSCAN1 overexpression represses the proliferation, invasion and metastasis of bladder cancer cells [Citation10]. CircPTK2 expression is down-regulated in NSCLC, and circPTK2 overexpression restrains the epithelial-mesenchymal transition (EMT) process and invasion of NSCLC cells [Citation11]. In the present work, bioinformatic analysis implies that circular RNA 0001955 (circ_0001955) expression is up-regulated in NSCLC. What’s more, some studies reports that circ_0001955 was up-regulated in various tumors, and circ_0001955 could promote the growth of tumor cells [Citation12,Citation13]. However, the molecular mechanism by which circ_0001955 exerts its effects in NSCLC has not been fully elucidated.
It has been reported that miR-769-5p can target epidermal growth factor receptor (EGFR) to restrain the growth of NSCLC cells [Citation14]. In this work, we report that miR-769-5p is a downstream target of circ_0001955, and circ_0001955 up-regulates EGFR by adsorbing miR-769-5p to promote the progression of NSCLC. These findings provide new therapeutic targets for NSCLC.
2. Materials and methods
2.1. Tissue samples collection
With the approval of the Ethics Committee of the Third Affiliated Hospital of Jinzhou Medical University and the informed consent of all subjects, 67 NSCLC patients were selected for this study. NSCLC tissues and para-cancer tissues were collected after the operation and preserved at −196°C in liquid nitrogen. The clinical information and clinicopathological information of all subjects were complete, and none of them received radiotherapy or chemotherapy before surgery.
2.2. Cell culture
Normal lung epithelial cells (BEAS-2B) and NSCLC cell lines (H520, A549, H358, H460 and HCC827) were obtained from the Cell Center of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA)+ 100 U/mL penicillin (Invitrogen, Carlsbad, CA, USA)+ 100 μg/mL streptomycin (Invitrogen, Carlsbad, CA, USA) at 37°C with 5% CO2 and 95% humidity.
2.3. Cell transfection
The empty vector (NC), circ_0001955 overexpression plasmid (circ_0001955), small interfering RNA targeting circ_0001955 (si-circ-1: 3’-dTdTGCCTCTTCGAAATCAGGTGAA-5’, and si-circ-2: 3’-dTdTTTCGAAATCAGGTGAAGGTCT-5’) and the negative controls (si-NC), miR-769-5p mimics (miR mimics: 3’-UCGAGUCUUGGGUCUCCAGAGU-5’) and its control (mimics NC), and miR-769-5p inhibitors (miR inhibitors: 3’-ACUCUGGAGACCCAAGACUCGA-5’) and its control (inhibitors NC) were synthesized by Genepharma (Shanghai, China). H520, H358 and HCC827 cells were cultured to the logarithmic growth phase, and cell concentration was diluted with the medium to 1 × 105 cells/mL, and then the cells were inoculated in 6-well plates. Then the cells were cultured to 70% confluence and subsequently, the transfection was performed using LipofectamineTM 2000 kit (Invitrogen, Carlsbad, CA, USA). 24 h after the transfection, the transfection efficiency was validated by quantitative real-time PCR (qRT-PCR).
2.4. qRT-PCR
Total RNA was extracted from tissues or cells by TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA was reverse transcribed into cDNA using the PrimeScript™ RT Reagent kit (Invitrogen, Carlsbad, CA, USA). Next, qRT-PCR was performed with SYBR GreenPCR reagent (Takara Bio, Inc., Otsu, Japan) on the ABI7500 real-time PCR system (Thermo Fisher Scientific, Inc., Foster City, CA, USA). Relative expression levels of circ_0001955, miR-769-5p and EGFR mRNA were calculated using the 2−ΔΔCt method. GAPDH and U6 were used as internal reference genes. All primer sequences were displayed in .
Table 1. Sequences used for qRT-PCR.
2.5. Cell counting kit-8 (CCK-8) experiment
The NSCLC cells were inoculated into 96-well plates at 2 × 103 cells/mL. Subsequently, the cells were culture for 24 h. After that, 10 μL of CCK-8 solution (MedChemExpress, Monmouth Junction, NJ, USA) was supplemented to each well, and the incubation was continued for 1 h. Finally, the absorbance (optical density, OD) value at 450 nm was measured using a microplate reader. Subsequently, the OD of the cells was measured on the 2nd and 3rd day.
2.6. BrdU experiment
H520 and H358 cells were inoculated in 24-well plates (with a coverslip placed inside) (2.5 × 105 cells/well). After 24 h of incubation, BrdU solution (Beyotime Biotechnology, Shanghai, China) was supplemented into each well, and the incubation was continued for 4 h. Following that, the cells were fixed with 4% paraformaldehyde for 30 min; the supernatant was then discarded, and 100 μL of 1× Apollo staining was supplemented to each well, and the cells were incubated for 30 min. The supernatant was discarded and 100 μL of 1× Hoechst 33342 reaction solution was supplemented to each well and the cells were incubated for 20 min in the darkness. After that, the total number of the cells and the number of BrdU-positive cells were observed and counted under the fluorescence microscope after phosphate buffer saline (PBS) washing, and the percentage of BrdU-positive cells was calculated.
2.7. Flow cytometry
The apoptosis was detected by AnnexinV-FITC/propidium iodide (PI) cell apoptosis double staining kit (Sigma-Aldrich, Louis, MO, USA). Transfected cells were trypsinized with trypsin, rinsed with pre-chilled PBS 2 times, and in each group, 1 × 105 cells were resuspended with 100 μL of 1× binding buffer. The cell suspensions were thoroughly mixed with 5 μL of AnnexinV-FITC staining solution and 5 μL of PI staining solution, incubated for 15 min, and the apoptosis was detected by a flow cytometer within 1 h. To analyze the cell cycle distribution of the cells, in each sample, a total of 1 × 105 cells were fixed by 75% ethanol overnight in an ice bath. Next, the cells were washed with PBS and centrifuged, followed by resuspension with 100 μL of RNaseA solution and incubation at 37°C for 30 min. Subsequently, PI staining solution was added to the cell suspension, followed by incubation for 30 min at 4°C. Next, the cell cycle analysis was performed with a flow cytometer.
2.8. Dual-luciferase reporter gene experiment
The binding sites between circ_0001955/miR-769-5p, and between miR-769-5p and EGFR 3ʹUTR were predicted by Circular RNA Interactome database and TargetScan database, respectively, and wild-type circ_0001955 (WT circ_0001955), wild-type EGFR (WT EGFR), mutant-type circ_0001955 (MUT circ_0001955), and mutant-type EGFR (MUT EGFR) dual-luciferase reporter vectors were constructed. The above reporter vectors were co-transfected into HEK-293 T cells with miR-769-5p mimics or miR-769-5p inhibitors, respectively. After 48 h of transfection, the luciferase activity was measured, and the ratio of the luminescence intensity of Renilla luciferase to the luminescence intensity of firefly luciferase was calculated.
2.9. RNA immunoprecipitation (RIP) assay
A Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) was utilized in RIP experiment to verify the combination between circ_0001955 and miR-769-5p. H520 and H358 cells were lysed in RIP lysis buffer, and 100 μL of cell lysates were incubated in RIP buffer with magnetic beads coupled with human anti-Argonaute2 (Ago2) or negative control IgG. The specimens were then incubated with Proteinase K to shake to remove proteins and then RNA precipitation was obtained. The purified RNA was subjected to qRT-PCR analysis for examining the contents of circ_0001955and miR-769-5p in the precipitation.
2.10. Western blot
NSCLC cells in logarithmic growth stage were fully lyzed by RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China). After centrifugation, the supernatant was collected. The protein quantification was performed using a BCA protein quantification kit (Beyotime Biotechnology, Shanghai, China). Next, protein denaturation was conducted in boiling water for 10 min. Then sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was executed. Subsequently, the proteins on the gel were transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA, USA). Next, the unspecific antigens on the PVDF membrane were blocked with 5% skim milk for 2 h. Next. the membrane was incubated with the primary antibody (Rabbit Anti-EGFR antibody, ab52894, 1:1000; Rabbit Anti-GAPDH antibody, ab245355, 1:1000) (Abcam, Cambridge, UK) at 4°C overnight. The membranes were then incubated with secondary anti-rabbit IgG (Goat Anti-Rabbit IgG, ab205718, 1:2000) (Abcam, Cambridge, UK) and incubated for 2 h at 37°C. Ultimately, the protein bands were visualized using an ECL luminescence kit (Beyotime Biotechnology, Shanghai, China).
2.11. Statistical analysis
SPSS 19.0 software was applied to analyze the experimental data. Measurement data were expressed as “mean ± standard deviation”. Student’s t-test was used for the comparison between two groups, and one-way ANOVA was used for comparison among multiple groups. Counting data or percentages (%) were tested using χ2 test. P < 0.05 signified statistical significance.
3. Results
3.1. The expression characteristics of circ_0001955 in NSCLC tissues
To identify the circRNAs that are implicated in the tumorigenesis of NSCLC, we analyzed the differentially expressed circRNAs by analyzing circRNA expression profile (GSE101684) and found that circ_0001955 expression was remarkably up-regulated in NSCLC tissue compared with that in the non-cancerous tissues ()). To further verify the abnormal expression of circ_0001955 in NSCLC, circ_0001955 expression was detected by qRT-PCR in 67 cases of NSCLC tissues and paracancerous tissues, and the data showed that circ_0001955 was markedly overexpressed in NSCLC ()). Additionally, it was revealed that circ_0001955 expression was associated with advanced TNM stage and lymph node metastasis of the patients with NSCLC ().
Figure 1. Circ_0001955 expression characteristics in NSCLC.
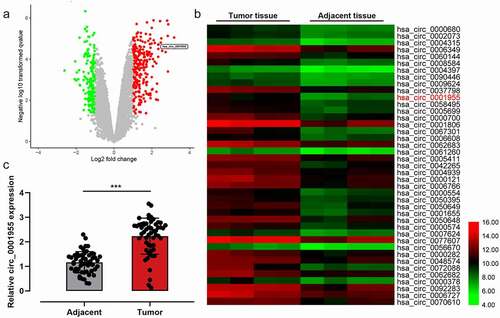
Table 2. Correlation between clinicopathological features and expression of circ_0001955 in NSCLC.
3.2. Effect of circ_0001955 on proliferation, apoptosis, migration, and cell cycle of NSCLC cells
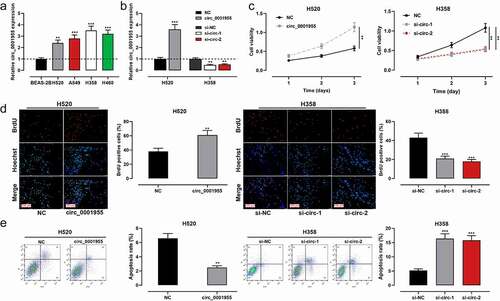
The cells were examined by qRT-PCR, and circ_0001955 expression was found to be remarkably up-regulated in all NSCLC cells compared with that in normal lung epithelial cells ()). To verify the biological function of circ_0001955 in NSCLC cells, circ_0001955 overexpression plasmid was transfected into H520 cells; si-circ_0001955-1 and si-circ_0001955-2 were transfected into H358 cells. qRT-PCR validated that the transfection was successful ()). Next, proliferation and apoptosis of NSCLC cells were detected by CCK-8 method, BrdU experiment and flow cytometry. As shown, circ_0001955 overexpression enhanced the proliferation of H520 cells and decreased the apoptosis rate of H520 cells compared with the control group, while knocking down circ_0001955 exerted the contrary effects ()). Scratch wound assays and flow cytometry assays also showed that overexpression of circ_0001955 promoted cell migration, and accelerated the cell cycle progression in H520 cells (Supplementary Figure S1A, B).
Figure 2. Regulatory effect of circ_0001955 on NSCLC cell proliferation and apoptosis.
3.3. Targeting relationship between circ_0001955 and miR-769-5p
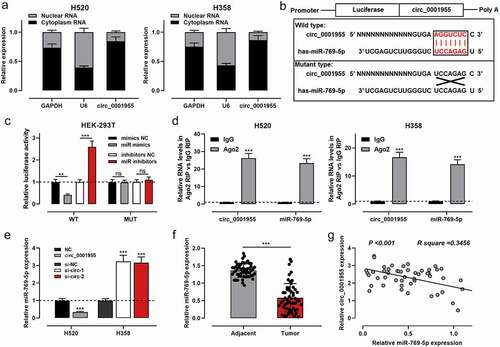
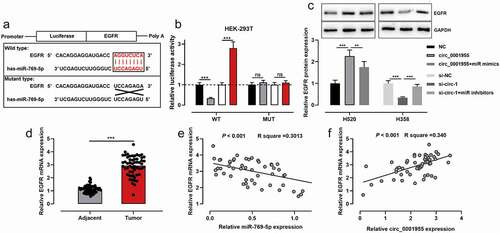
To investigate the role of circ_0001955, the subcellular localization of circ_0001955 in H520 and H358 cells was identified using qRT-PCR after the separation of subcellular fractions. The data implied that circ_0001955 was primarily located in the cytoplasm of NSCLC cells, hinting its role as a competitive endogenous RNA (ceRNA) ()). Next, a binding site between miR-769-5p and circ_0001955 was predicted by the Circular RNA Interactome database ()). As is well known, miR-769-5p was down-regulated in NSCLC, and miR-769-5p could inhibit the growth of NSCLC cells. Therefore, miR-769-5p was chosen as the potential downstream target of circ_0001955. Dual-luciferase reporter gene assay showed that transfection of miR-768-5p mimics reduced the luciferase activity of WT circ_0001955 reporter, and the transfection of miR-769-5p inhibitors increased the luciferase activity of WT circ_0001955 reporter, compared with the control group; however, neither miR-768-5p mimics nor inhibitors had a significant effect on the luciferase activity of MUT circ_0001955 reporter. RIP experiments showed that circ_0001955 and miR-769-5p were highly enriched in Ago2-containing microribonucleoproteins compared with IgG ()). qRT-PCR showed that circ_0001955 overexpression down-regulated miR-769-5p expression, while knocking down circ_0001955 exerted the opposite effect ()). Furthermore, it was observed that miR-769-5p expression was remarkably down-regulated in NSCLC tissues compared with paracancerous tissues, and circ_0001955 expression was negatively correlated with miR-769-5p expression in NSCLC samples ()).
Figure 3. Circ_0001955 directly targeted miR-769-5p.
3.4. Effects of Circ_0001955/miR-769-5p axis on NSCLC cell proliferation and apoptosis
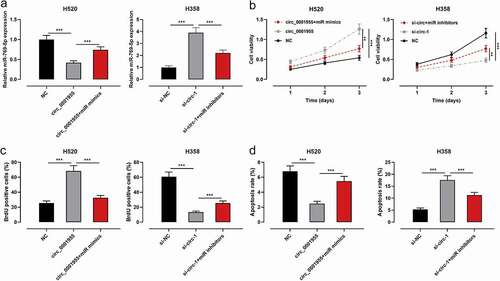
To elaborate on the effects of circ_0001955/miR-769-5p axis on the proliferation and apoptosis of NSCLC cells, circ_0001955 overexpression plasmid and miR-769-5p mimics were co-transfected into H520 cells; si-circ_0001955-1+ miR-769-5p inhibitors were co-transfected into H358 cells. qRT-PCR validated that the transfection was successful ()). CCK-8 method, BrdU experiment and flow cytometry showed that circ_0001955 overexpression enhanced proliferation and inhibited apoptosis, while transfection of miR-769-5p mimics reversed these effects ()); knocking down circ_0001955 inhibited proliferation and promoted apoptosis, while inhibition of miR-769-5p reversed these effects ()).
Figure 4. The effect of circ_0001955 on the proliferation and apoptosis of NSCLC cells by repressing miR-769-5p.
3.5. Circ_0001955 up-regulated EGFR expression through adsorbing miR-769-5p
To probe the downstream regulatory mechanisms of miR-769-5p, TargetScan database was searched, and it was predicted that EGFR was one of the potential downstream targets of miR-769-5p ()). The relationship between the two was validated by luciferase reporter gene experiment and the data revealed that transfection of miR-769-5p mimics reduced the luciferase activity of WT EGFR reporter, and transfection of miR-769-5p inhibitors enhanced the luciferase activity of WT EGFR reporter; neither miR-769-5p mimics nor inhibitors had a significant effect on the luciferase activity of MUT EGFR reporter ()). Western blot indicated that circ_0001955 overexpression increased EGFR protein expression, and the transfection of miR-769-5p mimics counteracted this effect in H520 and HCC827 cells; circ_0001955 knockdown decreased EGFR protein expression, while the transfection of miR-769-5p inhibitors reversed this effect in H358 cells (), Supplementary Figure S1C). Additionally, EGFR was revealed to be remarkably overexpressed in NSCLC compared with paracancerous tissues ()), and miR-769-5p was negatively correlated with EGFR expression, whereas circ_0001955 was positively correlated with EGFR expression in NSCLC tissues ()).
Figure 5. Circ_0001955 up-regulated EGFR expression through adsorption of miR-769-5p.
4. Discussion
CircRNAs are mainly derived from exons or introns and are produced by trans-splicing [Citation15]. CircRNAs are widespread in living organisms, and they are abundant, evolutionarily conserved and stable [Citation16]. These features endow circRNAs with diverse biological functions, such as acting as miRNA sponges to competitively bind with miRNAs, binding with RNA-binding proteins to form RNA-protein complexes, and modulating gene transcription [Citation17,Citation18]. Accumulating research reports that circRNAs participate in accelerating or impeding the development of tumors. For instance, circ-CTNNB1 activates the Wnt/β-catenin signaling pathway to enhance cancer cell proliferation, invasion, and metastasis through DDX3-mediated trans-activation of YY1 [Citation19]. Increasing research has reported that circRNAs are crucial in NSCLC tumorigenesis and progression [Citation20,Citation21]. For instance, high CircFGFR1 expression in NSCLC is linked to adverse pathological characteristics and prognosis of the patients, and CircFGFR1 can up-regulate CXCR4 via adsorption of miR-381-3p thereby accelerating the proliferation, migration, invasion and immune escape of NSCLC cells [Citation21]. In this work, circ_0001955 expression is revealed to be remarkably up-regulated in NSCLC tissues, and its high expression is markedly linked to unfavorable pathological characteristics in NSCLC patients, suggesting that circ_0001955 may be vital in the pathogenesis of NSCLC. Functional experiments validate that circ_0001955 overexpression remarkably enhances the proliferation and impedes the apoptosis of NSCLC cells, while knocking down circ_0001955 restrains the proliferation and enhances the apoptosis of NSCLC cells. The research implies that circ_0001955 is a novel oncogenic circRNA in NSCLC.
MiRNAs are endogenous single-stranded non-coding small RNAs of 18–24 nucleotides in length [Citation22]. MiRNAs can bind to the 3’ UTR of mRNAs by base complementary pairing and cause degradation or translational repression of target mRNAs, thereby participating in the regulation of post-transcriptional levels of genes [Citation23]. A lot of studies have reported that miRNAs are aberrantly expressed in NSCLC and are implicated in modulating NSCLC progression by targeting downstream genes. For instance, MiR-4326 enhances the proliferation of NSCLC cells by negatively modulating APC2 activation of the Wnt pathway [Citation24]. MiR-451a suppresses the proliferation and invasion of NSCLC cells by targeting ATF2 [Citation25]. Importantly, miR-769-5p is down-regulated in both bladder and oral squamous cell carcinoma and is linked to unfavorable prognosis; miR-769-5p overexpression impedes tumor cell proliferation, migration and invasion [Citation26,Citation27]. In NSCLC, miR-769-5p represses the proliferation, migration, and invasion of NSCLC cells by targeting TGFBR1 [Citation28]. Another study reports that, knockdown of LINC00460 up-modulates miR-769-5p expression and thus restrains EGFR expression, thereby increasing the chemoresistance of NSCLC cells [Citation29]. This work validates that miR-769-5p is a downstream target of circ_0001955, and the up-modulation of miR-769-5 reversed the growth-promoting effect of circ_0001955 overexpression on NSCLC cells, whereas inhibition of miR-769-5p exerts the opposite effects. Hence, we conclude that circ_0001955 facilitates NSCLC cell proliferation and impedes the apoptosis through the adsorption of miR-769-5p.
EGFR belongs to the tyrosine kinase family, consisting of an extracellular (ligand-binding), transmembrane domain, and an intracellular region (tyrosine kinase-containing domains) [Citation30]. Epidermal growth factor (EGF) binds to the extracellular region, causes phosphorylation of the intracellular tyrosine kinase domain, activates signaling pathways such as RAS/RAF/MAPK, STAT, and PI3K/AKT/mTOR, and promotes DNA transcription, thereby facilitating cell proliferation and metastasis, accelerating angiogenesis, and repressing apoptosis [Citation31]. EGFR overexpression is linked to the development and progression of diverse malignancies, including NSCLC [Citation32], breast cancer [Citation33], colorectal cancer [Citation34], and pancreatic cancer [Citation35]. In NSCLC, miR-19b activates the EGFR signaling pathway by targeting PP2A and BIM thereby enhancing NSCLC cell proliferation and impeding apoptosis [Citation32]. MiR-218-5p negatively modulates EGFR to repress the proliferation and migration of NSCLC cells and to suppress the growth of transplanted tumors in nude mice [Citation36]. Additionally, the down-modulation of EGFR expression caused by miR-769-5p increases the chemosensitivity of NSCLC cells [Citation29]. The studies mentioned above imply that EGFR dysregulation in NSCLC is related to miRNA. In this work, we validate that miR-769-5p specifically and negatively modulates EGFR expression and circ_0001955 overexpression enhances EGFR expression, whereas knocking down circ_0001955 impedes EGFR expression. These results imply that circ_0001955 can up-modulate EGFR expression by adsorbing miR-769-5p to facilitate NSCLC progression.
In conclusion, this work reports that circ_0001955 expression is up-regulated in NSCLC and implies a worse prognosis of the patients. Mechanistically, we validate that circ_0001955 accelerates NSCLC progression via modulating the miR-769-5p/EGFR molecular axis. This work lays a theoretical foundation for the diagnosis and treatment of NSCLC. Nonetheless, this work is limited to in vitro experiments and future animal experiments are needed to confirm our demonstration.
Ethics statement
Our study was approved by the Ethics Review Board of The Third Affiliated Hospital of Jinzhou Medical University.
Supplemental Material
Download MS Word (950.7 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data used to support the findings of this study are available from the corresponding author upon request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2100681
Additional information
Funding
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30.
- Osmani L, Askin F, Gabrielson E, et al. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52(Pt 1):103–109.
- Vijayvergia N, Mehra R. Clinical challenges in targeting anaplastic lymphoma kinase in advanced non-small cell lung cancer. Cancer Chemother Pharmacol. 2014;74(3):437–446.
- Zhang C, Wu W, Yang J, et al. Application of artificial intelligence in respiratory medicine. J Digital Health. 2022;1(1):30–39.
- Chang H, Li ZB, Wu JY, et al. Circ-100338 induces angiogenesis after myocardial ischemia-reperfusion injury by sponging miR-200a-3p. Eur Rev Med Pharmacol Sci. 2020;24(11):6323–6332.
- Huang SS, Guo WX, Ren MS. Circular RNA hsa_circ_103809 promotes cell migration and invasion of gastric cancer cells by binding to microRNA-101-3p. Eur Rev Med Pharmacol Sci. 2020;24(11):6064–6071.
- Xu T, Wu J, Han P, et al. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18(Suppl 6):680.
- Liang M, Huang G, Liu Z, et al. Elevated levels of hsa_circ_006100 in gastric cancer promote cell growth and metastasis via miR-195/GPRC5A signalling. Cell Prolif. 2019;52(5):e12661.
- Bi J, Liu H, Dong W, et al. Circular RNA circ-ZKSCAN1 inhibits bladder cancer progression through miR-1178-3p/p21 axis and acts as a prognostic factor of recurrence. Mol Cancer. 2019;18(1):133.
- Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol Cancer. 2018;17(1):140.
- Ding B, Yao M, Fan W, et al. Whole-transcriptome analysis reveals a potential hsa_circ_0001955/hsa_circ_0000977-mediated miRNA-mRNA regulatory sub-network in colorectal cancer. Aging (Albany NY). 2020;12(6):5259–5279.
- Yao Z, Xu R, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10(12):945.
- Xu Y, Li Y, Jin J, et al. LncRNA PVT1 up-regulation is a poor prognosticator and serves as a therapeutic target in esophageal adenocarcinoma. Mol Cancer. 2019;18(1):141.
- Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5(2):472–480.
- Abdelmohsen K, Kuwano Y, Kim HH, et al. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem. 2008;389(3):243–255.
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21.e7.
- Huang G, Li S, Yang N, et al. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017;404:8–18.
- Yang F, Fang E, Mei H, et al. Cis-Acting circ-CTNNB1 Promotes β-Catenin Signaling and Cancer Progression via DDX3-Mediated Transactivation of YY1. Cancer Res. 2019;79(3):557–571.
- Li C, Zhang L, Meng G, et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2019;145(12):2875–2889.
- Zhang PF, Pei X, Li KS, et al. Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol Cancer. 2019;18(1):179.
- Dong M, Ye Y, Chen Z, et al. MicroRNA 182 is a novel negative regulator of adipogenesis by targeting CCAAT/enhancer-binding protein α. Obesity (Silver Spring). 2020;28(8):1467–1476.
- Shukuya T, Ghai V, Amann JM, et al. Circulating MicroRNAs and extracellular vesicle-containing MicroRNAs as response biomarkers of anti-programmed cell death protein 1 or programmed death-ligand 1 therapy in NSCLC. J Thorac Oncol. 2020;15(11):1773–1781.
- Xu G, Zhang Z, Zhang L, et al. miR-4326 promotes lung cancer cell proliferation through targeting tumor suppressor APC2. Mol Cell Biochem. 2018;443(1–2):151–157.
- Shen YY, Cui JY, Yuan J, et al. MiR-451a suppressed cell migration and invasion in non-small cell lung cancer through targeting ATF2. Eur Rev Med Pharmacol Sci. 2018;22(17):5554–5561.
- Chen Y, Zhang W, Kadier A, et al. MicroRNA-769-5p suppresses cell growth and migration via targeting NUSAP1 in bladder cancer. J Clin Lab Anal. 2020;34(5):e23193.
- Zhou Y, Xu XM, Feng Y. MiR-769-5p inhibits cancer progression in oral squamous cell carcinoma by directly targeting JAK1/STAT3 pathway. Neoplasma. 2020;67(3):528–536.
- Yang Z, He J, Gao P, et al. miR-769-5p suppressed cell proliferation, migration and invasion by targeting TGFBR1 in non-small cell lung carcinoma. Oncotarget. 2017;8(69):113558–113570.
- Ma G, Zhu J, Liu F, et al. Long noncoding RNA LINC00460 promotes the gefitinib resistance of nonsmall cell lung cancer through epidermal growth factor receptor by sponging miR-769-5p. DNA Cell Biol. 2019;38(2):176–183.
- Cataldo VD, Gibbons DL, Pérez-Soler R, et al. Treatment of non-small-cell lung cancer with erlotinib or gefitinib. N Engl J Med. 2011;364(10):947–955.
- Akhtar S, Al-Zaid B, El-Hashim AZ, et al. Cationic polyamidoamine dendrimers as modulators of EGFR signaling in vitro and in vivo. PLoS One. 2015;10(7):e0132215.
- Baumgartner U, Berger F, Hashemi Gheinani A, et al. miR-19b enhances proliferation and apoptosis resistance via the EGFR signaling pathway by targeting PP2A and BIM in non-small cell lung cancer. Mol Cancer. 2018;17(1):44.
- Zhao Y, Ma J, Fan Y, et al. TGF-β transactivates EGFR and facilitates breast cancer migration and invasion through canonical Smad3 and ERK/Sp1 signaling pathways. Mol Oncol. 2018;12(3):305–321.
- Miyamoto Y, Suyama K, Baba H. Recent advances in targeting the EGFR signaling pathway for the treatment of metastatic colorectal cancer. Int J Mol Sci. 2017;18(4):752.
- Wei W, Liu Y, Lu Y, et al. LncRNA XIST promotes pancreatic cancer proliferation through miR-133a/EGFR. J Cell Biochem. 2017;118(10):3349–3358.
- Zhu K, Ding H, Wang W, et al. Tumor-suppressive miR-218-5p inhibits cancer cell proliferation and migration via EGFR in non-small cell lung cancer. Oncotarget. 2016;7(19):28075–28085.
