ABSTRACT
Accurate spatial and temporal regulation of cell cycle progression is essential for cell proliferation and organismic development. This review demonstrates the role of microspherule protein 58kD, commonly known as MCRS1, as a key cell cycle regulator of higher eukaryotic organisms. We discuss the isoforms and functional domains of MCRS1 as well as their subcellular localization at specific stages of the cell cycle. These molecular characteristics reveal MCRS1’s dynamic regulatory role in gene expression, genome stability, cell proliferation, and organismic development. Furthermore, we discuss the molecular details of its seemingly opposite, tumor-suppressive or tumor-promoting, role in different types of cancer.
1. Introduction
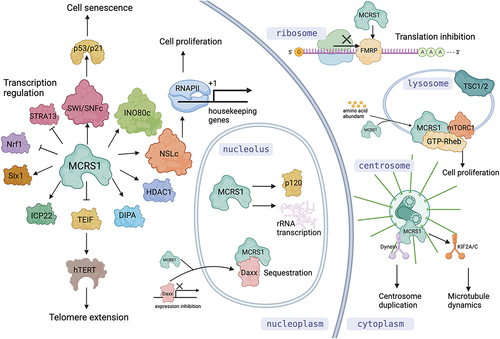
In eukaryotes, a cell cycle is composed of interphase, mitotic phase, and cytokinesis. Accurate regulation of cell cycle progression is essential for cell proliferation and organismic development. MCRS1 is one of the important cell cycle regulators. MCRS1 was first reported as a nucleolar protein whose overexpression was linked to tumorigenesis [Citation1,Citation2]. Since its discovery, many different binding partners of MCRS1 in multiple cellular pathways have been identified [Citation3–10]. The biological functions of MCRS1 are quite diverse because it participates in various pathways such as regulation of transcription factors for cell proliferation and stress response, histone posttranslational modification, mRNA targeting and translation, telomerase expression, senescence induction, mTOR pathway activation, centrosome integrity, and microtubule dynamics. ().
Figure 1. Diverse functions of MCRS1 and its isoforms in different subcellular compartments. In the nucleus, MCRS1 interacts with a variety of transcription factors, histone modifiers, and chromosome remodelling complexes to regulate cell proliferation and stress responses. In the nucleolus, it activates rRNA transcription and controls activity of transcription regulators by sequestration. In the cytosol, it binds to FMRP to regulate mRNA translation. On the cytoplasmic surface of lysosome, it interacts and activates mTOR pathway regulators. It also localizes at the centrosome for keeping its integrity and regulating k-fiber dynamics during mitosis. Figure produced by BioRender.
In this review, we summarize and discuss the studies of MCRS1 with its molecular characterization, major functions, regulatory mechanisms, and roles in developmental biology and tumorigenesis. We intend to provide a comprehensive understanding of the dynamic roles of MCRS1 and address the unanswered questions in the context of cell cycle regulation and genomic stability maintenance.
2. The molecular characteristics of MCRS1
Microspherule protein 58kD (MSP58) was first identified as a p120-associated protein localized at the nucleolar microspherules by the yeast two-hybrid screen [Citation2]. Since then, MSP58 has been reported to interact with a variety of factors functioning in gene expression, genome stability, cell proliferation, and tumorigenesis. The major functional domains of MSP58 include NLS (nuclear localization sequences), SANT (switching-defective protein 3, adaptor 2, nuclear receptor co-repressor, transcription factor TFIIIB), CC (coiled-coil), and FHA (forkhead-associated) domains (). These domains are highly conserved from zebrafish to humans (), and play important roles to regulate the function of MCRS1.
Figure 2. Functional domains of MCRS1. MCRS1 has NLS, SANT, CC, and FHA domains. The NLS1 regulate both nuclear and nucleolar localization of MCRS1. SANT domain binds directly to the histone H3 tail and recruit HDAC1 to control histone acetylation. CC and FHA domains mediate specific interaction with target proteins.
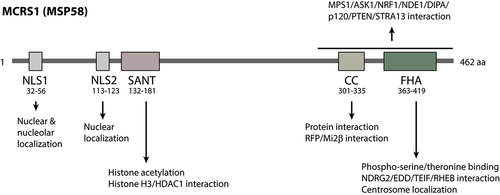
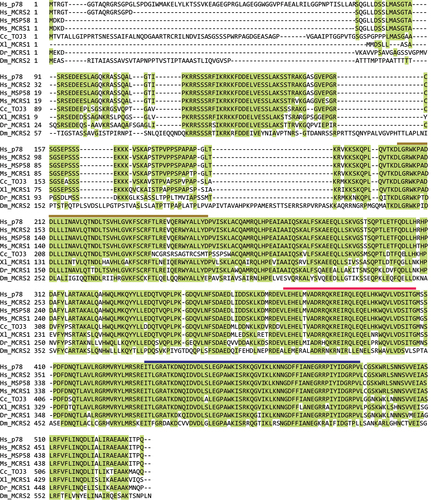
Figure 3. Multiple sequence alignment of MCRS1 orthologs by the MUSLCE algorithm of the SnapGene. Human MCRS1 isoforms (Hs_p78, Hs_MCRS2, and Hs_MSP58) and their orthologs (mouse Ms_MCRS1, quail Cc_TOJ3, frog Xl_MCRS1, zebrafish Dr_MCRS1, and fly Dm_MCRS2) are aligned based on sequence similarity. Highly conserved amino acid residues (>50%) are highlighted in green. SANT, CC, and FHA domains are marked with brown, red, and dark blue lines, respectively.
2.1. Isoforms of MCRS1 and their expression
In humans, MSP58 has three isoforms as a result of alternative splicing, and each transcript produces a polypeptide with a specific length [Citation8,Citation9]. The MSP58 transcript encodes a polypeptide of 462 amino acids, the p78 isoform contains 534 amino acids, and the MCRS2 isoform contains 475 amino acids (). MCRS1 is commonly used as the generic name for both MSP58 and p78, although some studies challenged the existence of p78 due to inconsistent sequencing data [Citation8,Citation9]. Thus, in this review, we use MCRS1 to specifically refer to MSP58.
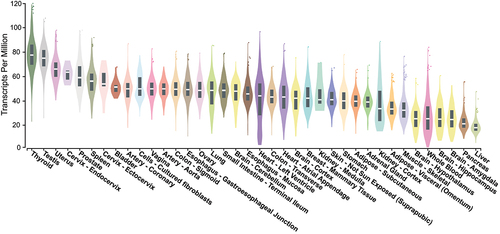
MCRS1 is ubiquitously expressed in most tissues () but the expression is differentially regulated in different cell cycle stages [Citation9,Citation11]. MCRS1 expression is induced to the maximal level during the early S phase and soon declines as cells progress to the G2 phase, suggesting its specific role in the S phase to promote cell cycle progression [Citation9,Citation11].
Figure 4. MCRS1 expression profile in different types of normal tissues. Tissue-specific expressions of MCRS1 from multiple studies were are as violin plots and boxplots (https://gtexportal.org). MCRS1 expression levels are relatively high in thyroid and testis tissues and low in livers and pancreas. The bottom of the box is the 25% quantile, the top of the box is the 75% quantile, and the middle line of the box is the median.
2.2. Subcellular localization of MCRS1
MCRS1 and its isoforms are localized to specific subcellular regions for distinct functions, including the nucleus, nucleolar microspherule, centrosome, centriolar satellite, lysosome, and cytoplasm. The nuclear localization is determined by the presence of nuclear localization sequences (NLS). MCRS1 contains two NLSs, of which NLS1 ranges from amino acids 32 to 56 and NLS2 ranges from amino acids 113 to 123 [Citation12]. Interestingly, NLS1 mutation leads to decreased rRNA expression, suggesting its alternative role as a nucleolar localization sequence (NoLS). However, in MCRS2, KRKK on amino acids 66–69 serves as an NLS and KKSK on amino acids 133–136 does as an NoLS [Citation13]. The CC and FHA domains near the C-terminus also seem to direct nucleolar localization as well. The centrosome localization of MCRS1 and its isoforms is mediated by the FHA domain [Citation3,Citation14,Citation15]. In humans, MCRS1 is specifically recruited to the minus end of mitotic spindles at the centrosome to regulate spindle dynamics [Citation14]. In Drosophila, dMCRS2 is localized at the centrosome as well as the telophase midbody [Citation16]. In zebrafish, MCRS1 is recruited to the centriolar satellite through the dynein complex [Citation17]. MCRS1 can also localize at the surface of late endosomes or lysosomes, which signals the activation of the mTORC1-Rheb GTPase axis when free amino acids are present [Citation18]. Other studies revealed the distribution of MCRS1 in the cytoplasm. In hippocampal neurons, MCRS1 colocalizes with FMRP in the cytoplasm, where they target mRNP to distal synapses for translation regulation [Citation5]. In HeLa cells, MCRS1 interacts with cytoplasmic ASK1 to regulate H2O2-mediated apoptosis [Citation19], and it also binds to Pkmyt1 for cell proliferation regulation in the cytoplasm of the gastric cancer cells [Citation20]. Overall, the spatial distribution in different subcellular compartments of MCRS1 and its isoforms demonstrates their diverse functions in the cells.
3. Cellular functions of MCRS1
3.1. MCRS1 in transcription and translation
Eukaryotic transcription by RNA polymerases is a complex process and is precisely controlled by DNA regulatory sequences and their associated transcription factors. Transcription factors specifically recognize and bind to regulatory sequences such as promoters and enhancers, which triggers the recruitment of coactivators to promote RNA polymerase binding and transcription initiation. Alternatively, the interaction between silencers and repressors recruits corepressors to inhibit the action of RNA polymerase [Citation21,Citation22]. Chromatin modifiers, chromosome remodeling complex, and mediator complex are also critical players to facilitate transcription initiation and elongation [Citation23,Citation24]. Chromatin modifiers covalently modify histone lysine residues by methylation, acetylation, ubiquitination, or sumoylation, and serine/threonine residues by phosphorylation [Citation25]. These modifications function as histone codes to direct recruitment of downstream regulators for controlling transcription [Citation23,Citation26]. After transcription, nascent RNAs are further spliced and transported to the cytosol for protein translation. All these processes require fine regulation, and MCRS1 is identified to play fundamental roles in them. Thus, we will discuss in detail how MCRS1 regulates various transcription factors, chromatin remodelling complex, histone acetyltransferases, rRNA transcription [Citation4,Citation6,Citation10,Citation11,Citation27–30], and mRNA translation [Citation5].
3.1.1 Regulating transcription factor activities
MCRS1 and MCRS2 regulate the activities of transcription factors and their binding partners in the nucleus and nucleolus. Lin et al. showed that MCRS1 inhibits Daxx activity by relocalization to the nucleolus [Citation10]. Daxx is a transcription factor for glucocorticoid receptor -dependent gene expression in the nucleus and Fas-induced apoptosis in the cytoplasm [Citation31]. When MCRS1 binds to Daxx, it redirects Daxx to the nucleolus for its inhibition. MCRS2 inhibits the transcription activity of RTA similarly [Citation32]. RTA promotes the expression of lytic genes during Epstein-Barr virus infections. MCRS2 relocates RTA to the nucleolus to prevent it from interacting with the viral genes in the nucleus.
Protein activities can be both positively and negatively regulated by MCRS1 and its isoforms in the nucleus. p78 binds to transcription factor ICP22 to regulate the expression of herpes simplex virus genes. Similarly, p78 binds to the nuclear protein DIPA to control the expression of the hepatitis delta virus genome [Citation4,Citation11]. MCRS2 binds to and inhibits Nrf1, a transcription factor involved in antioxidant response and embryonic development [Citation29]. Similarly, MCRS1 inhibits STRA13, a hypoxia-inducible bHLH transcription factor, to suppress various cell proliferation pathways [Citation8]. By contrast, MCRS1 binds to and activates transcription factor Six1 to promote proper craniofacial development during early embryogenesis [Citation33]. In addition, Xu et al. reported that MCRS1 interacts with ASK1 in the cytoplasm for its inhibition [Citation19].
The regulatory role of MCRS1 on rRNA transcription seems to be contradicted in two studies. Shimono et al. claimed that MCRS1 colocalizes with Mi2β, RFP, and UBF in the nucleolus to activate rRNA transcription [Citation6]. However, Yang et al. stated that MCRS1 inhibits rRNA transcription via RINT1, a Rad50-interacting protein that maintains genomic stability and cell homeostasis [Citation34]. The discrepancy may originate from the different genetic backgrounds of the cell lines used in these studies.
On the other hand, Andersen et al. showed that Drosophila dMCRS2 directly stimulates RNA polymerase II activity [Citation35]. dMCRS2 binds to both CDK11 and RNA polymerase II to enhance the expression of their target genes. The study further showed that dMCRS2 can specifically recruit MOF, an H4K16 acetyltransferase, to the promoter region of cyclin genes to enhance their expression [Citation35].
3.1.2 Regulating histone modifications
MCRS1 regulates histone tail modification and chromatin remodeling in the nucleus through several distinct pathways. In Drosophila, MOF is a major component of the dosage compensation complex that determines the male sex. Interestingly, mass spectrometric analysis of MOF identified the highly conserved NSL complex as a novel binding protein of MOF [Citation36]. The NSL subunit includes NSL1, NSL2, NSL3, WDS, and dMCRS2. It controls the expression of more than 4000 housekeeping genes by acetylating H4K16 within their promoters via MOF [Citation28,Citation37]. H4K16 acetylation, which is promoted by H4K20me2, activates gene expression via MLL4 H3K4 methyltransferase [Citation38,Citation39]. dMCRS2 depletion reduces the NSL complex stability and inhibits the recruitment of MOF and RNA polymerase II to the promoters of target genes [Citation28,Citation35]. Global genomic analyses identified the DNA replication-related element as the major target sequence of NSL-mediated RNA polymerase II recruitment [Citation40].
In humans, MCRS1 interacts with distinct chromosome remodeling units such as the INO80 complex, BRG1 ATPase of the SWI/SNF complex, and Mi-2β helicase of the NURD complex [Citation6,Citation27,Citation41]. Furthermore, the SANT domain of MCRS1 directly binds to histone H3 tail and recruits HDAC1 as well as Pontin/Reptin complex to regulate histone acetylation, which in turn controls bile acid transporter gene expression in liver cells [Citation42]. Pontin and Reptin are ATPases that function in various cellular activities including DNA replication, gene expression, DNA damage repair, telomere maintenance, mTOR activation, and microtubule dynamics [Citation43]. Interestingly, the HDAC1-mediated transcription regulation of MCRS1 seems to be independent of the INO80 or NSL-associated pathways. Altogether, these data suggest that MCRS1 interacts with distinct sets of chromatin modifiers to regulate gene expression.
3.1.3 Regulating mRNP targeting and translation
MCRS1 regulates the localization and translation of polyribosomal mRNP in neurons [Citation5]. FMRP is an mRNP binding protein exhibiting G-quartet binding activity in hippocampal neurons. It binds to mRNP to inhibit translation. MCRS1 is a binding partner of FMRP and it is proposed to escort FMRP-containing mRNP to the somato-dendritic region for spatial translation regulation. MCRS1 overexpression redistributes one of the FMRP isoforms to the nucleolus, where it becomes inactivated. In addition, MCRS1 also binds directly to mRNP to control its translation.
3.2 MCRS1 in chromosome segregation
Sister chromatids are evenly segregated in mitosis to form two genetically identical cells. This highly dynamic process needs to be precisely regulated by a set of sophisticated machinery. After nuclear envelope breakdown , chromosomes start to condense and the bipolar spindle apparatus start to assemble. Microtubules nucleated from centrosomes search and capture the kinetochores on the centromeres. The proper kinetochore-microtubule attachment allows chromosomes to be aligned along the metaphase plate [Citation44]. Then, sister chromatids are pulled apart toward the opposite poles in anaphase and cytokinesis separates the cytoplasm to generate two genetically identical daughter cells. The whole process is tightly governed by three types of microtubules, including polar microtubules, astral microtubules, and k-fibers [Citation44,Citation45]. The stability and dynamics of the spindle apparatus are regulated by various spindle assembly factors via Ran GTPase [Citation46]. MCRS1 is identified as one such factor that specifically regulates centrosome integrity and k-fiber dynamics [Citation14]. In addition, MCRS1 controls the expression of mitotic genes as a subunit of the NSL complex [Citation16].
3.2.1 Regulating centrosome integrity
Centrosome integrity is critical in maintaining microtubule architecture and normal cell cycle. p78/MCRS1 can regulate centrosome dynamics [Citation3,Citation17,Citation41]. Hirohashi et al. stated that siRNA-mediated p78 depletion inhibits cell proliferation, increases cell death, and decreases polyploid cells. However, Hsu et al. reported that apoptosis is elevated in cells with MCRS1 silencing, but so is the number of polyploid cells. It is unclear whether the discrepancy in the polyploid cell population is due to the knockdown of different isoforms or different efficiencies, but both studies revealed MCRS1’s effect on centrosome dynamics. In zebrafish, Lee et al. revealed that MCRS1 is localized to the centriole satellites and interacts with dynein motors to translocate the centriole satellites. MCRS1 inactivation greatly disrupts the distribution of centriole satellites, leading to increased apoptosis.
3.2.2 Regulating k-fiber dynamics
MCRS1 regulates mitotic spindle assembly [Citation14,Citation47]. Vernos et al. showed that MCRS1 facilitates microtubule assembly in vitro after being detached from importin-β. MCRS1 depletion perturbs chromosome alignment in metaphase, leading to significantly longer mitotic time. MCRS1 stabilizes the minus end of chromosome-driven microtubules in early prometaphase as well as k-fibers in metaphase by inhibiting MCAK/KIF2C kinesin complex. Furthermore, S35/S36 phosphorylation on MCRS1 by Aurora A kinase is required for the fine regulation of k-fiber dynamics [Citation48].
Yang et al. revealed that MCRS1 can be phosphorylated by Mps1, a key upstream kinase in the spindle assembly checkpoint during mitosis. This further demonstrates MCRS1’s function in chromosome segregation [Citation15]. MCRS1 phosphorylation is required to recruit the microtubule-destabilizing kinesin KIF2A to the centrosomes. Mutations on the phosphorylation site results in reduced KIF2A localization, increased inter-centrosome distance, and increased chromosome misalignments. Together, these studies reported that MCRS1 inactivation generates seemingly opposite effects on two similar microtubule-destabilizing kinesins, which needs further investigation for clarification.
3.2.3 Regulating mitotic gene expressions
MCRS1 also regulates cell cycle progression at the transcriptional level. Pavlova et al. showed that the NSL complex, the stability of which is dependent on MCRS1, controls the expression of many kinetochore and centrosome genes, and thus its inactivation generates various mitotic defects [Citation16,Citation28]. However, the role of NSL complex in cell cycle progression is complex because several subunits of the NSL complex are specifically localized to mitotic structures such as chromosomes, kinetochores, midbodies, and centrosomes, suggesting its additional role in mitotic processes. Thus, it is important to study the mitotic role of the NSL complex in both the transcription-dependent and transcription-independent manner in the future. This can be addressed by using an inducible protein degradation system to instantly break down MCRS1 upon mitotic entry [Citation49].
3.3 MCRS1 in cell proliferation
Growth factors signal cell proliferation in eukaryotes. Extracellular growth factors bind to cognitive membrane receptors to trigger signal transduction pathways promoting cell division, and mTOR is one such pathway regulated by MCRS1. mTOR pathway senses whether an adequate amount of nutrients is available to support cell proliferation [Citation50]. Specifically, sufficient amino acids, nucleotides, and growth factors can activate mTORC1 kinase on the cytosolic side of lysosomes via several small GTPases. Activated mTORC1 phosphorylates downstream effectors to facilitate protein translation, nucleotide synthesis, rRNA transcription, glycolysis, and autophagy inhibition. These processes collectively promote cell growth and proliferation. Studies revealed that MCRS1 promotes the mTOR pathway and subsequent cell proliferation [Citation18,Citation51]. On the other hand, MCRS1 also inhibits cell proliferation by regulating telomerase activities during DNA replication. As an essential prerequisite for mitosis, replication of linear eukaryotic DNA faces an intrinsic problem, which is telomere shortening. It particularly threatens genome integrity during extensive cell divisions. Thus, eukaryotic cells manage to protect telomeres by using telomerase to extend telomeric sequences [Citation52,Citation53]. MCRS1 was shown to be an essential component in telomerase regulation [Citation9,Citation54] and further to regulate senescence during DNA damage response [Citation41].
3.3.1 Promoting cell proliferation: mTOR activation
MCRS1 promotes cell proliferation via the mTOR pathway [Citation18,Citation51]. mTORC1 is a major kinase in the mTOR pathway that can be activated by Rheb GTPase and inhibited by TSC1/2 GAP. According to Fawal et al., when cells sense the presence of growth factors and nutrients, MCRS1 promotes the dissociation of Rheb from TSC1/2 GAP, which activates mTORC1 on the lysosomal surface for cell proliferation [Citation18]. On the other hand, Brandt et al. proved that MCRS1 activates the mTOR pathway to maintain intestinal homeostasis [Citation55]. The suppression of MCRS1 or mTORC1 in intestinal epithelial cells causes replication defects, leading to a high degree of DNA damage and chromosome instability. This is most likely caused by the inactivation of carbamoyl-phosphate synthase 2 (CAD), a target of mTORC1 responsible for nucleotide supply during the S phase [Citation56]. Brandt et al. also showed that MCRS1 perturbation promotes tumorigenesis when in association with inflammatory bowel disease, but prevents tumorigenesis when exposed to a genetic predisposition with familial adenomatous polyposis. The difference is likely caused by the distinct genetic context, specifically, different degrees of ongoing DNA damage, in two conditions.
3.3.2 Suppressing cell proliferation: telomerase inhibition and cell senescence
MCRS1 and MCRS2 can be tumor-suppressing in other scenarios through their interaction with telomerase. MCRS2 interacts with the telomerase-inhibitory protein LPTS/PinX1 to suppress telomerase activity in the nucleolus and other nuclear foci [Citation9]. MCRS2 also directly inhibits hTERT telomerase by binding to its N-terminus. As a result, MCRS2 overexpression significantly reduces telomere length, limiting long-term cell proliferation. On the other hand, MCRS1 regulates hTERT expression via TEIF [Citation54]. TEIF promotes hTERT expression but MCRS1 prevents TEIF from approaching the promoter region of hTERT. Overall, MCRS1 and MCRS2 can downregulate hTERT telomerase at the transcriptional level as well as the post-translational level to suppress the oncogenic proliferation of cells. However, shortening telomere length can be a risk factor for certain types of cancer, because it promotes the breakage-fusion-bridge cycle and increases chromosome instability [Citation52]. Therefore, MCRS1-induced hTERT inactivation may limit or facilitate tumorigenesis in a context-dependent manner. This relationship is supported by Brandt et al., which demonstrated MCRS1’s dual effects on tumorigenesis in the intestinal epithelium [Citation55].
In addition, MCRS1 regulates cell senescence via p53/p21-dependent pathways [Citation41]. MCRS1 overexpression activates p53-dependent DNA damage response and arrests cell growth. Loss of p53 allows cells to bypass senescence and become proliferative again. Thus, p53 is the key mediator of MCRS1-induced senescence.
4. Regulation of MCRS1 activity
We have demonstrated MCRS1’s complex regulatory role in transcription, translation, chromosome segregation, and cell proliferation. Here, we aim to address the expression regulation of MCRS1 itself and discuss how this drives MCRS1 to function properly in different pathways.
4.1. Modulating MCRS1 expression
MCRS1 expression is regulated by growth factor signaling and miRNA interference. TOJ3, quail MCRS1, is immediately expressed in response to v-Jun mediated growth factor signalling pathways [Citation1]. In human cells, MCRS1 expression is induced at the early S phase when cells are released from the G1 block upon serum addition [Citation9,Citation11]. Similarly, MCRS1 expression is upregulated in most cancer types because of their constitutively activated growth factor signalling [Citation41] (). Meanwhile, MCRS1 expression is repressed by miRNAs targeting its 3’-UTR. In lung cells, the reduced miR-129 level leads to MCRS1 upregulation and promotes tumorigenesis [Citation57]. In liver cells, the anti-tumorigenic miR-186 inhibits MCRS1 expression to suppress hepatocellular cancer development [Citation58].
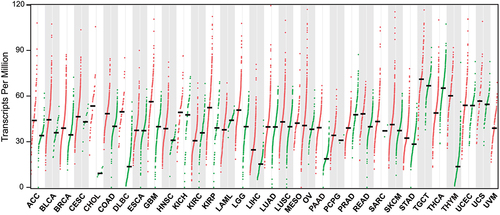
Figure 5. Expression profile of MCRS1 in different types of cancer tissues. MCRS1 expression across all tumor tissues (red) and their corresponding normal tissues (green) from the TCGA database are displayed by dot plots (https://gepia.cancer-pku.cn). Each dot represents the expression level of MCRS1 in one patient’s tissue sample. Most cancers overexpress MCRS1 compared to normal tissues. ACC: Adrenocortical carcinoma, BLCA: Bladder urothelial carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenoma, CHOL: Cholangio carcinoma, COAD: Colon adenocarcinoma, DLBC: Lymphoid neoplasm diffuse large B-cell lymphoma, ESCA: Esophageal carcinoma, GBM: Glioblastoma multiforme, HNSC: Head and neck squamous cell carcinoma, IKCH: Kidney chromophobe, KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute myeloid leukemia, LGG: Brain lower-grade glioma, LIHC: Liver hepatocellular carcinoma, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma, MESO: Mesothelioma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PCPG: Pheochromocytoma and paraganglioma, PRAD: Prostate adenocarcinoma, READ: Rectum adenocarcinoma, SARC: Sarcoma, SKCM: Skin cutaneous melanoma, STAD: Stomach adenocarcinoma, TGCT: Testicular germ cell tumors, THCA: Thyroid carcinoma, THYM: Thymoma, UCEC: Uterine corpus endometrial carcinoma, UCS: Uterine carcinosarcoma, UVM: Uveal melanoma.
4.2. Modulating MCRS1 degradation
MCRS1 degradation can be regulated by the ubiquitin-proteasome system in response to cell proliferation and DNA damage pathways. MCRS1 is targeted by E3 identified by Differential Display (EDD), a ubiquitin E3 ligase, before its destruction in proteasomes [Citation59]. Thus, inactivation of EDD stabilizes MCRS1 and causes it to accumulate. Perturbation of MCRS1 or EDD deregulates cyclin expressions, leading to abnormal cell cycle. This is consistent with the fact that MCRS1 regulates cyclin expressions [Citation35,Citation41]. However, it is unclear whether EDD directly stimulates MCRS1 ubiquitination, as RNAi-mediated EDD knockdown does not significantly decrease MCRS1 ubiquitination. Along the lines of ubiquitination, a yeast two-hybrid screen identified BRCA1-associated protein 1 (BAP1), a de-ubiquitination enzyme frequently mutated in clear cell renal cell carcinoma (ccRCC), as an MCRS1 binding partner [Citation60]. BAP1 deficiency upregulates MCRS1 ubiquitination and degradation, so cells with polyploidy or multi-lobed nuclei occur at a higher frequency. These studies validate ubiquitination as a key mechanism to control the protein level of MCRS1.
4.3. Modulating MCRS1 activity by phosphorylation and acetylation
MCRS1 activity can be regulated by phosphorylation and acetylation. In mitosis, MCRS1 S35/S36 is phosphorylated by Aurora A kinase [Citation48]. This modification maintains the integrity of the minus end of k-fibers in dividing cells while mutations on S35/S36 destabilize and shorten k-fibers. On the other hand, MCRS1 S65 can be phosphorylated by Mps1 kinase, a process that recruits KIF2A to the minus end of mitotic spindles to facilitate chromosome alignments in metaphase [Citation15]. Large-scale proteomic studies further revealed many putative sites subject to phosphorylation or acetylation. However, the corresponding kinases and acetyltransferases are to be determined [Citation61–65].
5. MCRS1 in development and tumorigenesis
Accurate regulation of cell proliferation and genomic stability is essential for organismic development. Thus, its deregulation inevitably results in pathological consequences such as birth defects or tumorigenesis. Here, we aim to address the critical role of MCRS1 in early embryonic development and its deregulation in tumorigenesis.
5.1. In early embryonic development
The developmental roles of MCRS1 have been widely studied in mice, fruit flies, frogs, and zebrafish. In mice, MCRS1 is essential during early embryonic development [Citation66]. MCRS1 mutant embryos grow normally up to the blastocyst stage but the epiblast lineage fails to develop functionally during the gastrulation stage. The hepatocyte-specific MCRS1 deletion perturbs the expression of key membrane transporter genes, leading to cirrhosis liver [Citation42]. In Xenopus, MCRS1 interacts with Six1, a homeodomain transcription factor essential for craniofacial development [Citation33]. An RNAi-mediated MCRS1 knockdown in their embryos results in defective otic vesicles in the neural ectoderm. In Drosophila, MCRS1 inactivation by the P element leads to early larval lethality (flybase.org/reports/FBgn0263832). In zebrafish, homozygous mcrs1 mutants remain viable but develop smaller brains and eyes due to enhanced apoptosis [Citation17]. These mutants also carry centriole satellite defects associated with reduced ciliogenesis in the olfactory placode. Together, these studies corroborate the essential roles of MCRS1 in early embryonic development.
5.2. In tumorigenesis
MCRS1 can promote the oncogenic transformation of cells in many organisms. In quail, TOJ3 is immediately expressed upon v-Jun activation for the neoplastic transformation of fibroblast cells [Citation1]. TOJ3 overexpression alone triggers the anchorage-independent growth of fibroblast cells. Similarly, MCRS1 overexpression transforms mice embryonic fibroblast cells to become cancerous, and this transformation can be repressed by the non-catalytic domain of PTEN on the C-terminus [Citation67]. It is interesting to see PTEN’s tumor-suppressing activity reside in the non-catalytic domain rather than the catalytic phosphatase domain, as the latter directly regulates the oncogenic PI3K signaling pathway.
In humans, MCRS1 overexpression is observed in various types of cancers and it is associated with poor prognosis (). MCRS1 is significantly enriched in glioblastoma and neuroblastoma tissues associated with higher malignant grades and poor prognosis [Citation68–71]. MCRS1 is also upregulated in colorectal tumors compared to their adjacent non-cancerous tissues [Citation72]. This upregulation is positively correlated with tumor invasion, local recurrence, tumor grade, and UICC stage, whereas downregulation of MCRS1 reduces the levels of cyclin D1, CDK4, and pRb and inhibits cell growth [Citation73]. Djouder group further showed that MCRS1 overexpression is correlated with poor prognosis and mTORC1 signaling in colon cancer [Citation18,Citation55]. Interestingly, unlike MCRS1, MCRS2 expression is significantly decreased in colorectal tumors compared to their corresponding non-tumor tissues, suggesting isoform-specific functions [Citation74]. MCRS1 is also overexpressed in esophageal squamous cell carcinoma (ESCC) cell lines and it regulates cell cycle progression by altering the levels of p21, CDK4, and cyclin D1 [Citation75]. In non-small cell lung cancer (NSCLC), MCRS1 is one of the upregulated genes on chromosome arm 12q13, a region with frequent chromosomal aberrations [Citation76]. MCRS1 promotes the tumorigenic epithelial-to-mesenchymal transition (EMT) via miRNAs [Citation57,Citation77,Citation78]. Specifically, MCRS1 overexpression directly upregulates miR-155, which inhibits the expression of tumor-suppressor Rb1. This pathway alters the expression profile of mitotic genes such as MYC, E2F2, PCNA, and Ki67; and downregulates many cell junction proteins to promote EMT, tumor invasion, and metastasis. In hepatocellular carcinoma (HCC), MCRS1 promotes cell proliferation and serves as a good prognostic marker for HCC patients [Citation79]. In renal cell carcinoma (RCC), MCRS1 depletion significantly inhibits cell proliferation, migration, and invasion [Citation80]. In gastric cancer (GC), MCRS1 overexpression is positively correlated with tumor invasiveness, differentiation grade, and metastatic stage, and it serves as an independent prognostic factor [Citation81]. However, Wang et al. reported contradicted data in gastric cancer, in which MCRS1 overexpression inhibits cell growth, migration, and invasion via Pkmyt1 interaction [Citation20].
MCRS1 can also play anti-tumorigenic roles. Brandt et al. showed that MCRS1 depletion promotes DNA damage and chromosome aberrations, which provides a selective advantage in the inflammatory milieu for tumorigenesis [Citation55]. According to the Cancer Profiling Array which examined cancer-specific MCRS1 expression levels, though MCRS1 is overexpressed in most cancer types, certain cancers have MCRS1 expression downregulated [Citation41]. Therefore, the effect of MCRS1 on tumorigenesis may highly depend on the genetic context of different cell types ().
6. Conclusion
MCRS1 is involved in the regulation of transcription factors, epigenetic histone modification, chromatin remodeling, mRNP transport, telomerase activity, DNA damage response, cell senescence, mTOR signaling, microtubule dynamics, and centrosome integrity. The striking functional diversity may be rooted in its transcriptional, translational, and post-translational regulation of a variety of downstream targets. In fact, the MCRS1-containing NSL complex can control the expression of thousands of housekeeping genes. Therefore, MCRS1 becomes a central player to regulate cell proliferation and multi-cellular organism development, and its deregulation causes devastating consequences in organisms such as cancer.
Many open questions remain to be answered before we can gain a more comprehensive understanding of MCRS1. First of all, it will be important to identify which functions are transcription-dependent or transcription-independent. Secondly, since MCRS1 can be both pro-tumorigenic and anti-tumorigenic, we are also interested in whether the isoforms of MCRS1 exhibit opposite effects during tumorigenesis. Thirdly, given MCRS1’s participation in many diverse cellular activities, we wonder whether it has one common biochemical feature to support all diverse activities. For instance, can MCRS1 control the assembly of different large complexes such as Pontin/Reptin ATPase does in various pathways? In addition, further studies are required to decipher how post-translational modifications of MCRS1 contribute to its activation, localization, and function. Lastly, its detailed regulatory machinery of centrosome and microtubule dynamics during chromosome segregation is to be revealed. Addressing these questions will deepen our understanding of the sophisticated regulations leading to organismic development and cancer.
Data availability
Data sharing does not apply to this article as no new data were created or analyzed in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bader AG, Schneider ML, Bister K, et al. TOJ3, a target of the v-Jun transcription factor, encodes a protein with transforming activity related to human microspherule protein 1 (MCRS1). Oncogene. 2001;20(51):7524–7535. DOI:10.1038/sj.onc.1204938
- Ren Y, Busch RK, Perlaky L, et al. The 58-kDa microspherule protein (MSP58), a nucleolar protein, interacts with nucleolar protein p120. Eur J Biochem. 1998;253(3):734–742. DOI:10.1046/j.1432-1327.1998.2530734.x
- Hirohashi Y, Wang Q, Liu Q, et al. P78/MCRS1 forms a complex with centrosomal protein Nde1 and is essential for cell viability. Oncogene. 2006;25(35):4937–4946. DOI:10.1038/sj.onc.1209500
- Du X, Wang Q, Hirohashi Y, et al. DIPA, which can localize to the centrosome, associates with p78/MCRS1/MSP58 and acts as a repressor of gene transcription. Exp Mol Pathol. 2006;81(3):184–190. DOI:10.1016/j.yexmp.2006.07.008
- Davidovic L, Bechara E, Gravel M, et al. The nuclear microspherule protein 58 is a novel RNA-binding protein that interacts with fragile X mental retardation protein in polyribosomal mRnps from neurons. Hum Mol Genet. 2006;15(9):1525–1538. DOI:10.1093/hmg/ddl074
- Shimono K, Shimono Y, Shimokata K, et al. Microspherule protein 1, Mi-2β, and RET finger protein associate in the nucleolus and up-regulate ribosomal gene transcription. J Biol Chem. 2005;280(47):39436–39447. DOI:10.1074/jbc.M507356200
- Okumura K, Zhao M, DePinho RA, et al. PTEN: a novel anti-oncogenic function independent of phosphatase activity. Cell Cycle. 2005;4(4):540–542. DOI:10.4161/cc.4.4.1614
- Ivanova AV, Ivanov SV, Lerman ML. Association, mutual stabilization, and transcriptional activity of the STRA13 and MSP58 proteins. Cell Mol Life Sci. 2005;62(4):471–484.
- Song H, Li Y, Chen G, et al. Human MCRS2, a cell-cycle-dependent protein, associates with LPTS/PinX1 and reduces the telomere length. Biochem Biophys Res Commun. 2004;316(4):1116–1123. DOI:10.1016/j.bbrc.2004.02.166
- Lin DY, Shih HM. Essential role of the 58-kDa microspherule protein in the modulation of Daxx-dependent transcriptional repression as revealed by nucleolar sequestration. J Biol Chem. 2002;277(28):25446–25456.
- Bruni R, Roizman B. Herpes simplex virus 1 regulatory protein ICP22 interacts with a new cell cycle-regulated factor and accumulates in a cell cycle-dependent fashion in infected cells. J Virol. 1998;72(11):8525–8531.
- Yang CP, Chiang C-W, Chen C-H, et al. Identification and characterization of nuclear and nucleolar localization signals in 58-kDa microspherule protein (MSP58). J Biomed Sci. 2015;22(1):33. DOI:10.1186/s12929-015-0136-0
- Kuo CW, Wang WH, Liu ST. Mapping signals that are important for nuclear and nucleolar localization in MCRS2. Mol Cells. 2011;31(6):547–552.
- Meunier S, Vernos I. K-fibre minus ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat Cell Biol. 2011;13(12):1406–1414.
- Yang H, Zhang F, Huang C-J, et al. Mps1 regulates spindle morphology through MCRS1 to promote chromosome alignment. Mol Biol Cell. 2019;30(9):1060–1068. DOI:10.1091/mbc.E18-09-0546
- Pavlova GA, Popova JV, Andreyeva EN, et al. RNAi-mediated depletion of the NSL complex subunits leads to abnormal chromosome segregation and defective centrosome duplication in Drosophila mitosis. PLoS Genet. 2019;15(9):e1008371. DOI:10.1371/journal.pgen.1008371
- Lee SH, Lee M-S, Choi T-I, et al. MCRS1 associates with cytoplasmic dynein and mediates pericentrosomal material recruitment. Sci Rep. 2016;6(1):27284. DOI:10.1038/srep27284
- Fawal MA, Brandt M, Djouder N. MCRS1 binds and couples Rheb to amino acid-dependent mTORC1 activation. Dev Cell. 2015;33(1):67–81.
- Xu M, Zhang F, Da L, et al. Microspherule protein 2 associates with ASK1 and acts as a negative regulator of stress-induced ASK1 activation. FEBS Lett. 2012;586(12):1678–1686. DOI:10.1016/j.febslet.2012.04.051
- Wang XM, Li Q-Y, Ren L-L, et al. Effects of MCRS1 on proliferation, migration, invasion, and epithelial mesenchymal transition of gastric cancer cells by interacting with Pkmyt1 protein kinase. Cell Signal. 2019;59:171–181.
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719.
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20(11):1405–1428.
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080.
- Grewal SIS, Jia S. Heterochromatin revisited. Nature Reviews Genetics. 2007;8(1):35–46.
- Zhao Y, Garcia BA. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb Perspect Biol. 2015;7(9):a025064.
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45.
- Chen L, Cai Y, Jin J, et al. Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J Biol Chem. 2011;286(13):11283–11289. DOI:10.1074/jbc.M111.222505
- Raja SJ, Charapitsa I, Conrad T, et al. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol Cell. 2010;38(6):827–841. DOI:10.1016/j.molcel.2010.05.021
- Wu JL, Lin Y-S, Yang C-C, et al. MCRS2 represses the transactivation activities of Nrf1. BMC Cell Biol. 2009;10(1):9. DOI:10.1186/1471-2121-10-9
- Zhang J, Liu J, Li X, et al. The physical and functional interaction of NDRG2 with MSP58 in cells. Biochem Biophys Res Commun. 2007;352(1):6–11. DOI:10.1016/j.bbrc.2006.10.141
- Ko YG, Kang Y-S, Park H, et al. Apoptosis signal-regulating kinase 1 controls the proapoptotic function of death-associated protein (Daxx) in the cytoplasm. J Biol Chem. 2001;276(42):39103–39106. DOI:10.1074/jbc.M105928200
- Hwang SP, Huang L-C, Wang W-H, et al. Expression of Rta in B lymphocytes during Epstein–Barr virus latency. J Mol Biol. 2020;432(19):5227–5243. DOI:10.1016/j.jmb.2020.07.011
- Neilson KM, Keer S, Bousquet N, et al. Mcrs1 interacts with Six1 to influence early craniofacial and otic development. Dev Biol. 2020;467(1–2):39–50. DOI:10.1016/j.ydbio.2020.08.013
- Yang CP, Kuo Y-L, Lee Y-C, et al. RINT-1 interacts with MSP58 within nucleoli and plays a role in ribosomal gene transcription. Biochem Biophys Res Commun. 2016;478(2):873–880. DOI:10.1016/j.bbrc.2016.08.044
- Andersen DS, Raja SJ, Colombani J, et al. Drosophila MCRS2 associates with RNA polymerase II complexes to regulate transcription. Mol Cell Biol. 2010;30(19):4744–4755. DOI:10.1128/MCB.01586-09
- Mendjan S, Taipale M, Kind J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21(6):811–823. DOI:10.1016/j.molcel.2006.02.007
- Sheikh BN, Guhathakurta S, Akhtar A. The non-specific lethal (NSL) complex at the crossroads of transcriptional control and cellular homeostasis. EMBO Rep. 2019;20(7):e47630.
- Zhang Y, Jang Y, Lee J-E, et al. Selective binding of the PHD6 finger of MLL4 to histone H4K16ac links MLL4 and MOF. Nat Commun. 2019;10(1):2314. DOI:10.1038/s41467-019-10324-8
- Wang T, Holt MV, Young NL. Early butyrate induced acetylation of histone H4 is proteoform specific and linked to methylation state. Epigenetics. 2018;13(5):519–535.
- Lam KC, Mühlpfordt F, Vaquerizas JM, et al. The NSL complex regulates housekeeping genes in Drosophila. PLoS Genet. 2012;8(6):e1002736. DOI:10.1371/journal.pgen.1002736
- Hsu CC, Lee Y-C, Yeh S-H, et al. 58-kDa microspherule protein (MSP58) is novel Brahma-related gene 1 (BRG1)-associated protein that modulates p53/p21 senescence pathway. J Biol Chem. 2012;287(27):22533–22548. DOI:10.1074/jbc.M111.335331
- Garrido A, Kim E, Teijeiro A, et al. Histone acetylation of bile acid transporter genes plays a critical role in cirrhosis. J Hepatol. 2021;76(4):850–861. DOI:10.1016/j.jhep.2021.12.019
- Mao Y-Q, Houry WA. The role of pontin and reptin in cellular physiology and cancer etiology. Front Mol Biosci. 2017;4. DOI:10.3389/fmolb.2017.00058
- Prosser SL, Pelletier L. Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol. 2017;18(3):187–201.
- Meunier S, Vernos I. Acentrosomal microtubule assembly in mitosis: the where, when, and how. Trends Cell Biol. 2016;26(2):80–87.
- Cavazza T, Vernos I. The RanGTP pathway: from nucleo-cytoplasmic transport to spindle assembly and beyond. Front Cell Dev Biol. 2015;3:82.
- Petry S, Vale RD. A new cap for kinetochore fibre minus ends. Nat Cell Biol. 2011;13(12):1389–1391.
- Meunier S, Timon K, Vernos I. Aurora-A regulates MCRS1 function during mitosis. Cell Cycle. 2016;15(13):1779–1786.
- Nishimura K, Fukagawa T, Takisawa H, et al. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6(12):917–922. DOI:10.1038/nmeth.1401
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293.
- Garrido A, Brandt M, Djouder N. Transport to Rhebpress activity. Small GTPases. 2016;7(1):12–15.
- Tanaka H, Watanabe T. Mechanisms underlying recurrent genomic amplification in human cancers. Trends Cancer. 2020;6(6):462–477.
- Shay JW, Wright WE. Telomeres and telomerase: three decades of progress. Nat Rev Genet. 2019;20(5):299–309.
- Hsu CC, Chen C-H, Hsu T-I, et al. The 58-kda microspherule protein (MSP58) represses human telomerase reverse transcriptase (hTERT) gene expression and cell proliferation by interacting with telomerase transcriptional element-interacting factor (TEIF). Biochim Biophys Acta. 2014;1843(3):565–579. DOI:10.1016/j.bbamcr.2013.12.004
- Brandt M, Grazioso TP, Fawal M-A, et al. mTORC1 inactivation promotes colitis-induced colorectal cancer but protects from APC loss-dependent tumorigenesis. Cell Metab. 2018;27(1):118–135 e8. DOI:10.1016/j.cmet.2017.11.006
- Ben-Sahra I, Howell JJ, Asara JM, et al. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339(6125):1323–1328. DOI:10.1126/science.1228792
- Liu MX, Zhou KC, Cao Y. MCRS1 overexpression, which is specifically inhibited by miR-129*, promotes the epithelial-mesenchymal transition and metastasis in non-small cell lung cancer. Mol Cancer. 2014;13(1):245.
- Wang H, Ou J, Jian Z, et al. miR-186 modulates hepatocellular carcinoma cell proliferation and mobility via targeting MCRS1-mediated Wnt/β-catenin signaling. J Cell Physiol. 2019;234(12):23135–23145. DOI:10.1002/jcp.28878
- Benavides M, Chow-Tsang L-F, Zhang J, et al. The novel interaction between microspherule protein Msp58 and ubiquitin E3 ligase EDD regulates cell cycle progression. Biochim Biophys Acta. 2013;1833(1):21–32. DOI:10.1016/j.bbamcr.2012.10.007
- Peng J, Ma J, Li W, et al. Stabilization of MCRS1 by BAP1 prevents chromosome instability in renal cell carcinoma. Cancer Lett. 2015;369(1):167–174. DOI:10.1016/j.canlet.2015.08.013
- Zhou H, Di Palma S, Preisinger C, et al. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res. 2013;12(1):260–271. DOI:10.1021/pr300630k
- Rigbolt KT, Prokhorova TA, Akimov V, et al. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4(164):rs3. DOI:10.1126/scisignal.2001570
- Olsen JV, Vermeulen M, Santamaria A, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3(104):ra3. DOI:10.1126/scisignal.2000475
- Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. DOI:10.1126/science.1175371
- Dephoure N, Zhou C, Villén J, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105(31):10762–10767. DOI:10.1073/pnas.0805139105
- Cui W, Cheong A, Wang Y, et al. MCRS1 is essential for epiblast development during early mouse embryogenesis. Reproduction. 2020;159(1):1–13. DOI:10.1530/REP-19-0334
- Okumura K, Zhao M, DePinho RA, et al. Cellular transformation by the MSP58 oncogene is inhibited by its physical interaction with the PTEN tumor suppressor. Proc Natl Acad Sci U S A. 2005;102(8):2703–2706. DOI:10.1073/pnas.0409370102
- Lin W, Zhang J, Zhang J, et al. RNAi-mediated inhibition of MSP58 decreases tumour growth, migration and invasion in a human glioma cell line. J Cell Mol Med. 2009;13(11–12):4608–4622. DOI:10.1111/j.1582-4934.2008.00499.x
- Wu L, Zhang Z-G, Qin H-Z, et al. Downregulation of MSP58 suppresses cell proliferation in neuroblastoma cell lines. Neuroreport. 2012;23(16):932–936. DOI:10.1097/WNR.0b013e328359566e
- Lin W, Li X-M, Zhang J, et al. Increased expression of the 58-kD microspherule protein (MSP58) is correlated with poor prognosis in glioma patients. Med Oncol. 2013;30(4):677. DOI:10.1007/s12032-013-0677-6
- Lin W, Dai S-H, Chen T, et al. Expression of 58-kD microspherule protein (MSP58) is highly correlated with PET imaging of tumor malignancy and cell proliferation in glioma patients. Cell Physiol Biochem. 2016;38(2):635–645. DOI:10.1159/000438656
- Shi H, Chen S, Jin H, et al. Downregulation of MSP58 inhibits growth of human colorectal cancer cells via regulation of the cyclin D1-cyclin-dependent kinase 4-p21 pathway. Cancer Sci. 2009;100(9):1585–1590. DOI:10.1111/j.1349-7006.2009.01223.x
- Shi H, Li S-J, Zhang B, et al. Expression of MSP58 in human colorectal cancer and its correlation with prognosis. Med Oncol. 2012;29(5):3136–3142. DOI:10.1007/s12032-012-0284-y
- Li C, Chen M, Zhao P, et al. Expression of MCRS1 and MCRS2 and their correlation with serum carcinoembryonic antigen in colorectal cancer. Exp Ther Med. 2016;12(2):589–596. DOI:10.3892/etm.2016.3424
- Xu CS, Zheng J-Y, Zhang H-L, et al. MSP58 knockdown inhibits the proliferation of esophageal squamous cell carcinoma in vitro and in vivo. Asian Pac J Cancer Prev. 2012;13(7):3233–3238. DOI:10.7314/APJCP.2012.13.7.3233
- Liang Y, Liu M, Wang P, et al. Analysis of 20 genes at chromosome band 12q13: rACGAP1 and MCRS1 overexpression in nonsmall-cell lung cancer. Genes Chromosomes Cancer. 2013;52(3):305–315. DOI:10.1002/gcc.22030
- Zhang Y, An J, Lv W, et al. miRNA-129-5p suppresses cell proliferation and invasion in lung cancer by targeting microspherule protein 1, E-cadherin and vimentin. Oncol Lett. 2016;12(6):5163–5169. DOI:10.3892/ol.2016.5372
- Liu M, Zhou K, Huang Y, et al. The candidate oncogene (MCRS1) promotes the growth of human lung cancer cells via the miR–155–rb1 pathway. J Exp Clin Cancer Res. 2015;34(1):121. DOI:10.1186/s13046-015-0235-5
- Zhong M, Zhang X, Li B, et al. Expression of MSP58 in hepatocellular carcinoma. Med Oncol. 2013;30(2):539. DOI:10.1007/s12032-013-0539-2
- Zhai X, Wu Y, Zhang D, et al. Knockdown of MSP58 inhibits the proliferation and metastasis in human renal cell carcinoma cells. Biomed Pharmacother. 2017;91:54–59.
- Cui J, Xi H, Cai A, et al. Increased expression of 58-kDa microspherule protein (MSP58) in human gastric cancer promotes cell proliferation and correlates with poor patient survival. Clin Lab. 2016;62(6):993–1001. DOI:10.7754/Clin.Lab.2015.150828
