ABSTRACT
Circular RNAs (circRNAs) have shown pivotal regulatory roles in multiple human ocular diseases, including age-related cataract (ARC). Here, we explored the role of circRNA mitogen-activated protein kinase kinase kinase 4 (circMAP3K4, hsa_circ_0078619) in ARC pathology and its associated mechanism. The expression of RNAs and proteins was examined by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot assay. Cell viability, senescence, proliferation, and apoptosis were analyzed by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, senescence-associated-β-galactosidase (SA-β-Gal) staining, 5-ethynyl-20-deoxyuridine (EdU) assay, and flow cytometry. The oxidative stress status of SRA01/04 cells was analyzed using the commercial kits. The interaction between microRNA-193a-3p (miR-193a-3p) and circMAP3K4 or phospholipase C delta 3 (PLCD3) was verified by dual-luciferase reporter assay, RNA immunoprecipitation (RIP) assay, and RNA-pull down assay. CircMAP3K4 was significantly down-regulated in ARC patients and H2O2-induced SRA01/04 cells. H2O2 treatment restrained the viability and proliferation and promoted the senescence, apoptosis, and oxidative stress of SRA01/04 cells, and circMAP3K4 overexpression protected SRA01/04 cells from H2O2-induced dysfunction. MiR-193a-3p was a direct target of circMAP3K4, and circMAP3K4 overexpression-mediated protective effects in H2O2-induced SRA01/04 cells were largely reversed by the accumulation of miR-193a-3p. MiR-193a-3p interacted with the 3’ untranslated region (3’UTR) of PLCD3, and PLCD3 knockdown largely overturned miR-193a-3p silencing-induced protective effects in H2O2-induced SRA01/04 cells. CircMAP3K4 up-regulated the expression of PLCD3 via sponging miR-193a-3p in SRA01/04 cells. In conclusion, circMAP3K4 protected SRA01/04 cells from H2O2-induced dysfunction in ARC through mediating miR-193a-3p/PLCD3 axis.
Introduction
Cataract is a vital cause of visual impairment and blindness [Citation1]. The pathology of age-related cataract (ARC) is related to multiple factors, containing aging, diabetes, genetics, oxidative stress, and UV exposure [Citation2]. Surgical intervention is the most effective therapeutic strategy for ARC. Nevertheless, surgical intervention may lead to severe postoperative complications [Citation3]. Therefore, understanding the pathogenesis of ARC is important to prevent or delay the progression of this disease. Accumulating evidence have pointed out that the senescence, apoptosis, and oxidative injury of human lens epithelial cells (HLECs) are important pathological foundation of non-congenital cataracts [Citation4–6]. Here, we explored the molecular mechanism behind ARC pathology using H2O2-induced HLECs.
Circular RNAs (circRNAs), a class of non-coding RNA with circular structure, have been confirmed to play important roles in human diseases [Citation7,Citation8]. Accumulating studies demonstrated that circRNAs can regulate cell biological behaviors via acting as sponges for microRNAs (miRNAs) [Citation9,Citation10]. Meanwhile, miRNAs can modulate gene expression by inducing messenger RNA (mRNA) degradation or suppressing mRNA translation [Citation11]. Previous studies have associated the aberrant expression of circRNAs with the pathology of several ocular diseases [Citation12–14]. Reduced expression of circRNA mitogen-activated protein kinase 4 (circMAP3K4, hsa_circ_0078619) has been reported in ARC lens samples in comparison with that in the transparent lens samples [Citation15]. However, the role of circMAP3K4 in ARC pathology remains to be disclosed.
The purpose of this study is to explore the role and mechanism of circMAP3K4 in ARC progression. Through predicting the downstream miRNA/mRNA axis of circMAP3K4 by bioinformatics analysis, we found that circMAP3K4 might interact with miR-193a-3p, and miR-193a-3p could bind to phospholipase C delta 3 (PLCD3) 3’UTR. Therefore, we proposed the hypothesis of circMAP3K4/miR-193a-3p/PLCD3 axis and tested it. Our study hopes to provide a potential molecular target for the treatment of ARC.
Materials and methods
Human lens epithelium tissues
Human lens epithelium tissues were collected from normal eyes (n = 15; free of ocular diseases) within 8 h after donor death and ARC patients (n = 15; free of other ocular diseases; grade 4 to 6). These lens epithelium tissues were obtained during cataract surgery at The Fifth Affiliated Hospital of Zhengzhou University by an experienced surgeon. All samples were immediately stored at −80°C. Clinical study was authorized by the Ethical Committee of The Fifth Affiliated Hospital of Zhengzhou University. All the participants or their families had signed the written informed consent before sample collection. The clinical features of ARC patients and normal controls are listed in .
Table 1. Clinical features of ARC patients and normal controls.
Cell culture
HLEC SRA01/04 was obtained from Shanghai Academy of Life Science (Shanghai, China) and cultured in DMEM (Sigma, St. Louis, MO, USA) added with 10% FBS (Sigma) and 1% penicillin/streptomycin (Sigma) mixture under the standard condition (37°C; 5% CO2). For mimic ARC cell model in vitro, SRA01/04 cells were treated with different concentrations (10, 20, 50 and 100 µM) of H2O2 for 24 h or 50 µM H2O2 for different times (6, 12, 24 and 48 h).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA samples were prepared with TRIzol reagent (Invitrogen, Waltham, MA, USA). M-MLV reverse transcriptase kit (Invitrogen) and miRNA cDNA Synthesis Kit (GeneCopoeia, Rockville, MD, USA) were adopted to synthesize cDNA. Afterward, cDNA was amplified via the Power SYBR-Green PCR master mix (Applied Biosystems, Foster, CA, USA). The relative abundance was determined by the 2−ΔΔCt formula=ΔCt value for target gene-ΔCt value for the average of internal gene (β-actin or U6). The primers are shown in .
Table 2. Specific primers for RT-qPCR.
3-(4,5-Dimethylthiazol-2-Yl)-2,5-Diphenyltetrazolium bromide (MTT) assay
SRA01/04 cells were seeded into 96-well plates (1 × 105 cells). After cultured for 48 h, cells were mixed with 20 µL MTT solution (Sigma) for 4 h. Subsequently, 150 µL DMSO (Sigma) was pipetted to the wells to dissolve the formazan crystals. The absorbance was examined at 570 nm.
RNase R treatment
RNA samples (2 µg) isolated from SRA01/04 cells were mixed with 3 U/µg RNase R (Epicenter Technologies, Madison, WI, USA) for 15 min, and then RNA samples were then subjected to RT-qPCR assay.
Actinomycin D treatment
SRA01/04 cells were incubated with 2 mg/mL Actinomycin D (Sigma) for the indicated time points (0, 4, 8, 12, and 24 h). Then, RNA samples were isolated with TRIzol reagent and were subjected to RT-qPCR analysis.
Cell transfection
CircMAP3K4 overexpression plasmid (circMAP3K4), empty vector (Vector), miR-193a-3p mimics (miR-193a-3p), miRNA negative control (miR-NC), miR-193a-3p inhibitor (in-miR-193a-3p), in-miR-NC, PLCD3 overexpression plasmid (PLCD3), empty vector (pcDNA), small interfering (si)RNA targeting PLCD3 (si-PLCD3), and si-NC were acquired from GenePharma (Shanghai, China). LipofectamineTM 3000 (Invitrogen) was used to introduce 40 nM small RNAs or 2 µg plasmids into SRA01/04 cells. After transfection for 24 h, cells were stimulated with 50 µM H2O2 for 24 h.
Senescence-associated-β-Galactosidase (SA-β-Gal) staining
According to the instructions of SA-β-gal staining kit (Beyotime, Shanghai, China) SRA01/04 cells were seeded on 6-well plates and treated with 50 µM H2O2 for 96 h. Cells were incubated with 4% paraformaldehyde, incubated with freshly prepared SA-β-Gal stain solution overnight. Stained cells were analyzed by microscopy to count SA-β-gal positive cell rate.
5-ethynyl-2’-deoxyuridine (EdU) assay
SRA01/04 cells were seeded onto the 96-well plates (1 × 105 cells). After cultured for 24 h, cells were mixed with 50 μM EdU reagent (Ribobio, Guangzhou, China). After fixing with 4% formaldehyde for 30 min and incubating with 0.1% Triton X-100 solution for 20 min, the nucleus of SRA01/04 cells was stained with DAPI (Sigma). The fluorescent images were captured under a fluorescence microscope (Olympus, Tokyo, Japan).
Flow cytometry
Annexin V-FITC/PI kit (BD Biosciences, Shanghai, China) was adopted in this assay. Transfected SRA01/04 cells (5 × 105 cells) were simultaneously incubated with 10 µL Annexin V-FITC and PI for 15 min. The apoptosis of SRA01/04 cells was assessed.
Western blot assay
SRA01/04 cells were disrupted with RIPA solution (Beyotime). A total of 30 µg protein samples were loaded onto the separating gel and then shifted to a PVDF membrane (Millipore, Billerica, MA, USA). After blocked with non-skimmed milk, the membrane was labeled with the primary antibodies overnight at 4°C, including anti-Bcl-2 (ab182858; Abcam, Cambridge, MA, USA), anti-Bax (ab32503; Abcam), anti-PLCD3 (ab151594; Abcam), and anti-β-actin (ab8226; Abcam). After washing, the membrane was labeled with the secondary antibody (Abcam). Finally, the immuno-reactive bands were detected using an ECL kit (Pierce, Waltham, MA, USA).
Oxidative stress analysis
The level of MDA and the activities of two oxidative stress-associated key enzymes (SOD and GSH-PX) were examined using the commercial kits (Jiancheng Biotech, Nanjing, China) according to the manufacturer’s instructions.
Dual-luciferase reporter assay
The interacted miRNAs of circMAP3K4 and the interacted mRNAs of miR-193a-3p were sought by starbase database (http://starbase.sysu.edu.cn). The partial sequence of circMAP3K4 or PLCD3, including the WT or MUT base-pairing sequence with miR-193a-3p, was inserted into the pmirGLO vector (Promega, Shanghai, China). SRA01/04 cells at 24-well plates were co-introduced with WT or MUT luciferase reporter and miR-193a-3p mimics/miR-NC for 48 h. The luciferase activities were determined using a commercial kit (Promega).
RNA immunoprecipitation (RIP) assay
SRA01/04 cells were lyzed with RIP buffer, and then cell extracts were co-immuno-precipitated with magnetic beads that pre-coated with anti-AGO2 or anti-IgG. The immuno-precipitated RNAs were isolated and subjected to RT-qPCR assay.
RNA pull-down assay
The biotinylated-circMAP3K4 probe was incubated with C-1 magnetic beads (Life Technologies, Carlsbad, CA, USA) to obtain probe-covered beads. Subsequently, cell extracts were incubated with probe-covered beads overnight at 4°C. The immuno-precipitated RNAs were subjected to RT-qPCR assay.
Statistical analysis
Figures were produced using GraphPad Prism 7.0 software and the results were expressed as mean ± standard deviation (SD). The differences were assessed by Student’s t-test and analysis of variance (ANOVA). P < 0.05 was set as the threshold of significance.
Results
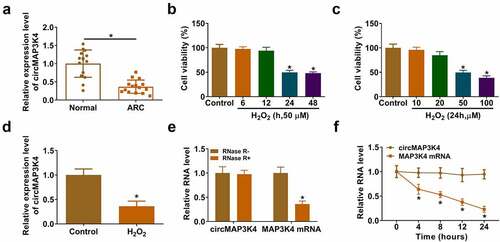
CircMAP3K4 expression is down-regulated in ARC patients and H2O2-induced SRA01/04 cells
To explore the biological role of circMAP3K4 in ARC pathology, we first analyzed the expression pattern of circMAP3K4 in the lens epithelium tissues of ARC patients (n = 15) and normal controls (n = 15). We found that circMAP3K4 expression was down-regulated in ARC patients compared with normal controls (). Then, we analyzed the optimal concentration and time of H2O2 treatment in SRA01/04 cells. The results showed that the cell viability was significantly decreased at 24 h and 48 h after treatment with 50 µM H2O2 for different times (). After 24 h of treatment with different concentrations of H2O2, the cell viability was significantly decreased at 50 µM and 100 µM (). Therefore, the cells treated with 50 µM of H2O2 for 24 h had the best effect. The expression of circMAP3K4 was decreased in SRA01/04 cells treated with 50 µM H2O2 for 24 h (). We analyzed the stability of circMAP3K4 with exonuclease RNase R and transcriptional inhibitor Actinomycin D. The results showed that circMAP3K4 was resistant to RNase R (), and the expression of circMAP3K4 was stable upon Actinomycin D treatment (), suggesting that circMAP3K4 was a circular transcript. Taken together, circMAP3K4 expression might be associated with ARC pathology.
Figure 1. CircMAP3K4 expression is down-regulated in ARC patients and H2O2-induced SRA01/04 cells. (a) RT-qPCR was conducted to determine the level of circMAP3K4 in the lens epithelium tissues of ARC patients (n = 15) and normal controls (n = 15). (b and c) SRA01/04 cells were treated with H2O2 at different doses or at different time points. Cell viability was analyzed by MTT assay. (d) The level of circMAP3K4 was detected by RT-qPCR in SRA01/04 cells treated with 50 µm H2O2 for 24 h. (e) RNA samples were treated with RNase R or not, and the RNA levels of circMAP3K4 and its linear counterpart MAP3K4 were measured by RT-qPCR. (f) SRA01/04 cells were treated with Actinomycin D for different time points, and the expression of circMAP3K4 and MAP3K4 was measured by RT-qPCR. *P < 0.05.
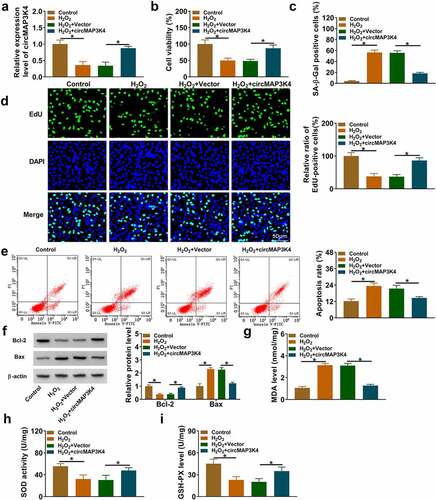
CircMAP3K4 overexpression protects SRA01/04 cells from H2O2-induced dysfunction
To investigate the molecular mechanism behind the abnormal down-regulation of circMAP3K4 in H2O2-induced SRA01/04 cells, we performed gain-of-function experiments. H2O2 treatment reduced the expression of circMAP3K4, and the addition of circMAP3K4 overexpression plasmid largely rescued the level of circMAP3K4 in SRA01/04 cells (). MTT assay revealed that H2O2 treatment reduced the viability of SRA01/04 cells, which was largely recovered by the addition of circMAP3K4 plasmid (). In addition, H2O2 treatment enhanced SA-β-Gal positive cell rate, while this effect could be abolished by circMAP3K4 overexpression (). EdU assay showed that H2O2 treatment suppressed the percentage of EdU-positive cells, and the introduction of circMAP3K4 plasmid largely recovered the proliferation ability of SRA01/04 cells (). H2O2 treatment-induced cell apoptosis, and circMAP3K4 overexpression attenuated the apoptosis of SRA01/04 cells (). We also determined the expression of anti-apoptotic protein Bcl-2 and pro-apoptotic protein Bax. Western blot assay showed that H2O2 stimulation-mediated effects on the expression of Bax and Bcl-2 were largely overturned by the addition of circMAP3K4 plasmid (), further demonstrating that circMAP3K4 overexpression alleviated H2O2-induced apoptosis in SRA01/04 cells. H2O2 stimulation promoted the production of MDA and the suppressed the activities of SOD and GSH-PX, and these effects were largely reversed by the overexpression of circMAP3K4 (). Overall, circMAP3K4 overexpression protected SRA01/04 cells from H2O2-induced dysfunction.
Figure 2. CircMAP3K4 overexpression protects SRA01/04 cells from H2O2-induced dysfunction. (a-i) SRA01/04 cells were transfected with Vector or circMAP3K4 plasmid followed by 50 µm H2O2 treatment. (a) RT-qPCR was conducted to measure the expression of circMAP3K4 in SRA01/04 cells. (b) Cell viability was analyzed by MTT assay. (c) SA-β-Gal staining was used to measure cell senescence. (d) EdU assay was conducted to analyze the proliferation ability of SRA01/04 cells. (e) Flow cytometry was conducted to assess the apoptosis of SRA01/04 cells. (f) Western blot assay was performed to determine the protein expression of anti-apoptotic protein Bcl-2 and pro-apoptotic protein Bax in SRA01/04 cells. (g-i) the oxidative stress of SRA01/04 cells was analyzed using the commercial kits. *P < 0.05.
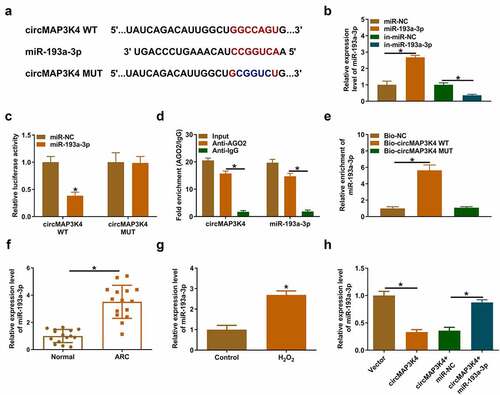
CircMAP3K4 acts as a molecular sponge for miR-193a-3p in SRA01/04 cells
It is well established that circRNAs can act as miRNA sponges to regulate cell biological behaviors [Citation9,Citation16]. To unveil the working mechanism of circMAP3K4, we predicted the downstream miRNAs of circMAP3K4 using bioinformatics database starbase. MiR-193a-3p was one of the predicted target of circMAP3K4, and the putative binding sites between circMAP3K4 and miR-193a-3p are shown in . RT-qPCR confirmed the overexpression and knockdown efficiencies of miR-193a-3p mimics and in-miR-193a-3p in SRA01/04 cells (). Then, we conducted dual-luciferase reporter assay, RIP assay, and RNA-pull down assay to verify the target relationship between circMAP3K4 and miR-193a-3p. As shown in , the overexpression of miR-193a-3p markedly reduced the luciferase activity of wild-type vector (circMAP3K4 WT) but not that of mutant vector (circMAP3K4 MUT), suggesting that circMAP3K4 interacted with miR-193a-3p via the predicted sites. RIP assay displayed that circMAP3K4 and miR-193a-3p expression were enriched in AGO2 antibody-associated RISC (), suggesting the spatial interaction between circMAP3K4 and miR-193a-3p. RNA-pull down assay showed that miR-193a-3p was pulled down when using biotin-labeled circMAP3K4WT probe (), further demonstrating the interaction between circMAP3K4 and miR-193a-3p. The expression of miR-193a-3p was notably up-regulated in ARC patients and H2O2-induced SRA01/04 cells (). CircMAP3K4 overexpression reduced the expression of miR-193a-3p, and the addition of miR-193a-3p mimics rescued the expression of miR-193a-3p level in SRA01/04 cells (). Taken together, miR-193a-3p was a direct target of circMAP3K4 in SRA01/04 cells.
Figure 3. CircMAP3K4 acts as a molecular sponge for miR-193a-3p in SRA01/04 cells. (a) The interacted miRnas of circMAP3K4 were predicted by starbase database, and miR-193a-3p was predicted as one of the candidate targets of circMAP3K4. (b) RT-qPCR was conducted to analyze the overexpression and knockdown efficiencies of miR-193a-3p and in-miR-193a-3p, respectively. (c-e) the interaction between circMAP3K4 and miR-193a-3p was verified by dual-luciferase reporter assay, RIP assay, and RNA-pull down assay. (f) RT-qPCR was conducted to determine the expression of miR-193a-3p in the lens epithelium tissues of ARC patients (n = 15) and normal controls (n = 15). (g) the level of miR-193a-3p in SRA01/04 cells treated with 50 µm H2O2 for 24 h was examined by RT-qPCR. (H) SRA01/04 cells were transfected with circMAP3K4 plasmid alone or together with miR-193a-3p mimics. The expression of miR-193a-3p was examined by RT-qPCR. *P < 0.05.
CircMAP3K4 overexpression-mediated protective effects in H2O2-induced SRA01/04 cells by sponging miR-193a-3p
Considering the target relationship between circMAP3K4 and miR-193a-3p, we further explored their functional correlation in H2O2-induced SRA01/04 cells. CircMAP3K4 overexpression reduced the expression of miR-193a-3p, and the addition of miR-193a-3p mimics largely recovered this in H2O2-induced SRA01/04 cells (). CircMAP3K4 overexpression protected SRA01/04 cells from H2O2-induced dysfunction (). Furthermore, we observed that the addition of miR-193a-3p reduced the viability, enhanced SA-β-Gal positive cell rate, and suppressed the proliferation of SRA01/04 cells again (). The introduction of miR-193a-3p also induced the apoptosis and oxidative stress of SRA01/04 cells (). These results suggested that circMAP3K4-mediated protective effects in H2O2-induced SRA01/04 cells were largely based on the down-regulation of miR-193a-3p.
Figure 4. CircMAP3K4 overexpression-mediated protective effects in H2O2-induced SRA01/04 cells by miR-193a-3p. (a–i) SRA01/04 cells were transfected with circMAP3K4 plasmid alone or together with miR-193a-3p mimics followed by 50 µm H2O2 treatment for 24 h. (a) RT-qPCR was conducted to determine the expression of miR-193a-3p in SRA01/04 cells. (b) Cell viability was analyzed by MTT assay. (c) SA-β-Gal staining was performed to assess cell senescence. (d) Cell proliferation ability was assessed by EdU assay. (e) Flow cytometry was conducted to analyze the apoptosis of SRA01/04 cells. (f) the protein levels of Bax and Bcl-2 were determined by western blot assay. (g-i) the level of MDA and the activities of SOD and GSH-PX were assessed using the commercial kits. *P < 0.05.
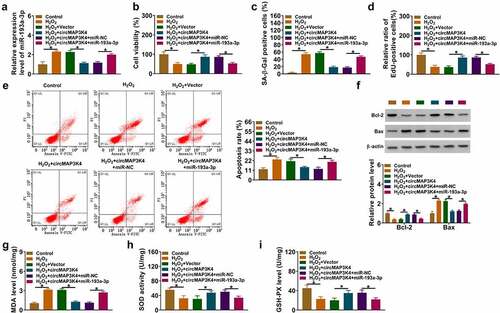
MiR-193a-3p interacts with the 3’UTR of PLCD3 in SRA01/04 cells
Previous articles have demonstrated that miRNAs can interact with the 3”UTR of mRNAs, thereby leading to their degradation or translational repression [Citation11,Citation17]. To explore the working mechanism by which miR-193a-3p regulated the biological behaviors of H2O2-induced SRA01/04 cells, we predicted the possible mRNA targets of miR-193a-3p using starbase database. PLCD3 was predicted as one of the candidate target of miR-193a-3p, and their putative binding sequences were shown in . The luciferase activity of wild-type plasmid (PLCD3 3”UTR WT) was dramatically reduced by the overexpression of miR-193a-3p (), indicating the direct interaction between miR-193a-3p and the 3’UTR of PLCD3. The mRNA and protein expression of PLCD3 was decreased in ARC patients compared with normal controls (). H2O2 stimulation down-regulated the protein level of PLCD3 in SRA01/04 cells (). MiR-193a-3p overexpression reduced the protein level of PLCD3, and this effect could be abolished by PLCD3 overexpression plasmid (). CircMAP3K4 overexpression up-regulated the protein level of PLCD3, and the addition of miR-193a-3p reduced the protein expression of PLCD3 in SRA01/04 cells (). These results demonstrated that PLCD3 was a direct target of miR-193a-3p, and its expression was positively regulated by circMAP3K4 in SRA01/04 cells.
Figure 5. MiR-193a-3p interacts with the 3’UTR of PLCD3 in SRA01/04 cells. (a) Bioinformatics database starbase was used to predict the interacted mRnas of miR-193a-3p, and PLCD3 was predicted as a possible target of miR-193a-3p. (b) the target relationship between miR-193a-3p and PLCD3 was verified by dual-luciferase reporter assay. (c and d) The mRNA and protein expression of PLCD3 was determined in the lens epithelium tissues of ARC patients and normal controls (n = 15) by RT-qPCR and Western blot assay. (e) Western blot assay was performed to analyze the protein level of PLCD3 in SRA01/04 cells treated with 50 µm H2O2 for 24 h. (f) SRA01/04 cells were transfected with miR-193a-3p alone or together with PLCD3 plasmid, and the protein level of PLCD3 was determined by western blot assay. (g) SRA01/04 cells were transfected with circMAP3K4 plasmid alone or together with miR-193a-3p mimics, and the protein level of PLCD3 was examined by Western blot assay. *P < 0.05.
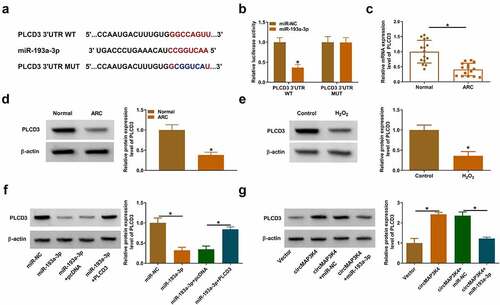
MiR-193a-3p knockdown protects SRA01/04 cells from H2O2-induced dysfunction largely by up-regulating PLCD3
To investigate the functional correlation between miR-193a-3p and PLCD3 in H2O2-induced SRA01/04 cells, we performed rescue experiments. The silence of miR-193a-3p up-regulated PLCD3 protein level, which was reduced by the addition of si-PLCD3 in SRA01/04 cells (). The knockdown of miR-193a-3p enhanced cell viability, suppressed SA-β-Gal positive cell rate, and promoted the proliferation of H2O2-induced SRA01/04 cells, while these effects were eliminated by PLCD3 knockdown (). The introduction of si-PLCD3 also reversed the suppressive effects of miR-193a-3p inhibitor on cell apoptosis rate, Bax protein level, MDA level, as well as the increasing effect on Bcl-2 protein level, SOD activity and GSH-PX level in H2O2-induced SRA01/04 cells (). Overall, these findings suggested that miR-193a-3p silencing protected SRA01/04 cells from H2O2-induced dysfunction largely by up-regulating PLCD3.
Figure 6. MiR-193a-3p knockdown protects SRA01/04 cells from H2O2-induced dysfunction largely by up-regulating PLCD3. (a-i) SRA01/04 cells were transfected with in-miR-193a-3p alone or together with si-PLCD3 followed by 50 µm H2O2 treatment for 24 h. (a) Western blot assay was conducted to measure the protein level of PLCD3 in SRA01/04 cells. (b) MTT assay was conducted to assess the viability of SRA01/04 cells. (c) Cell senescence was assessed using SA-β-Gal staining. (d) EdU assay was performed to analyze the proliferation of SRA01/04 cells. (e) Cell apoptosis was analyzed by flow cytometry. (f) Western blot assay was performed to detect the protein expression of Bcl-2 and Bax in SRA01/04 cells. (g-i) the oxidative stress of SRA01/04 cells was analyzed using the commercial kits. *P < 0.05.
Discussion
CircRNAs have attracted a lot of attention due to their important modulatory activities in many physiological and pathological processes, including ocular diseases [Citation18,Citation19]. ARC is featured by the accumulation of insoluble crystalline proteins and the abnormal death and oxidative damage of HLECs [Citation20]. Previous articles have demonstrated the functions of several circRNAs in ARC pathology. Cui et al. demonstrated that circHIPK3 exhibited a protective role in HLECs by absorbing miR-221-3p in ARC pathology [Citation21]. Xu et al. reported that circZNF292 overexpression alleviated H2O2-induced damage in HLECs through mediating the miR-222-3p/E2F3 axis [Citation22]. A previous article reported that circMAP3K4 abundance was decreased in ARC lens samples compared with the transparent lens samples [Citation15]. However, its role and molecular mechanism in regulating ARC pathology remain to be clarified. Consistent with former study [Citation15], we observed that circMAP3K4 abundance was markedly reduced in the lens epithelium samples of ARC cases than that in normal controls.
Lens epithelial cell monolayer is responsible for lens proliferation, differentiation and damage repair, which exerts vital protective effects in the transparency and the homeostasis in the lens [Citation23]. It has also to be remind that senescence is a leading cause of ARC that plays an important role in the pathological process of ARC [Citation24,Citation25]. Here, we investigated the function of circMAP3K4 in H2O2-induced HLECs injury in vitro. We found that H2O2 treatment reduced circMAP3K4 level in SRA01/04 cells. H2O2 exposure inhibited cell viability and proliferation capacity and triggered cell senescence, apoptosis and oxidative stress in SRA01/04 cells, and gain-of-function experiments showed that circMAP3K4 overexpression protected SRA01/04 cells from H2O2-induced dysfunction.
CircRNAs have shown pivotal regulatory roles in gene expression by acting as miRNA sponges [Citation16]. We identified miR-193a-3p as a direct target of circMAP3K4 for the first time. Liu et al. demonstrated that circHIPK3 exerts a protective role in HLECs by enhancing CRYAA expression via absorbing miR-193a in ARC [Citation15], indicating that miR-193a facilitates the pathological progress of ARC. We observed that miR-193a-3p abundance was elevated in the lens epithelium tissues of ARC patients and H2O2-induced SRA01/04 cells. CircMAP3K4 negatively regulated miR-193a-3p expression in SRA01/04 cells. The silence of miR-193a-3p protected SRA01/04 cells from H2O2-induced dysfunction, suggesting that miR-193a-3p contributed to ARC pathology. Furthermore, we found that circMAP3K4 overexpression-mediated protective effects in H2O2-induced SRA01/04 cells were largely reversed by miR-193a-3p overexpression, indicating that circMAP3K4 protected SRA01/04 cells from H2O2-induced dysfunction largely by down-regulating miR-193a-3p.
miRNAs can interact with the 3’UTR of mRNAs to induce their degradation or translational repression [Citation11]. Here, PLCD3 was identified as a downstream target of miR-193a-3p for the first time. PLCD3 abundance was decreased in the lens epithelium samples of ARC cases and H2O2-exposed SRA01/04 cells. PLCD3 is a member of the PLC family, a vital enzyme in the phosphoinositide signaling. PLC isozymes are involved in the pathology of multiple human diseases, including schizophrenia [Citation26], diabetes [Citation27], obesity [Citation28], and tumorigenesis [Citation29,Citation30]. Liu et al. demonstrated that PLCD3 plays a pro-tumor activity in nasopharyngeal carcinoma by facilitating cell proliferation and motility [Citation31]. Xiang et al. demonstrated that upregulation of PLCD3 expression restrained the progression of ARC [Citation32]. We found that PLCD3 was reversely modulated by miR-193a-3p in SRA01/04 cells. In addition, circMAP3K4 can positively regulate the expression of PLCD3 by serving as miR-193a-3p sponge. The results of compensation assays demonstrated that miR-193a-3p silencing-mediated protective effects in H2O2-induced SRA01/04 cells were largely counteracted by PLCD3 interference, indicating that miR-193a-3p silencing protected SRA01/04 cells from H2O2-induced dysfunction largely by up-regulating PLCD3.
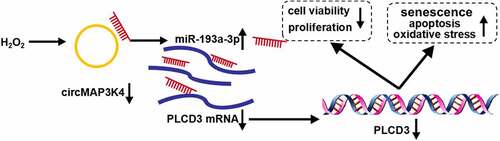
In conclusion, circMAP3K4 was down-regulated in ARC patients and H2O2-induced SRA01/04 cells. CircMAP3K4 overexpression protected SRA01/04 cells from H2O2-induced dysfunction. Furthermore, we found that circMAP3K4 overexpression-mediated protective role was largely based on miR-193a-3p/PLCD3 axis (). This study provided several novel potential targets for the prevention and therapy of ARC.
Supplemental Material
Download PDF (2.2 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2114587
Additional information
Funding
References
- Congdon NG, Friedman DS, Lietman T. Important causes of visual impairment in the world today. JAMA. 2003;290(15):2057–2060. Epub 2003/10/16 PubMed PMID: 14559961. DOI:10.1001/jama.290.15.2057.
- Asbell PA, Dualan I, Mindel J, et al. Age-related cataract. Lancet. 2005;365(9459):599–609. Epub 2005/02/15 PubMed PMID: 15708105. DOI:10.1016/s0140-6736(05)17911-2.
- Watkinson S, Seewoodhary R. Cataract management: effect on patients’ quality of life. Nursing standard (Royal College of Nursing (Great Britain) : 1987). 2015;29(21):42–48. Epub 2015/01/22 PubMed PMID: 25605115. DOI:10.7748/ns.29.21.42.e9222.
- Gattoussi S, Cougnard-Grégoire A, Delyfer MN, et al. Vitreomacular adhesion and its association with age-related macular degeneration in a population-based setting: the alienor study. Invest Ophthalmol Visual Sci. 2017;58(4):2180–2186. Epub 2017/04/12 PubMed PMID: 28399268. DOI:10.1167/iovs.16-20741.
- Spector A. Review: oxidative stress and disease. J Ocul Pharmacol Ther. 2000;16(2):193–201. Epub 2000/05/10 PubMed PMID: 10803430. DOI:10.1089/jop.2000.16.193.
- Yan Y, Yu H, Sun L, et al. Laminin alpha4 overexpression in the anterior lens capsule may contribute to the senescence of human lens epithelial cells in age-related cataract. Aging (Albany NY). 2019;11(9):2699–2723. PubMed PMID: 31076560; PubMed Central PMCID: PMC6535067. DOI:10.18632/aging.101943.
- Zhao X, Cai Y, Xu J. Circular RNAs: biogenesis, mechanism, and function in human cancers. Int J Mol Sci. 2019;20(16):3926. PubMed PMID: 31412535; PubMed Central PMCID: PMC6720291. DOI:10.3390/ijms20163926.
- Zhou WY, Cai ZR, Liu J, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172. PubMed PMID: 33317550; PubMed Central PMCID: PMC7734744. DOI:10.1186/s12943-020-01286-3.
- Panda AC. Circular RNAs act as miRNA sponges. advances in experimental medicine and biology. 2018;1087: 67–79. Epub 2018/09/28 PubMed PMID: 30259358. DOI:10.1007/978-981-13-1426-1_6.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. Epub 2019/08/10 PubMed PMID: 31395983. DOI:10.1038/s41576-019-0158-7.
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRnas. Annu Rev Biochem. 2010;79(1):351–379. Epub 2010/06/11 PubMed PMID: 20533884. DOI:10.1146/annurev-biochem-060308-103103.
- Shan K, Liu C, Liu BH, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136(17):1629–1642. Epub 2017/09/02 PubMed PMID: 28860123. DOI:10.1161/circulationaha.117.029004.
- Zhang SJ, Chen X, Li CP, et al. Identification and characterization of circular RNAs as a new class of putative biomarkers in diabetes retinopathy. Invest Ophthalmol Visual Sci. 2017;58(14):6500–6509. PubMed PMID: 29288268. DOI:10.1167/iovs.17-22698.
- Han J, Gao L, Dong J, et al. The expression profile of developmental stage-dependent circular RNA in the immature rat retina. Mol Vis. 2017;23: 457–469. Epub 2017/08/02. PubMed PMID: 28761319; PubMed Central PMCID: PMCPmc5524268.
- Liu X, Liu B, Zhou M, et al. Circular RNA HIPK3 regulates human lens epithelial cells proliferation and apoptosis by targeting the miR-193a/cryaa axis. Biochem Biophys Res Commun. 2018;503(4):2277–2285. PubMed PMID: 29959922. DOI:10.1016/j.bbrc.2018.06.149.
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. Epub 2014/01/17 PubMed PMID: 24429633; PubMed Central PMCID: PMCPmc4113481. DOI10.1038/nature12986.
- Cannell IG, Kong YW, Bushell M. How do microRnas regulate gene expression? Biochem Soc Trans. 2008;36(Pt 6):1224–1231. PubMed PMID: 19021530. DOI:10.1042/BST0361224.
- Zhang C, Hu J, Yu Y. CircRNA is a rising star in researches of ocular diseases. Frontiers in cell and developmental biology. 2020;8:850. PubMed PMID: 33015046; PubMed Central PMCID: PMC7494781. DOI:10.3389/fcell.2020.00850.
- Guo N, Liu XF, Pant OP, et al. Circular RNAs: novel promising biomarkers in ocular diseases. Int J Med Sci. 2019;16(4):513–518. Epub 2019/06/07 PubMed PMID: 31171902; PubMed Central PMCID: PMCPmc6535655. DOI:10.7150/ijms.29750.
- Michael R, Bron AJ. The ageing lens and cataract: a model of normal and pathological ageing. Philos Trans R Soc London Ser B Biol Sci. 2011;366(1568):1278–1292. PubMed PMID: 21402586; PubMed Central PMCID: PMC3061107. DOI:10.1098/rstb.2010.0300.
- Cui G, Wang L, Huang W. Circular RNA HIPK3 regulates human lens epithelial cell dysfunction by targeting the miR-221-3p/pi3k/akt pathway in age-related cataract. Exp Eye Res. 2020;198:108128. PubMed PMID: 32681842.
- Xu X, Gao R, Li S, et al. Circular RNA circZNF292 regulates H(2) O(2) -induced injury in human lens epithelial HLE-B3 cells depending on the regulation of the miR-222-3p/e2f3 axis. Cell Biol Int. 2021;45(8):1757–1767. Epub 2021/04/15 PubMed PMID: 33851769. DOI:10.1002/cbin.11615.
- Che X, Zhao Q, Li D. Expression of thioredoxin-2 in human lens epithelial cells with oxidative damage and its significance]. Zhong nan da xue xue bao Yi xue ban =. J Central South Univ Med Sci. 2018;43(3):253–259. Epub 2018/04/28 PubMed PMID: 29701186. DOI:10.11817/j.issn.1672-7347.2018.03.004.
- Liu J, Zhang J, Zhang G, et al. CircMRE11A_013 binds to UBXN1 and integrates ATM activation enhancing lens epithelial cells senescence in age-related cataract. Aging (Albany NY). 2021;13(4):5383–5402. PubMed PMID: 33508783; PubMed Central PMCID: PMC7950295. DOI:10.18632/aging.202470.
- Chen M, Zhang C, Zhou N, et al. Metformin alleviates oxidative stress-induced senescence of human lens epithelial cells via AMPK activation and autophagic flux restoration. J Cell Mol Med. 2021;25(17):8376–8389. PubMed PMID: 34296521; PubMed Central PMCID: PMC8419182. DOI:10.1111/jcmm.16797.
- Del Caño G G, Montaña M, Aretxabala X, et al. Nuclear phospholipase C-β1 and diacylglycerol LIPASE-α in brain cortical neurons. Adv Biol Regul. 2014;54: 12–23. Epub 2013/10/01 PubMed PMID: 24076015. DOI:10.1016/j.jbior.2013.09.003.
- Barker CJ, Li L, Köhler M, et al. β-Cell Ca(2+) dynamics and function are compromised in aging. Adv Biol Regul. 2015;57:112–119. Epub 2014/10/06 PubMed PMID: 25282681.
- Nakamura Y, Kanemarum K, Fukami K. Physiological functions of phospholipase Cδ1 and phospholipase Cδ3. Adv Biol Regul. 2013;53(3):356–362. Epub 2013/08/21 PubMed PMID: 23948486. DOI:10.1016/j.jbior.2013.07.003.
- Arteaga CL, Johnson MD, Todderud G, et al. Elevated content of the tyrosine kinase substrate phospholipase C-gamma 1 in primary human breast carcinomas. Proc Natl Acad Sci USA. 1991;88(23):10435–10439. Epub 1991/12/01 PubMed PMID: 1683701; PubMed Central PMCID: PMCPmc52943. DOI:10.1073/pnas.88.23.10435.
- Fu L, Qin YR, Xie D, et al. Characterization of a novel tumor-suppressor gene PLC delta 1 at 3p22 in esophageal squamous cell carcinoma. Cancer Res. 2007;67(22):10720–10726. Epub 2007/11/17 PubMed PMID: 18006814. DOI:10.1158/0008-5472.can-07-2411.
- Liu W, Liu X, Wang L, et al. PLCD3, a flotillin2-interacting protein, is involved in proliferation, migration and invasion of nasopharyngeal carcinoma cells. Oncol Rep. 2018;39(1):45–52. Epub 2017/11/09 PubMed PMID: 29115528; PubMed Central PMCID: PMCPmc5783603. DOI:10.3892/or.2017.6080.
- Xiang J, Chen Q, Kang L, et al. LncRNA PLCD3-OT1 functions as a CeRNA to prevent age-related cataract by sponging miR-224-5p and regulating PLCD3 expression. Invest Ophthalmol Visual Sci. 2019;60(14):4670–4680. Epub 2019/11/15 PubMed PMID: 31725166. DOI:10.1167/iovs.19-27211.