ABSTRACT
Long non-coding RNAs (LncRNA) play important roles in multiple types of cancers. We addressed the role of LINC02535 by regulating the miR-30a-5p /GalNAc Transferase 3 (GALNT3) axis to promote the proliferation, migration, and invasion in lung adenocarcinoma (LUAD) cells. The Cancer Genome Atlas (TCGA) database screened differentially expressed lncRNAs. Quantitative real-time PCR analysis (qRT-PCR) confirmed that LINC02535 is highly expressed in LUAD tissues and cells. In vitro experiments showed that LINC02535 promotes the proliferation, migration, and invasion of LUAD cells. A xenograft mouse model was used to show that LINC02535 promotes tumor growth in vivo. RNA immunoprecipitation (RIP) and Dual-luciferase reporter assay results confirmed that LINC02535 targets miR-30a-5p. The Vicia villosa lectin (VVA) pull-down assay indicated that MUC1 is the glycosylation target of GALNT3, and western blot verified that NF-κB is the downstream signaling pathway of MUC1. We found that LINC02535 was increased in LUAD tissues and cells, and LINC02535 was correlated with the poor prognosis of LUAD patients. miR-30a-5p acts as a tumor suppressor in LUAD by targeting GALNT3. We also demonstrated that LINC02535 might function as the sponge of miR-30a-5p to up-regulate GALNT3, and consequently promote the proliferation and metastasis of LUAD. LINC02535 acts as a competing endogenous RNA (ceRNA) to interact with miR-30a-5p, thereby upregulating the expression of GALNT3, enhancing the function of MUC1, and activating the NF-κB signaling pathway, promoting the malignant progression of LUAD cells.
Abbreviations: LncRNA:long non-coding RNA; LUAD: lung adenocarcinoma; TCGA: The Cancer Genome Atlas; GALNT3: GalNAc Transferase 3; qRT-PCR: quantitative real-time PCR analysis; RIP: RNA immunoprecipitation; SPF: specific pathogen-free; VVA: Vicia villosa lectin; ceRNA: competing endogenous RNA; MiRNAs: microRNAs; FBS: fetal bovine serum; PBS: Phosphate buffered saline; CCK-8: Cell Counting Kit-8; NSCLC: non-small cell lung cancer; OC: ovarian cancer; HCC: hepatocellular carcinoma.
KEYWORDS:
Introduction
LUAD is the most common and fatal malignant tumor [Citation1]. The current treatments for LUAD are mainly surgery, chemotherapy, radiotherapy, molecular targeted therapy, and immunotherapy [Citation2]. However, most patients have lost the opportunity for surgical resection when diagnosed [Citation3]. Even if they can receive chemotherapy and radiotherapy, patients will gradually develop resistance during the treatment process [Citation4]. Targeted therapy requires driver gene testing and can only be treated if mutations occur. Immunotherapy also needs to be examined. As a result, any of these treatments have limitations and ultimately lead to extremely high mortality in LUAD patients [Citation5].
LncRNA is a non-coding RNA with a length of more than 200 nucleotides, which can regulate gene function at the levels of epigenetics, transcription, and post-transcriptional processing [Citation6]. Some studies have shown that lncRNA plays an important role in tumor cell activity and participates in multi-gene regulatory networks, suggesting that the detection of lncRNA may be a promising biomarker for early tumor screening and diagnosis [Citation7]. Current research on LINC02535 is still lacking, and its function in the tumorigenesis and progression of LUAD is still unknown [Citation8].
MiRNA (microRNA) is a small non-coding RNA composed of 20 nucleotides [Citation9]. MiR-30a-5p belongs to the miR-30 family, inhibiting tumor development by regulating tumor proliferation and metastasis [Citation10].
The concept of ceRNA holds that endogenous RNA has the same binding sites as some miRNAs [Citation11]. These endogenous RNAs can competitively bind with common miRNAs to weaken the latter’s inhibitory effect on their target genes and then participate in tumor progression.
Glycosylation is the process of adding glycosyls to proteins under the control of glycosyltransferase [Citation12]. As a glycosyltransferase, GALNT3 is highly expressed in various malignant tumors. In colorectal carcinoma, GALNT3 regulates the proliferation, migration, and invasion of tumor cells [Citation13]. However, the role of GALNT3 in LUAD has not been reported.
We aim to explore the role of LINC02535 in LUAD and its internal biological mechanisms to provide experimental evidence for further research and clinical practice.
Materials and methods
Patient samples
With the ethics committee’s approval of the First Affiliated Hospital of Soochow University, Fresh tumor tissues and adjacent normal tissues were collected and confirmed by pathology from 2020 to 2021. The patients informed all the samples and did not receive radiotherapy or chemotherapy.
Analysis of differentially expressed transcripts in LUAD tissue using TCGA database
Differentially expressed transcripts expression in LUAD and complete patient clinical data were downloaded from the TCGA database. We use R studio software (3.1.3) to analyze the data.
Cell culture
BEAS-2B, A549, H1299, and H1650 cells were purchased from the Cell Bank of the Chinese Academy of Sciences. The cells were cultured in an incubator at 37°C and 5% CO2 with RPMI 1640 (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (Gibco, Carlsbad, CA, USA).
Cell transfection
The experimental procedures of transfection assays followed the instructions of lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). si-NC, si-LINC02535, si-GALNT3 and si-MUC1 (GenePharma, Shanghai, China) (The sequences were: si-LINC02535-1,5’-GCCGATTGCTCACAAAGAT-3′,si-LINC02535-2,5′-GCATACAATGGGACAGTTT-3′;si-GALNT3-1,5′-CCATAGATCTGAACACGTT-3′,si-GALNT3-2,5′-GCAAGGATATTATACAGCA-3′,si-MUC1,5′-GCGUGAGUGAUGUGCCAUUTT-3′) and miR-NC, miR-30a-5p mimics, inhibitor NC and miR-30a-5p inhibitor (RiboBioCorp, Guangzhou, China) were transfected into the cells. After 6 hours, the media were changed with RPMI 1640 supplemented with 10% FBS (fetal bovine serum) to continue culturing for 48 hours.
Establishment of stable LINC02535-knockout cells
sh-LINC02535(sh-LINC02535-1,5′-GCCGATTGCTCACAAAGAT-3′,sh-LINC02535-2,5′-GCATACAATGGGACAGTTT-3′) virus and sh-NC virus stock solution (GenePharma, Shanghai, China) was added to the A549 and H1299 cells of the 6-well plate. After 48 hours, the cells were transferred to a culture flask, and then drug screening was performed with G418 (Amresco, Solon, OH, USA).
Construction of LINC02535 overexpressed cell line
To construct the plasmid, the fragment of the human LINC02535 sequence was cloned into the PLVX-Puro vector between XhoI and SmaI. H1650 cell line was transfected with control PLVX-Puro-Vector and PLVX-Puro-LINC02535, respectively. lipofectamine 2000 was used, and the DNA: liposome ratio was 1:4. The cells were further cultured for two days, and then drug screening was performed with G418.
CCK-8 (Cell Counting Kit-8) assay
Cells were seeded at a density (3.0 × 103 cells per well) in a 96-well plate, and each group had three replicate wells. 10 μL of CCK-8 (Sigma-Aldrich, St. Louis, MO, USA) was added to each well at 0 hours, 24 hours, 48 hours, and 72 hours. After incubation for 4 hours, the absorbance at 450 nm of each well was measured on a microplate reader.
Colony-forming assay
Cells were seeded at a density (3.0 × 103 cells per well) in a 6-well plate. After culturing for about 7 days, the cells were fixed with methanol for 30 minutes and then stained with 0.1% crystal violet (Beyotime, Shanghai, China) for 12 hours. After gentle rinsing twice with PBS (Phosphate buffered saline), we took pictures and counted the number of formed clones.
Transwell assays
Matrigel (BD Science, Sparks, MD, USA) was pre-spread (the migration experiment does not have this step). The cells were added into the upper chamber of a transwell, a 24 well plate containing 200 μL of 1% serum medium, with 3.0 × 104 cells per well. Then, 800 μL 10% serum medium of RPMI 1640 culture medium was added into the lower chamber. After culture for 48 hours, the upper chamber was removed and rinsed twice. Then, we gently wiped the upper layer of the chamber with a wet cotton swab without passing through the cells. The cells were fixed with methanol for 30 minutes and then stained with 0.1% crystal violet (Beyotime, Shanghai, China) for 12 hours. After gentle rinsing twice with PBS, three fields were randomly selected for counting the cells out of the chamber.
RNA extraction and qRT-PCR
Total RNA was extracted using RNAiso Plus (Takara, Osaka, Japan) and reverse transcribed to cDNA using a reverse transcription kit (Takara, Osaka, Japan) according to the manufacturer’s protocol. qRT-PCR was performed following the instructions of qRT-PCR reagents (Takara, Osaka, Japan). Primers are as follows: LINC02535,5′-GGCTGGTTGTGGTGGCTCATG-3(forward)and5′-TTGCGATATTGCCCAGGCTTGTC-3′(reverse); GALNT3, 5′-AGGAGCAACAGTCGCAACAGC-3′ (forward)and5′-CGACAGCCGTGTAGTTCTCAGC-3′(reverse);β-actin,5′-CACAGAGCCTCGCCTTTGC-3′(forward)and5′-ACCCATGCCCACCATCACG-3′(reverse);
miR-30a-5p,5′-TGTAAACATCCTCGACTGGAAG-3′(forward)and5′-GCAGGGTCCGAGGTATTC-3′(reverse); U6, 5′-GGAACGATACAGAGAAGATTAGC-3′(forward) and 5′-TGGAACGCTTCACGAATTTGCG-3′(reverse).
RIP assay
Cells were seeded in a culture dish. When approaching 90% confluence, cells were lysed and collected with RIP lysis buffer (Millipore, Bedford, MA, USA). Then, 100 μL of lysis buffer and 900 μL of RIP buffer with anti-Ago2 or anti-IgG antibody (Millipore, Bedford, MA, USA) for coupling magnetic beads (Millipore, Bedford, MA, USA) were added. Cells were incubated overnight and then centrifuged at 800rpm for 3 minutes. The lower layer of precipitate was the desired RNA.
Dual-luciferase reporter assay
Wild-type (WT) and mutant-type (MUT) LINC02535/GALNT3 luciferase reporter gene plasmids (Genewiz, Suzhou, China), miR-NC and miR-30a-5p mimics were co-transfected into the cell by using lipofectamine 2000, and each group was set up with three replicate wells to continue the culture. After transfection for 48 hours, the relative luciferase activity was measured by the instructions.
Western blot
Total protein was extracted from cells using RIPA lysis buffer and protease inhibitors. Use of antibodies and concentrations as follows: anti-GALNT3 (ProteinTech Group, Chicago, IL, USA, lot: 16,716-1-AP) (at a 1:1,000 dilution), anti-MUC1 (ProteinTech Group, Chicago, IL, USA, lot: 19,976-1-AP) (at a 1:1,000 dilution), anti-MUC1-C (ProteinTech Group, Chicago, IL, USA, lot: 23,614-1-AP) (at a 1:1,000 dilution), anti-NF-κB (Cell Signaling Technology, Danvers, MA, USA, lot: 8242S) (at a 1:1,000 dilution), anti-p-NF-κB (Cell Signaling Technology, Danvers, MA, USA, lot: 3033S) (at a 1:1,000 dilution), anti-Lamin B1 (ProteinTech Group, Chicago, IL, USA, lot: 12,987-1-AP) (at a 1:5,000 dilution) and anti-β-actin (Cell Signaling Technology, Danvers, MA, USA, lot: 3700) (at a 1:2,000 dilution); anti-mouse (Cell Signaling Technology, Danvers, MA, USA, lot: 91186S) (at a 1:2,000 dilution) and anti-rabbit (Cell Signaling Technology, Danvers, MA, USA, lot: 98164S) (at a 1:2,000 dilution) secondary antibodies.
In vivo tumor xenograft mouse model
Twelve female BALB/c nude mice (age: 4 weeks old, weighting 16–20 g) were acquired from GemPharmatech (Nanjing, China). Mice were group-housed (3 per cage) and maintained group-housed on a 12 h/12 h light dark cycle at 25°C under specific pathogen-free (SPF) conditions. Care was taken to minimize pain and discomfort for the animals. A549 cells that stable LINC02535-knockout and the control group were diluted into a cell suspension of 4.0 × 107 cells/mL. 100 μL of cell suspension was taken and inoculated into the armpit of the left upper limb of BALB/c nude mice. The body weight and tumor size were measured every day, and tumor volume was calculated according to the following formula: Tumor volume = shortest tumor diameter2 × longest tumor diameter/2. Twenty-six days later, the mice were anesthetized by 2% sodium pentobarbital (B005, Jiancheng, Nanjing) at 50 mg/kg and sacrificed by cervical dislocation, subsequent to which the tumor tissues of all mice were harvested and weighed. In order to minimize the subjective bias, we will ensure that evaluation and statistical analysis are performed by the researchers who were unaware of the grouping. All animal experiments were conducted following the guidelines for the care and use of experimental animals issued by the experimental animal center of Soochow University.
Lectin pull-down assay
The Tn antigen in glycoprotein was detected using Vicia lectin (EY Laboratories, San Mateo, CA, USA) agarose beads (Sigma-Aldrich, St. Louis, MO, USA). Cell lysate protein (0.5 mg) was incubated with 30 μL VVA-conjugated agarose beads at 4°C for 16 hours. The lectin/glycoprotein complex was collected by centrifugation (10,000 rpm, 1 minute) and then boiled for 5 minutes. The protein was precipitated for western blot.
Cellular fractionation
A protein extraction kit was purchased from TransGen Biotech (TransGen Biotech, Beijing, China) for use in cellular fraction experiments. Generally, cells were first lysed with cytoplasmic protein extraction buffer and then with extraction buffer to obtain the fraction.
Statistical analysis
GraphPad prism 8.0 software was used for statistical analysis. All data were compared as the mean ± standard deviation for continuous variable data between the two groups, and a t-test was used for comparison.
Results
Bioinformatics analysis and expression verification of LINC02535
TCGA-LUAD data (https://portal.gdc.cancer.gov/repository) showed that LINC02535 was significantly overexpressed in LUAD tissues ()) (normal = 54, tumor = 497), and a high expression level was significantly related to the poor prognosis of patients ()). Differential expression analysis and survival analysis of lncRNAs in the heatmap show that LINC02535 is one of the most potential carcinogens (). In addition, the expression level of LINC02535 increased with the progression of TNM in LUAD patients ()) and was closely related to stage grading ()). The results showed that the expression level of LINC02535 in LUAD tissues was higher than that in adjacent normal tissues ()). qRT-PCR showed that compared with the BEAS-2B cell line, the expression level of LINC02535 was significantly increased ()). These results indicate that LINC02535 is highly expressed in LUAD tissues and cells.
Figure 1. Bioinformatic analysis and expression verification of LINC02535 in LUAD. (a) A heat map was built based on the TCGA database: red indicates high expression, green indicates low expression, black means no difference. (b) Survival analysis of LINC02535 in LUAD. (c) LINC02535 expression in different T stages of LUAD. (d) LINC02535 expression in different N stages of LUAD. (e) LINC02535 expression in different M stages of LUAD. (f) LINC02535 expression in different stages of LUAD. (g) LINC02535 mRNA expression levels in patient samples (adjacent normal tissues = 40, tumor tissues = 40). (h) Expression of LINC02535 in BEAS-2B, A549, H1299 and H1650 cells (**p < 0.01, ***p < 0.001).
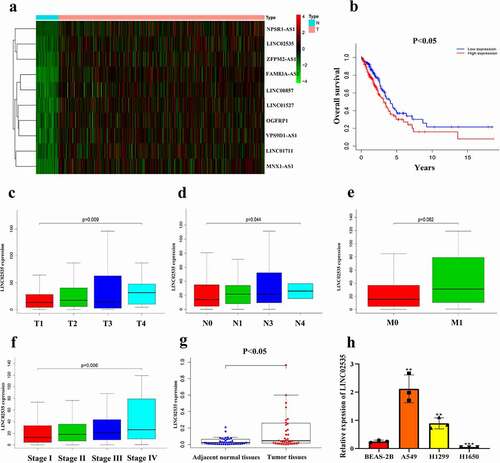
Table 1. Differential expression and survival analysis analysis of lncRNAs in the heatmap (FC = Fold change, HR = Hazard Rate).
Effect of LINC02535 on the malignant progression of LUAD cells in vitro
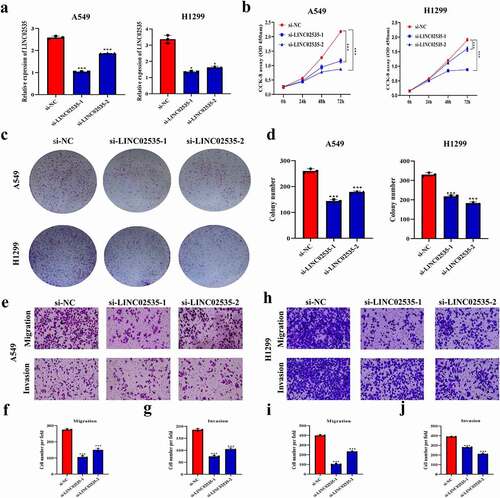
qRT-PCR showed that the expression level of LINC02535 decreased significantly after transfection of si-LINC02535 ()). CCK-8 and colony-forming assays showed that the cell proliferation ability significantly reduced after transfection with si-LINC02535 ()). Transwell assays showed that the migration and invasion ability of cells transfected with si-LINC02535 decreased significantly ()).
Figure 2. Effect of LINC02535 on the malignant progression of LUAD cells in vitro. (a) Interference with LINC02535 in A549 and H1299 cells (*p < 0.05, ***p < 0.001). (b) CCK-8 assay detecting the proliferation of LUAD cells (***p < 0.001). (c,d) Colony-forming assay detecting the proliferation of LUAD cells (***p < 0.001). (e-j) Transwell assays detect the migration and invasion of LUAD cells (***p < 0.001).
LINC02535 regulates miR-30a-5p as ceRNA
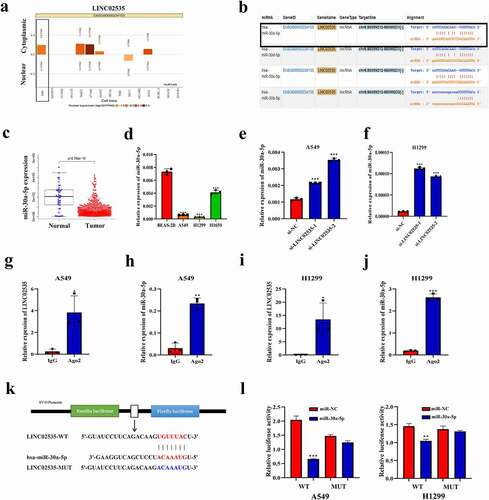
The LncATLAS database (http://lncatlas.crg.eu/) predicts that LINC02535 is mainly located in the cytoplasm of the A549 cell line ()). StarbaseV3.0 database (http://starbase.sysu.edu.cn/) prediction shows that LINC02535 has binding sites with the miR-30 family ()). Through the analysis of TCGA data, we found that miR-30a-5p was downregulated in LUAD and that the fold change was the largest (), Supplementary Figure S1(a-c)) (normal = 45, tumor = 483). qRT-PCR showed that compared with the BEAS-2B cell, the expression level of miR-30a-5p in A549 and H1299 cells was significantly decreased ()). After interference with LINC02535, the expression level of miR-30a-5p increased significantly ()). qRT-PCR showed that the expression level of miR-30a-5p increased or decreased substantially after transfection of miR-30a-5p mimics or inhibitor (Supplementary Figure S1(d,e)). After upregulation or downregulation with miR-30a-5p, the expression level of LINC02535 was significantly decreased or increased (Supplementary Figure S1(f,g)). RIP assay showed that LINC02535 binds to miR-30a-5p through Ago ()). According to the Starbase V3.0 database, plasmids containing wild-type sequences of LINC02535 binding site to miR-30a-5p and mutation sequence corresponding to mutation site were designed ()). Compared with the negative control group, the luciferase activity of the co-transfected LINC02535 WT plasmid decreased significantly by upregulating the expression level of miR-30a-5p ()).
Figure 3. LINC02535 regulates miR-30a-5p as ceRNA. (a) The subcellular location of LINC02535. (b) The binding sites of LINC02535 and miRNA. (c) TCGA-LUAD data analysis of the differential expression of miR-30a-5p. (d) Expression of miR-30a-5p in cells (***p < 0.001). (e,f) Expression of miR-30a-5p detected by qRT-PCR after siRNA-mediated knockdown of LINC02535 (***p < 0.001). (g-j) RIP assay proving that LINC02535 and miR-30a-5p bind to each other (*p < 0.05, **p < 0.01, ***p < 0.001). (k) Construction of the wild-type sequence plasmid (LINC02535l-WT) and mutant sequence plasmid (LINC02535-MUT) containing binding sites of LINC02535 and miR-30a-5p. (l) Dual-luciferase reporter assay proving that LINC02535 and miR-30a-5p bind to each other (**p < 0.01, ***p < 0.001).
miR-30a-5p inhibits LUAD progression by regulating GALNT3
Different online databases (TargetScan: http://www.targetscan.org/; PicTar: http://pictar.mdc-berlin.de/;miRTarBase:http://mirtarbase.mbc.nctu.edu.tw/index.html;miRanda:http://www.microrna.org/microrna/home.do;PITA:http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html.) were used to predict the possible target genes of miR-30a-5p ()). Co-expression analysis with LINC02535 (cor>0.3, p < 0.05) to screen mRNAs by using TCGA-LUAD data, combined with the possible target genes of miR-30a-5p, we got 17 candidate genes (Supplementary Figure S1(h)). Differential expression analysis and survival analysis of candidate genes are shown in . The expression level of GALNT3 in LUAD tissues was higher than that in adjacent normal tissues (Supplementary Figure S1(i)). There was a significant correlation between the expression of LINC02535 and GALNT3 in patient samples (r = 0.44, p < 0.05) (Supplementary Figure S1(j)). The binding site of miR-30a-5p to GALNT3 was predicted using the Starbase V3.0 database ()). According to the UALCAN database (http://ualcan.path.uab.edu/index.html), the results showed that the mRNA and protein expression levels of GALNT3 in LUAD tissues were higher than those in adjacent tissues ()). The Kaplan-Meier plotter online database (http://www.kmplot.com) suggested that the expression level of GALNT3 increases with the metastasis of LUAD ()). TCGA-LUAD data showed that high expression was significantly correlated with poor prognosis ()). The results of qRT-PCR and western blot showed that the expression level of GALNT3 in A549 and H1299 cells was significantly higher than that in the BEAS-2B cell line ()). In addition, miR-30a-5p mimics significantly inhibited the expression level of GALNT3 ()), and miR-30a-5p inhibitor significantly promoted the expression of GALNT3 ()). According to the StarbaseV3.0 online database, a plasmid containing the wild-type sequence of miR-30a-5p and the GALNT3 binding site and the mutation sequence corresponding to the mutation site was designed ()). Compared with the negative control group, the luciferase activity co-transfected with GALNT3-WT plasmid decreased significantly by upregulating the expression level of miR-30a-5p ()).
Figure 4. miR-30a-5p inhibits LUAD progression by regulating GALNT3. (a) Online database predicting possible downstream target genes of miR-30a-5p. (b) The binding sites of miR-30a-5p and GALNT3. (c) Online database predicting the differences in mRNA and protein expression levels of GALNT3 (p < 0.05). (d) Changes in the expression of GALNT3 with LUAD metastasis were analyzed by the online database (p < 0.05). (e) Survival analysis of GALNT3 in LUAD. (f) GALNT3 expression was detected by qRT-PCR and western blot (***p < 0.001). (g-j) miR-30a-5p regulated the mRNA and protein expression verification of GALNT3 (*p < 0.05,***p < 0.001). (k) Construction of the wild-type sequence plasmid (GALNT3-WT) and mutant sequence plasmid (GALNT3-MUT) containing miR-30a-5p and GALNT3 binding sites. (l) Dual-luciferase reporter assay proving that miR-30a-5p and GALNT3 bind to each other (*p < 0.05, ***p < 0.001).
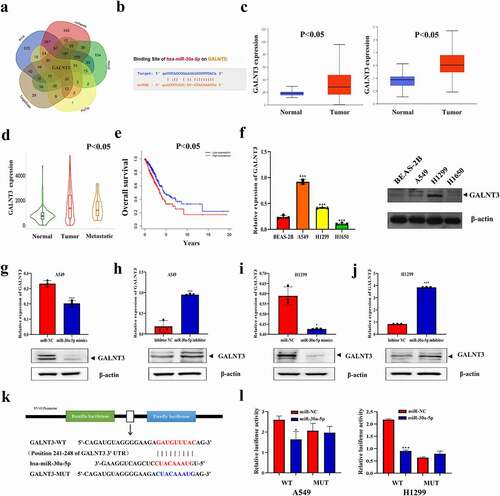
Table 2. Differential expression and survival analysis of 17 target genes (FC = Fold change, HR = Hazard Rate).
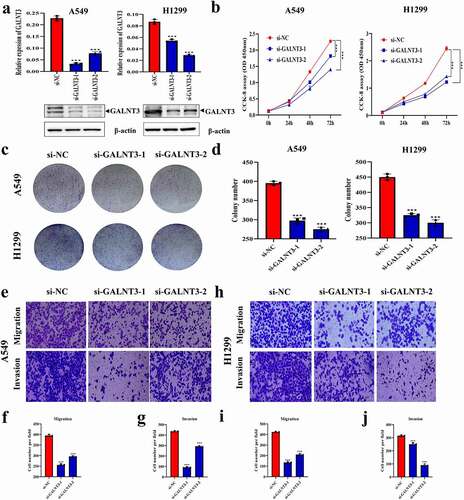
Effect of GALNT3 on the malignant progression of LUAD cells in vitro
qRT-PCR and western blot showed that the expression level of GALNT3 in A549 and H1299 cells decreased significantly after transfection of si-GALNT3 ()). CCK-8 and colony-forming assays showed that the proliferation of A549 and H1299 cells reduced considerably after transfection with si-GALNT3 ()). Transwell assays showed that the motility of A549 and H1299 cells was significantly inhibited after transfection with si-GALNT3 ()).
Figure 5. Effect of GALNT3 on the malignant progression of LUAD cells in vitro. (a) Interference with GALNT3 in A549 and H1299 cells (***p < 0.001). (b) CCK-8 assay detecting the changes in proliferation of LUAD cells (***p < 0.001). (c,d) Colony-forming assay detecting the changes in proliferation of LUAD cells (***p < 0.001). (e-j) Transwell assays detect the changes in migration and invasion of LUAD cells (***p < 0.001).
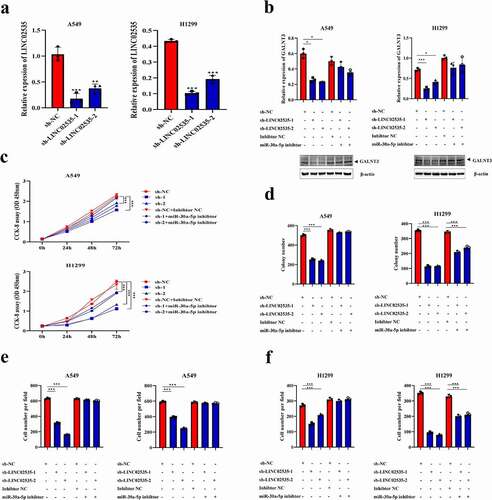
LINC02535/miR-30a-5p/GALNT3 axis contributes to LUAD malignant progression
LINC02535 stable knockdown cells were established ()). qRT-PCR and western blot showed that transfection of miR-30a-5p inhibitor could reverse the down-regulation of GALNT3 induced by sh-LINC02535 ()). CCK-8 ()), colony-forming () and Supplementary Figure S2(a)), and transwell assays () and Supplementary Figure S2(b)) showed that miR-30a-5p inhibitor reversed the inhibitory effect of sh-LINC02535 on cell proliferation, migration, and invasion. The expression level of LINC02535 in the OE-LINC02535 group was significantly higher than that in the PLVX group (Supplementary Figure S3(a)). Overexpression of LINC02535 resulted in down-regulation of miR-30a-5p expression (Supplementary Figure S3(b)). qRT-PCR and western blot showed that transfection of miR-30a-5p mimics could reverse the upregulation of GALNT3 induced by OE-LINC02535 (Supplementary Figure S3(c)). qRT-PCR showed that overexpression of miR-30a-5p resulted in decreased LINC02535 expression (Supplementary Figure S3(d)). CCK-8 (Supplementary Figure S3(e)) and transwell assays (Supplementary Figure S3(f,h)) showed that miR-30a-5p mimics reversed the promotion of cell proliferation, migration, and invasion induced by OE-LINC02535.
Figure 6. LINC02535/miR-30a-5p/GALNT3 axis contributes to LUAD malignant progression. (a) Construction of the LINC02535 was stably interfered with in A549 and H1299 cells (**p < 0.01, ***p < 0.001). (b) The changes in GALNT3 mRNA and protein expression levels in LUAD cells with different expression states of LINC02535 and miR-30a-5p (*p < 0.05, ***p < 0.001). (c) CCK-8 assay detecting the changes in proliferation of LUAD cells (***p < 0.001). (d) Colony-forming assay detecting the changes in proliferation of LUAD cells (***p < 0.001). (e,f) Transwell assays detect the changes in migration and invasion of LUAD cells (***p < 0.001).
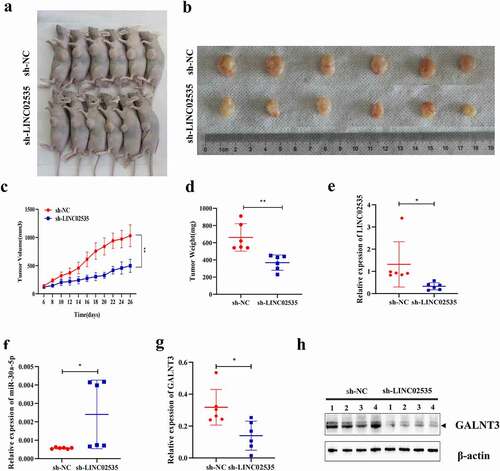
Effect of LINC02535 on the tumorigenesis ability of A549 cell line in vivo
Tumor formation experiments in nude mice showed that sh-LINC02535 significantly inhibited the tumor formation ability of the A549 cell line in nude mice ()). The tumor volume and weight in the sh-LINC02535 group were significantly smaller than those in the sh-NC group ()). qRT-PCR showed that LINC02535 and GALNT3 decreased considerably in the sh-LINC02535 group ()), and the expression of miR-30a-5p was significantly increased ()). Western blot showed that the expression of GALNT3 decreased substantially in the sh-LINC02535 group ()).
Figure 7. Effect of LINC02535 on the tumorigenesis ability of A549 cell line in vivo. (a,b) A549-LINC02535 stable interference cells and control cells were used to perform subcutaneous tumor formation assays in nude mice. Then, the nude mice were sacrificed to take pictures, and the tumors were removed to take pictures. (c) Growth curve of nude mice (**p < 0.01). (d) Weigh the tumor of nude mice (**p < 0.01). (e) qRT-PCR was detecting the LINC02535 expression level (*p < 0.05). (f) qRT-PCR was detecting the miR-30a-5p expression level (*p < 0.05). (g) qRT-PCR detecting the GALNT3 expression level (*p < 0.05). (h) Western blot detecting the GALNT3 expression level. N = 4 per group.
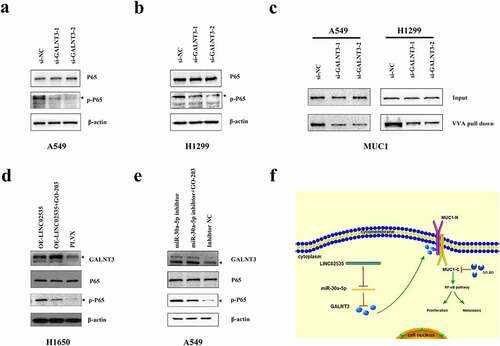
GALNT3 activates the NF-κB signaling pathway through glycosylation of MUC1
TCGA-LUAD data suggested that GALNT3 may regulate the NF-κB signaling pathway (Supplementary Figure S4(a)). After transfection with si-GALNT3, the expression of p-NF-κB in LUAD cells was significantly reduced ()). The STRING database (https://string-db.org/) indicated that GALNT3 might bind to MUC1 (Supplementary Figure S4(b)). O-glycosylated glycan-binding lectin VVA agarose pull-down was performed, and western blot was performed. A549 and H1299 cells transfected with si-GALNT3 showed decreased VVA binding MUC1 ()). GO-203, a specific inhibitor of MUC1-C. Using GO-203 based on OE-LINC02535, we found that p-NF-κB expression could be partially down-regulated ()). Using GO-203 based on miR-30a-5p inhibitor, we found that p-NF-κB expression could be partially down-regulated () and Supplementary Figure S4(c)). To further confirm the effect of MUC1 on NF-κB signaling pathway, we transfected si-MUC1 into A549 and H1299 cells. After transfection with si-MUC1, the expression of C-terminus of MUC1 (MUC1-C) and p-NF-κB in LUAD cells was significantly reduced (Supplementary Figure S4(d)). After transfection with si-MUC1, the cellular fractionation experiment was performed, and the results showed that inhibition of MUC1 significantly inhibited the expression of NF-κB in the nuclear compared with the control group (Supplementary Figure S4(d)). LINC02535 is highly expressed in LUAD tissues and cells, as a ceRNA, it interacts with miR-30a-5p to up-regulate the expression of GALNT3, enhance the function of MUC1 and activate the NF-κB signaling pathway, promoting proliferation, migration, and invasion of LUAD ()).
Figure 8. GALNT3 activates the NF-κB signaling pathway through glycosylation of MUC1. (a,b) Western blot detecting the effects of GALNT3 on the NF-κB signaling pathway. (c) Western blot detecting the effects of GALNT3 on the MUC1. (d) GO-203 was used based on the overexpression LINC02535. (e) GO-203 was used based on inhibition of miR-30a-5p. (f) Pattern diagram.
Discussion
LncRNA is a promising therapeutic target found in recent years, which is closely related to the occurrence and progression of tumors [Citation14]. With the discovery of new lncRNA and the study of its internal mechanism, lncRNA is expected to provide clues for follow-up research and clinical practice. Exploring the mechanism of lncRNAs in malignant tumors is conducive to the early diagnosis, treatment, and improvement of the prognosis of malignant tumors [Citation15].
The function of lncRNA in tumors depends on its subcellular localization. Determining whether lncRNA is located in the cytoplasm or nuclear is of great significance for the follow-up study of the mechanism of lncRNA [Citation16]. Generally, if lncRNA is mainly located in the cytoplasm, it adsorbs the corresponding miRNA through the sponge and then regulates the expression of downstream mRNA to act as an oncogene or tumor suppressor gene [Citation17]; if lncRNA is mainly located in the nuclear, it generally works by binding to specific proteins [Citation18]. According to the online database, we found that LINC02535 is mainly located in the cytoplasm of the A549 cell line. The database indicates that the miR-30a family is most likely to bind to LINC02535. Considering our previous research results, miR-30a-5p was selected [Citation10]. Ago2 is a key component of the RNA-induced silencing complex, and miRNAs play a role in regulating gene silencing by binding Ago2 [Citation19]. The RIP assay indicated that LINC02535 and miR-30a-5p could interact with each other.
The mechanism of ceRNA belongs to the most common mechanism of lncRNA. Han X et al. reported a new lncRNA, UPLA1, which is highly expressed in non-small cell lung cancer (NSCLC). UPLA1 promotes the proliferation, migration, and invasion of NSCLC cells. UPLA1 activates the Wnt/β-catenin signal pathway. YY1 acts as a transcription factor to inhibit the expression of UPLA1 [Citation20]. Ma P et al. found that knockdown of LINC00152 can inhibit the proliferation and invasion of hepatocellular carcinoma (HCC) cells. Mechanistically, LINC00152 regulates Cyclin D1 by sponging miR-193a/b-3p to act as an oncogene [Citation21]. There are three other common modes of action of lncRNAs. The first mode of action is to regulate the transcription of downstream genes. From the organism’s perspective, the use of RNA to regulate transcription has obvious advantages because the use of RNA regulation does not involve protein translation, so it has a better response speed. Certain acute reactions in organisms may make the response faster. LncRNA TPT1-AS1 has been proven to be mainly distributed in the nuclear of ovarian cancer (OC) cells and positively regulates the activity and transcription of the TPT1 promoter, and TPT1-AS1 induces the growth and metastasis of OC through TPT1 [Citation22]. The second mode of action of lncRNA is the binding of lncRNA to protein and then the localization of the protein complex to a specific DNA sequence. LncRNA HOXD-AS3 promotes downstream HDAC5 transcription by binding to YBX1 and plays a key role in the pathogenesis of gastric cancer [Citation23]. LncRNA can also serve as a “central platform”; in tumor cells, a variety of transcription factors can bind to this lncRNA molecule. On the other hand, when multiple signal pathways are activated simultaneously, these downstream effectors can bind to the same lncRNA molecule so that the information between different signal pathways can be cross integrated. This is conducive to the cell and organism producing feedback and regulating external signals and stimuli. In HCC, lncRNA SPRY4-IT1 also acts as a molecular platform to connect the transcription factor SP1 with the downstream EZH2/LSD1/DNMT1 complex and miR-101-3p, playing an important role in the malignant progression of HCC [Citation24].
To further investigate the detailed regulatory mechanism downstream of miR-30a-5p, we used a public database to predict the target genes of miR-30a-5p. Through literature research, we found that GALNT3 has not been reported in the LUAD. GALNT3 belongs to the glycosyltransferase family [Citation25]. There are few studies on GALNT3 in tumors [Citation26].
The online database predicted that MUC1 might be the glycosylation target of GALNT3. MUC1 is a mucin-like transmembrane glycoprotein, a type I transmembrane protein encoded by MUC1 [Citation27]. After translation, MUC1 is cut into two fragments in the endoplasmic reticulum, and the N-terminus is secreted to the outside of the cell membrane to form an extracellular region, modified with O-type glycosidic bonds [Citation28]. The C-terminus includes the extracellular region, the transmembrane region, and the intracellular region [Citation29]. MUC1 plays an important role in epithelial renewal and differentiation, maintaining of epithelial integrity, tumorigenesis, and metastasis [Citation30]. In many malignancies, changes in the glycosylation of MUC1 lead to its highly abnormal expression [Citation31]. The transmembrane C-terminus of MUC1 (MUC1-C) acts as an oncoprotein that can interact with multiple signaling pathways [Citation32]. MUC1 can affect the NF-κB signaling pathway and promote the occurrence and development of a variety of malignant tumors, but no relevant reports have been reported in LUAD [Citation33]. We preliminarily demonstrated the role of MUC1 in NF-κB signaling pathway in LUAD. Hopefully, we will explore its deeper mechanism in the future.
Conclusions
LINC02535, as a ceRNA, inhibits the expression of miR-30a-5p, up-regulates the expression of GALNT3, enhances the function of MUC1, activates the NF-κB signaling pathway.
Author contributions
JAH and ZYL designed the experiments. YL, JZ, and MJ analyzed the data and wrote the manuscript. WJZ, AQW, XC, and JJZ provided helpful discussion and reviewed the manuscript. All authors reviewed the manuscript.
Ethics approval and consent to participate
This study was approved by the Academic Advisory Board of Soochow University under approval number 2020-375. All animal experiments were carried out under the Guide for the Care and Use of Experimental Animals from the Experimental Animal Center of Soochow University.
Consent for publication
Consent for publication was obtained from the participants.
Supplemental Material
Download Zip (55.1 MB)Acknowledgments
We thank all patients who participated in this study for their cooperation and Dr. Hua-Long Qin and Dr. Jun Chen of the First Affiliated Hospital of Soochow University, China, for their clinical sample collection.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2101336
Additional information
Funding
References
- Relli V, Trerotola M, Guerra E, et al. Abandoning the notion of non-small cell lung cancer. Trends Mol Med. 2019;25(7):585–594.
- Zhang Z, Gu M, Gu Z, et al. Role of long non-coding RNA polymorphisms in cancer chemotherapeutic response. J Pers Med. 2021;12(1):11.
- Qiu B, Wang D, and Li Q, et al. Concurrent chemoradiation therapy with or without nimotuzumab in locally advanced squamous cell lung cancer: a phase 2 randomized trial. Int J Radiat Oncol Biol Phys. 2021;111(4):917–925.
- Chi A, He X, Hou L, et al. Classification of non-small cell lung cancer’s tumor immune micro-environment and strategies to augment its response to immune checkpoint blockade. Cancers (Basel). 2021;14(1):13.
- Sung H, Ferlay J, Siegel R, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249.
- Kozłowska J, Kolenda T, Poter P, et al. Long intergenic non-coding RNAs in HNSCC: from “Junk DNA” to important prognostic factor. Cancers (Basel). 2021;14(1):13.
- Wang W, Min L, Qiu X, et al. Biological function of long non-coding RNA (LncRNA) xist. Front Cell Dev Biol. 2021;9:645–647.
- Wen D, Huang Z, Li Z, et al. LINC02535 co-functions with PCBP2 to regulate DNA damage repair in cervical cancer by stabilizing RRM1 mRNA. J Cell Physiol. 2020;235(10):7592–7603.
- Leão R, Albersen M, Looijenga L, et al. Circulating MicroRNAs, the next-generation serum biomarkers in testicular germ cell tumors: a systematic review. Eur Urol. 2021;80(4):456–466.
- Zhu J, Zeng Y, Li W, et al. CD73/NT5E is a target of miR-30a-5p and plays an important role in the pathogenesis of non-small cell lung cancer. Mol Cancer. 2017;16(1):34.
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358.
- Zhao M, Feng J, Tang L. Competing endogenous RNAs in lung cancer. Cancer Biol Med. 2021;18(1):1–20.
- Sahasrabudhe N, Lenos K, van der Horst J, et al. Oncogenic BRAFV600E drives expression of MGL ligands in the colorectal cancer cell line HT29 through N-acetylgalactosamine-transferase 3. Biol Chem. 2018;399(7):649–659.
- Di Fiore R, Suleiman S, Felix A, et al. An overview of the role of long non-coding RNAs in human choriocarcinoma. Int J Mol Sci. 2021;23(1):22.
- Statello L, Guo C, Chen L, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118.
- Mosca L, Vitiello F, Borzacchiello L, et al. Mutual correlation between non-coding RNA and S-adenosylmethionine in human cancer: roles and therapeutic opportunities. Cancers (Basel). 2021;14(1):13.
- Li S, Zhu Y, Li R, et al. LncRNA Lnc-APUE is repressed by HNF4α and promotes G1/S phase transition and tumor growth by regulating MiR-20b/E2F1 axis. Adv Sci( Weinheim, Baden-Wurttemberg, Germany). 2021;8(7):2003094.
- Luo J, Wang H, Wang L, et al. lncRNA GAS6-AS1 inhibits progression and glucose metabolism reprogramming in LUAD via repressing E2F1-mediated transcription of GLUT1. Mol Ther Nucleic Acids. 2021;25:11–24.
- Tien J, Chugh S, and Goodrum A, et al. AGO2 promotes tumor progression in KRAS-driven mouse models of non-small cell lung cancer. Proc Natl Acad Sci USA. 2021;118(20).
- Han X, Jiang H, Qi J, et al. Novel lncRNA UPLA1 mediates tumorigenesis and prognosis in lung adenocarcinoma. Cell Death Dis. 2020;11(11):999.
- Ma P, Wang H, Sun J, et al. LINC00152 promotes cell cycle progression in hepatocellular carcinoma via miR-193a/b-3p/CCND1 axis. Cell Cycle (Georgetown, Tex.). 2018;17(8):974–984.
- Wu W, Gao H, Li X, et al. Lnc RNA TPT 1-AS1 promotes tumorigenesis and metastasis in epithelial ovarian cancer by inducing TPT1 expression. Cancer Sci. 2019;110(5):1587–1598.
- Shi S, Jiang J, Zhang W, et al. A novel lncRNA HOXC-AS3 acts as a miR-3922-5p sponge to promote breast cancer metastasis. Cancer Invest. 2020;38(1):1–12.
- Xu Y, Yao Y, Jiang X, et al. SP1-induced upregulation of lncRNA SPRY4-IT1 exerts oncogenic properties by scaffolding EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma. J Exp Clin Cancer Res. 2018;37(1):81.
- Lixin S, Wei S, Haibin S, et al. miR-885-5p inhibits proliferation and metastasis by targeting IGF2BP1 and GALNT3 in human intrahepatic cholangiocarcinoma. Mol Carcinog. 2020;59(12):1371–1381.
- Sheta R, Bachvarova M, and Macdonald E, et al. The polypeptide GALNT6 displays redundant Functions upon suppression of its closest homolog GALNT3 in mediating aberrant O-Glycosylation, associated with ovarian cancer progression. Int J Mol Sci. 2019;20(9):2264.
- Chen W, Zhang Z, Zhang S, et al. MUC1: structure, function, and clinic application in epithelial cancers. Int J Mol Sci. 2021;23(1):22.
- Mao Y, Zhang Y, Fan S, et al. GALNT6 promotes tumorigenicity and metastasis of breast cancer cell via β-catenin/MUC1-C signaling pathway. Int J Biol Sci. 2019;15(1):169–182.
- Khodabakhsh F, Merikhian P, Eisavand M, et al. Crosstalk between MUC1 and VEGF in angiogenesis and metastasis: a review highlighting roles of the MUC1 with an emphasis on metastatic and angiogenic signaling. Cancer Cell Int. 2021;21(1):200.
- Supruniuk K, Czarnomysy R, Muszyńska A, et al. Combined action of anti-MUC1 monoclonal antibody and pyrazole-platinum(II) complexes reveals higher effectiveness towards apoptotic response in comparison with monotherapy in AGS gastric cancer cells. Pharmaceutics. 2021;14(1):13.
- Hagiwara M, Fushimi A, Yamashita N, et al. MUC1-C activates the PBAF chromatin remodeling complex in integrating redox balance with progression of human prostate cancer stem cells. Oncogene. 2021;40(30):4930–4940.
- Yamashita N, Long M, Fushimi A, et al. MUC1-C integrates activation of the IFN-γ pathway with suppression of the tumor immune microenvironment in triple-negative breast cancer. J Immunother Cancer. 2021;9(1):e002115.
- Ahmad R, Raina D, Trivedi V, et al. MUC1 oncoprotein activates the IkappaB kinase beta complex and constitutive NF-kappaB signalling. Nat Cell Biol. 2007;9(12):1419–1427.
