ABSTRACT
Pluripotent stem cells (PSCs) are a promising source of stem cells for regenerative therapies. Stem cell function depends on telomere maintenance mechanisms that provide them with the proliferative capacity and genome stability necessary to multiply and regenerate tissues. We show here that established human embryonic stem cells (hESCs) have stable telomere length that is dependent on telomerase but not on alternative mechanisms based on homologous recombination pathways. Here, we show that human-induced pluripotent stem cells (hiPSCs) reprogrammed from somatic cells show progressive telomere lengthening until reaching a length similar to ESCs. hiPSCs also acquire telomeric chromatin marks of ESCs including decreased abundance of tri-methylated histone H3K9 and H4K20 and HP1 heterochromatic marks, as well as of the shelterin component TRF2. These chromatin features are accompanied with increased abundance of telomere transcripts or TERRAs. We also found that telomeres of both hESCs and hiPSCs are well protected from DNA damage during telomere elongation and once full telomere length is achieved, and exhibit stable genomes. Collectively, this study highlights that hiPSCs acquire ESC features during reprogramming and reveals the telomere biology in human pluripotent stem cells (hPSCs).
AUTHOR SUMMARY
We show that established human embryonic stem cells (hESCs) have a maximum and stable telomere length that is dependent on telomerase but not on the alternative homologous recombination pathway or ALT. Human-induced pluripotent stem cells (hiPSCs) reprogrammed from somatic cells show progressive telomere lengthening until reaching a length similar maximum telomere length than ESCs, suggesting that telomere length is regulated by epigenetic mechanisms in human cells. In this regard, hiPSCs acquire telomeric chromatin marks characteristic of an “open chromatin” including increased abundance of telomere transcripts or TERRAs. Telomeres of both hESCs and hiPSCs are well protected during telomere elongation and exhibit stable genomes. Collectively, this study highlights that hiPSCs acquire ESC features during reprogramming and reveals the telomere biology in human pluripotent stem cells (hiPSCs).
Keywords:
Introduction
Telomeres consist of tandem repeats of the (TTAGGG)n sequence at the ends of linear chromosomes, which are essential for genome integrity [Citation1]. A protein complex known as shelterin binds and protects telomeres, preventing chromosome ends from being recognized as DNA breaks by the DNA repair machinery [Citation2]. Telomeres shorten gradually with increasing cell division cycles due to the end replication problem [Citation3]. Telomerase, a eukaryotic ribonucleoprotein (RNP) complex [Citation4], as well as recombination-based alternative lengthening of telomeres (ALT) are responsible for de novo telomere synthesis [Citation5]. These telomere lengthening mechanisms however are only active in certain cell types, such as cancer and pluripotent stem cells [Citation6,Citation7]. In the adult organism, telomerase is absent in most somatic cells with the exception of some activity in the adult stem cell compartments, although this is insufficient to prevent progressive telomere shortening with age [Citation8]. Critically short telomeres eventually trigger a persistent DNA damage response at chromosome ends that leads to cellular senescence or cell death, and impairs stem cell function [Citation6,Citation9]. Indeed, mutations in telomere maintenance related genes have been associated with different human diseases known as telomeropathies that include dyskeratosis congenita, aplastic anemia, and idiopathic pulmonary fibrosis [Citation10–12]. In turn, we have shown that longer telomeres owing to telomerase activation in the adult organism result in delayed aging and improved health span [Citation13,Citation14].
Telomeres are enriched in epigenetic marks that are characteristic of heterochromatin. In particular, the telomeric chromatin is enriched in H3K9me3, H4K20me3 and HP1 heterochromatic marks, in agreement with a repressed heterochromatic structure [Citation15–17]. In addition, the DNA of subtelomeric is heavily methylated [Citation18]. These epigenetic marks contribute to a higher order of telomere length regulation, and loss of any of these heterochromatic marks leads to telomere elongation and activation of telomere DNA recombination [Citation15,Citation16,Citation18]. Telomeres can be transcribed into the so-called telomeric RNAs or TERRAs [Citation19,Citation20]. TERRAs bind to telomeres and have an important role in telomere maintenance and protection, as well as facilitate the assembly of the telomeric heterochromatin by recruiting the Polycomb complex [Citation21]. In turn, TERRA transcription is regulated by the status of the telomeric chromatin and a more “open” chromatin results in increased TERRA levels [Citation19].
Telomere length regulation starts as early as during the formation of the oocytes. Telomeres are short in oocytes but are rebuilt during early embryo development until the blastocyst stage by activation of either telomerase or a recombination-based ALT mechanisms. In particular, activation of DNA recombination-based ALT pathway has been described at the cleavage stage and gradually substituted by telomerase-based telomere elongation at the blastocyst stage [Citation22,Citation23]. Thereafter, telomere length is partially maintained at post-implantation stages but telomeres start to gradually shorten after birth [Citation24,Citation25].
We have previously shown that cell derivation and in vitro expansion of Embryonic Stem Cells (ESCs) derived from the Inner Cell Mass (ICM) of the blastocyst results in de novo telomere elongation reaching a telomere length which is beyond the normal length of the ICM of the species [Citation24]. We reported a similar phenomenon in the establishment of Induced Pluripotent Stem Cells (iPSCs) [Citation26]. In particular, IPSCs can be derived from somatic cells by the expression of Oct3/4, Sox2, Klf4 and c-Myc, the so-called Yamanaka factors [Citation27]. IPS cells have been shown to acquire ESC properties, and are able to contribute to mouse embryonic development and the mouse germline [Citation27,Citation28]. We have shown that in the mouse, telomeres are elongated de novo in the iPSC in a telomerase-dependent manner, and that this is essential for the genomic stability and pluripotency potential of these cells [Citation26,Citation29]. In particular, mouse ESCs (mESCs) and iPSCs (miPSCs) with longer telomeres show higher efficiency in generating chimeras than those with short telomeres [Citation26,Citation30]. Interestingly, chimeric mice derived from ESCs with hyper-long telomeres, have longer telomeres and less DNA damage in the adult organism, delayed aging and increased longevity, as well as less metabolic aging and less cancer [Citation25,Citation31].
During reprogramming, miPSCs also acquire similar epigenetic marks of ESCs, characterized by a lower density of H3K9me3 and H4K20me3 at telomeres as compared to the parental somatic cells and results in TERRA upregulation [Citation26]. Similar to miPSCs, TERRA level is upregulated in hiPSCs, while the levels of epigenetic marks at telomeres remain unknown [Citation32].
The development of iPSCs has provided a new source of stem cells for the customized transplantation therapies. In the case of humans, hiPSCs have been shown to have longer telomere length compared to parental somatic cells. However, there is a significant variability in telomere length among various hiPSC lines originated from the same tissue or parental cell type [Citation33]. The dynamics of telomere length in hESCs and hiPSCs during long-term in vitro expansion has not been well documented. It remains unknown if telomere lengthening of hiPSCs remain stable once reached the length of hESCs or continue to elongate further.
Here, we studied the telomere length in both established hESCs and reprogrammed hiPSCs in vitro throughout early and prolonged culture. Our results show that established hESCs have stable telomere length and do not show progressive telomere elongation, while the hiPSCs progressively lengthen the telomeres to a similar length of ESCs at late passage and then remain stable. Telomeres of hESCs and hiPSCs are well protected during in vitro expansion. In addition, the telomeric chromatin of iPSCs reset into a more ‘’open’’ state with decreased levels of H3K9me3, H4K20me3 and HP1 deposition on telomeres and show increased TERRA transcription. These findings deepen our knowledge on telomere biology in hPSCs and provide evidences for their potential use in regenerative medicine.
Results
Stable telomere length in established ESCs maintained by telomerase
Telomeres are elongated de novo in mouse ESCs (mESCs) during their in vitro derivation and expansion. Once a maximum telomere length is established, it is maintained throughout later passages [Citation24,Citation25]. To address telomere length dynamics in human ESCs (hESCs), here, we used three independent hES cell lines from the National Cellular Biobank of Spain of the lowest possible passages (p8-p10) to study telomere length changes from early to late passages. To this end, we cultured the different hES cells up to passage 40 and measured telomere length every 5 passages by using Southern blot-based telomere restriction analysis (TRF) (Figure S1A) up to passage 40 and every 10 passages by quantitative telomere FISH (Q-FISH) up to passage 30 (Figures S1B, C). None of the three established hES cell lines under study underwent telomere length changes up to passage 40. We confirmed that these hESCs maintained the expression of different pluripotent markers throughout all passages, including NANOG, OCT4, SOX2, and also TRF1, the later previously shown by us to be overexpressed in pluripotent stem cells [Citation34,Citation35] (Figures S2A-B). We also observed expression of the catalytic subunit of telomerase (TERT) at the protein level, as well as presence of telomerase activity (Figures S2A-D). None of the human pluripotent cell lines showed differential expression of different pluripotent markers and activity of telomerase throughout the passages, indicating that the pluripotency of the hESCs was well maintained during the culture. These results suggest that the maximum telomere length of these hESCs had been achieved before passages 8–10 and then stably maintained by telomerase during the following passages. This notion is in agreement with previous reports showing that telomere elongation only takes place during early expansion [Citation36].
Next, we set to address the mechanisms responsible for telomere length maintenance in the hESCs studied here. First, we assessed the role of telomerase by knocking down telomerase expression using lentiviral vectors harboring two independent shRNAs targeting TERT mRNA. Successful knockdown of TERT mRNA was confirmed by using quantitative real-time PCR (qRT-PCR), which was concomitant with decreased telomerase activity as detected by using the telomeric repeat amplification protocol (TRAP) assay (Figures S3A, B). A progressive telomere shortening was observed from passage 3 to 8 in hESCs with TERT knockdown (Figure S3C), demonstrating that telomerase is required for telomere length maintenance in established hESCs.
Telomere recombination is another mechanism of telomere elongation, the so-called ALT, which has been shown to be important for telomere lengthening during early embryonic development [Citation5,Citation22]. To address this possibility in hESCs, we studied the presence of a hallmark of ALT, the so-called ALT-associated promyelocytic leukemia (PML) nuclear bodies (APB) as well as the frequency of telomere sister chromatid exchanges (T-SCE) by using the chromosome orientation-FISH assay. As a positive control for ALT activation, we used the U2OS cell line, known to maintain telomeres by ALT. hESCs showed significantly fewer APBs compared to U2OS and only few of them were co-localized with telomeres (Figure S3D). In addition, T-SCE events were rare in hESCs compared to U2OS (Figure S3E). Together, these results indicate that telomere maintenance in hESCs is mainly dependent on telomerase, as features of ALT were rare.
Telomere elongation in iPSCs during in vitro expansion
iPSCs share similar features with ESCs [Citation27,Citation28]. We showed in the past that telomeres are elongated in both mouse ESCs and iPSCs with increasing passages [Citation26]. Reprograming of human somatic cells also results in telomere lengthening [Citation33]. As we did not observe telomere elongation in the four hESCs studied here, we set to address telomere dynamics during in vitro expansion of hiPSCs. To this end, we used human BJ fibroblasts for the generation of human iPSCs. To investigate the role of telomerase, we generated BJ fibroblasts that were knock-out by CRISPR/cas9 technology for TERT and TERC, TERT-/- and TERC-/- respectively (see Material and Methods). Two independent TERT−/− clones and one TERC−/− clone were obtained. The introduction of frameshift mutations of TERT and deletion of the TERC were confirmed by DNA sequence analysis (). Next, we generated iPSCs by introducing the four Yamanaka factors into wild-type (WT) and telomerase knock-out BJ fibroblasts. BJ WT fibroblasts were successfully reprogrammed with the establishment of six independent hiPS clones, while the BJ TERT-/- and TERC-/- fibroblasts failed to be reprogrammed ().
Figure 1. BJ deficient for telomerase fail to be reprogrammed into iPSCs.
a. Generation of TERT knock-out BJ fibroblasts by targeting the exon2 of TERT using the CRISPR/Cas9 technology. The sequences of BJ clones with successful TERT knockout are indicated. b. Generation of TERC knock-out BJ fibroblasts. Two sgRNAs flanking TERC were designed to delete TERC. Sequence of BJ clone with successful TERC deletion is indicated. Image of DNA electrophoresis gel showing the successful deletion of TERC is shown. c. Reprogramming of BJ fibroblasts with (WT) or without telomerase genes (TERT−/−&TERC−/−). Representative images of alkaline phosphatase (AP) staining showing iPS colonies are shown. Bright field images showing iPS colonies (yellow arrow) on feeders (top) and iPSCs established on feeder-free culture (bottom). d. Representative images of western blot analyzing NANOG, OCT4, TERT and TRF1 in different iPS clones at indicated passages by western blot. e. Protein level quantification. Bars represent mean values and error bars the standard deviation. One-way ANOVA was used for statistical analysis. n=number of independent clones.
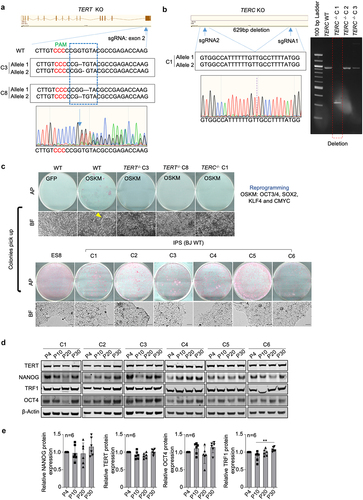
Next, we addressed whether the established cells were bona fide iPSCs. The six established iPS cell clones formed ES cell-like compacted colonies with high nuclear cytoplasm ratio and were positive for alkaline phosphatase (AP) (). The Yamanaka transgenes were silenced while the endogenous pluripotent markers, OCT4, SOX2 and NANOG were reactivated at RNA (Figure S4A) and protein levels (). In addition, TERT and TRF1 were also reactivated and no differences in protein levels were observed among passages ().
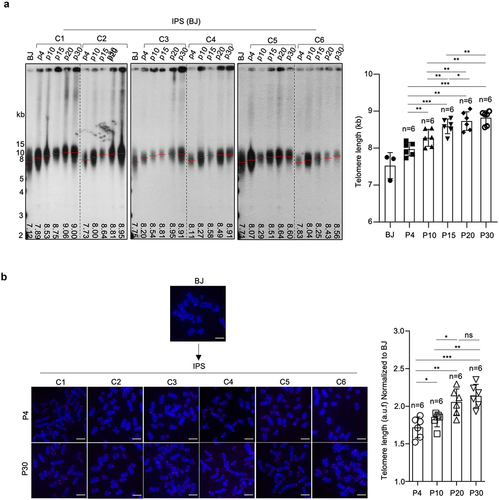
Finally, we addressed whether telomeres were elongated during in vitro expansion of the different hiPSCs. Telomere lengthening was observed in all iPS clones compared to the parental BJ fibroblasts (). Of note, iPSCs at passage 4 had much longer telomeres than BJ fibroblasts, suggesting telomere length was mainly rebuilt during reprogramming. Subsequently, progressive telomere elongation was observed at successive passages until passage 20 from which further expansion did not result in telomere lengthening, suggesting that a maximum telomere length was achieved by that passage. This fully established telomere length of hiPSCs between 8–9 Kb was comparable to that of the established hESCs studied here (Figure S1A).
Figure 2. Progressive telomere elongation in reprogrammed hiPSCs.
Telomerase is reactivated in iPSCs and maintained during in vitro expansion
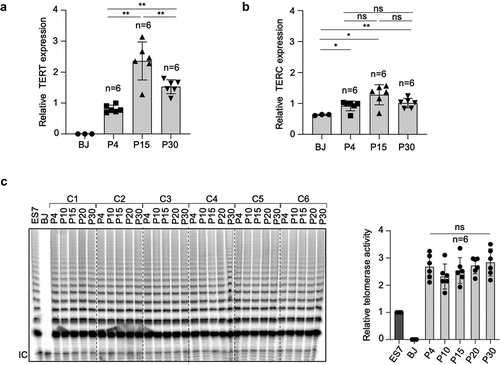
The transcriptional expression of both components of telomerase, the catalytic subunit and the RNA component, TERT and TERC respectively, was analyzed at different passages in all hiPS clones generated here (). TERC but not TERT expression was detected in BJ fibroblasts (). hiPSCs reactivated TERT and upregulated TERC expression compared to parental BJ fibroblasts (). Of note, TERT mRNA expression levels increased with passages with highest mRNA expression level at passage 15, while no differences in TERC expression levels were observed at different passages (). These results indicate that TERT but not TERC is the limiting component of telomerase activation during reprogramming. Next, we analyzed telomerase activity in all hiPS clones at different passages. As expected, telomerase was reactivated in all hiPS clones compared to the parental BJ fibroblasts and its activity maintained across different passages (). The six hiPS cell clones showed similar levels of telomerase activity ().
Figure 3. Telomerase is reactivated and maintained in hiPSCs.
Telomeres are protected and genome stability is maintained in iPSCs
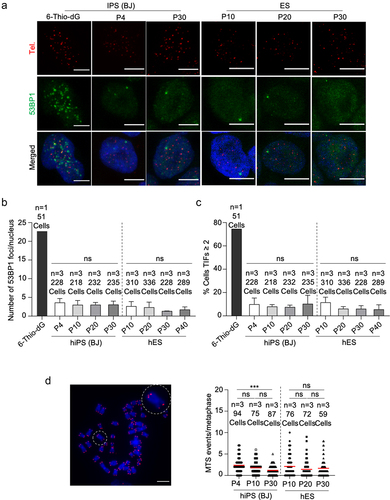
It has been previously described that hPSCs frequently acquire chromosomal abnormalities, including gains or losses of whole chromosomes [Citation37,Citation38]. Thus, we next performed karyotype analysis at both early (p4) and late (p30) passages in the six hiPS cell clones under study. We did not observe chromosomal abnormalities in any of the hiPS cell clones indicating that these clones do not present genomic instability and preserve normal karyotype with passages (Figure S4B), in agreement with the fact that they have a proper telomere maintenance. Next, we asked whether telomeres of hESCs and hiPSCs were well protected during in vitro culture as increased DNA damage and dysfunctional telomeres have been reported in mESCs at later passages [Citation25]. To check telomeric DNA damage, we performed immunofluorescence for the DNA damage marker 53BP1 together with a quantitative telomeric FISH assay to detect telomeres. Treatment with 6-thio-dG was used as a positive control for telomeric DNA damage as this nucleotide analogue is selectively incorporated into the telomeres generating dysfunctional telomeres [Citation39]. We did not observe increased global DNA damage foci or telomere induced foci (TIFs) in hESCs and hiPSCs up to passage 40 and passage 30, respectively (). In contrast, cells treated with 6-thio-dG showed increased DNA damage foci that colocalized with telomeres (). Finally, we checked the presence of chromosome aberrations on metaphase spreads. We did not observe end-to-end telomere fusions or the presence of telomere-signal free ends, aberrations commonly observed when telomeres are short or dysfunctional (). Similarly, we did not observe an increase in multitelomeric signals (MTS), indicative of telomere fragility, with accumulative passages (). These results demonstrate that telomeres are well protected during telomere elongation in hiPSCs and that genome stability is well maintained up to passage 30, when maximum telomere length is already reached.
Figure 4. Telomeres of hiPSCs and hESCs are protected and genomically stable throughout successive passages.
Telomeric chromatin is more “open” and telomere transcription is upregulated in iPSCs
During cells reprogrammed to achieve pluripotency, the telomeres of miPSCs acquire ESC features with decrease abundance of H3K9me3 and H4K20me3 heterochromatic histone marks at telomeres [Citation26]. Thus, we next addressed the abundance of heterochromatin marks and of the TRF2 shelterin protein in hESCs and hiPSCs at different passages during in vitro cell expansion. We observed a decrease of H3K9me3, H4K20me3 and HP1 heterochromatic histone marks at the telomeres of iPSCs compared to the parental BJ fibroblasts by using Chromatin Immune Precipitation (ChIP) analysis (). We also observed a decrease of the TRF2 shelterin protein at telomeres, suggesting a lower density of the shelterin complex in iPSCs compared to BJ fibroblasts (). Of note, the abundance of these heterochromatin marks and of TRF2 in hiPSCs were similar to those detected in hESCs, confirming that hiPSCs acquire ESC features during reprogramming. Of interest, no differences in the density of these marks were found in hiPSCs at the different passages studied here (), again suggesting that these events occurred at earlier passages.
Figure 5. Telomeric chromatin is more “open” and telomeric RNA is upregulated in hiPSCs.
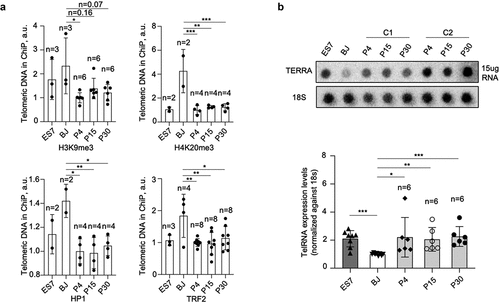
TERRA abundance has been previously shown to correlate with telomere length and to be regulated by the status of telomeric heterochromatin [Citation19]. It has also been shown that TERRA levels are upregulated during reprogramming both in mouse and human cells compared to the parental differentiated cells [Citation26,Citation32]. TERRA levels of hiPSCs in comparison to hESCs have not been documented. Here, we studied TERRA levels in six independent hiPS clones at different passages and found that TERRA levels were upregulated in all six hiPS clones compared to the parental BJ fibroblasts. Furthermore, TERRA levels of hiPSCs were similar to that of the hESCs, and TERRA expression did not show differential expression across passages ().
IPSCs with different telomere length show equal efficiency in teratoma formation
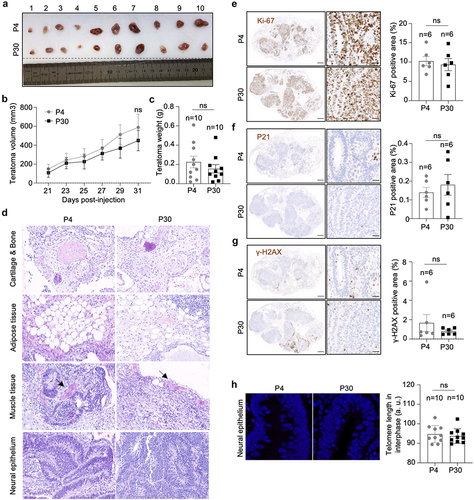
In order to address whether telomere length in hiPSCs correlates with differential developmental pluripotency, we performed in vivo differentiation assay by checking the teratoma formation. We used iPSCs at passage 4 and at passage 30. IPSCs with different telomere length showed similar teratoma growth rate and tumor weight (). They were also able to differentiate into different cell types including neural epithelium, muscle, adipose and cartilage/bone tissues (). Furthermore, we did not observe any difference in the percentage of ki67, p21 and γ-H2AX positive cells indicating similar cell proliferation, cell cycle arrest and DNA damage burden, respectively (). Finally, we checked telomere length in the teratomas, specifically in the neural epithelium cells within the rosette structure (). We found similar telomere length in teratomas originated from iPSCs at passage 4 and from iPSCs at passage 30, indicating that during in vivo differentiation telomere length is stabilized.
Figure 6. hiPSCs with different telomere length show equal teratoma formation efficiency.
Discussion
We previously showed that mouse-induced pluripotent stem cells (miPSC) undergo a de novo progressive telomerase-dependent telomere elongation which is concomitant with a more “open” telomeric chromatin structure with decreased heterochromatic histone marks and increased telomere transcription [Citation26]. A similar observation was made by us in the case of mouse embryonic stem cells (mESCs) derived from the blastocyst Inner Cell Mass (ICM), which also show a progressive telomere lengthening concomitant with a decrease in telomere heterochromatic marks [Citation24].
Here, we study the dynamics of telomere length and telomere chromatin in human embryonic stem cells (hESC) as well as in human induced pluripotent stem cells (hiPSC). In particular, we show loss of histone heterochromatic marks (H3K9me3, H4K20me3 and HP1) at telomeres in hiPSCs compared to those of the parental BJ fibroblasts, reaching similar levels as those observed in independent hESCs clones. Furthermore, we show that telomeres are progressively elongated with increasing passages of hiPSCs until reaching a maximum length comparable to that of the previously established hESCs studied here. Importantly, we also show that telomere transcripts or TERRAs are upregulated in hiPSCs to similar levels to those of hESCs.
We failed to observe telomere elongation in hESCs during in vitro culture starting from passage 8–10, suggesting that full telomere length had been already established during early passages before passage 8–10. In contrast, in mESCs, telomere length increases until passage 24 and then is maintained until passage 60 [Citation25]. Both human and mouse iPSCs also lengthen their telomeres during and post-reprogramming [Citation26,Citation40,Citation41]. However, while the greatest telomere lengthening in hiPSCs is achieved during reprogramming, in miPSCs the greatest telomere lengthening is observed during post-reprogramming [Citation26], suggesting a more efficient telomere elongation in human cells. In line with this, telomeres of hES/iPSCs are better protected than in mouse as few telomeric DNA damage foci are observed up to passage 40 and 30, respectively. In contrast, a dramatic increase in telomeric DNA damage foci can be observed in mESCs at passage 24 [Citation25]. Finally, we observed that not only telomeric histone heterochromatic marks in hiPSCs can be reprogrammed to acquire similar features as the hESCs, but also the TRF2 shelterin protein is upregulated in hiPSC, something which has not been observed in miPSCs reprogrammed from mouse embryonic fibloblasts [Citation26], and which could explain better telomere protection in the hiPSC compared to the miPSC.
A telomerase-mediated de novo telomere elongation has been associated with pluripotency in both iPSCs and ESCs. Furthermore, we have shown that telomerase is essential for the generation of fully functional pluripotent stem cells, as both telomerase-deficient mESCs and miPSCs failed to contribute to chimeras owing to the presence of short telomeres and telomeric DNA damage [Citation26]. miPSCs with different telomere length acquired during in vitro expansion showed differential developmental pluripotency, i.e. the longer the telomere the higher efficiency to generate chimeras [Citation30]. In hESCs, telomere length has been reported to correlate with the expression of pluripotent markers [Citation42]. Here, we did not observe an association between telomere length and the pluripotency of hiPSCs. hiPSCs at different passages with different telomere length show similar levels of pluripotent markers, including NANOG, OCT4, SOX2, and also TRF1. Furthermore, hiPSCs with different telomere length show similar teratoma formation. These results suggest that pluripotency impairment might only be detected when telomeres are either critically short or dysfunctional.
We show here that hESCs use telomerase to elongate and maintain telomeres, and TERT-knockout hESCs show progressive telomere shortening [Citation36,Citation43]. Telomere elongation in miPSCs is also mediated by telomerase since miPSCs derived from TERC-/- MEF have similar telomere length as the parental wild-type MEF [Citation26]. Here, we successfully generated BJ TERT-/- and TERC-/-, however we failed to reprogram these cells into iPSCs. In contrast, reprogramming of G1 TERC-/- MEFs rendered miPSCs at similar efficiency as the WT MEF, although these miPSCs were not fully functional (Marion et al., 2009a). DNA damage response and activation of Ink4/Arf locus in aged parental cells restrict the iPS cell generation [Citation29,Citation44]. Thus, the failure of reprogramming BJ TERT-/- and TERC-/- is probably not directly caused by telomerase deficiency itself but rather to BJ TERT-/- and TERC-/- becoming senescent during the gene editing procedure, limiting the derivation to iPSCs.
Development of iPSCs has provided unprecedented opportunities for regenerative medicine, and telomere status is closely linked to stem cell pluripotency and function. In this work, we have revealed for the first-time telomere dynamics during hiPSCs generation and long-term in vitro expansion. We found that hiPSCs lengthened their telomeres at early stages during reprogramming, and progressive acquired similar telomere length as hESCs. Telomere lengthening was concomitant with a decrease in heterochromatic markers at telomeres, and an increase of telomere transcription. This study expands the knowledge of telomere biology in hiPSCs and expedite the development of cell therapies using hiPSCs.
Experimental procedures
Ethical statement
The hESCs were obtained from the National Cellular Biobank of Spain, and the use of the hESCs and hiPSCs were approved by the RESEARCH ETHICS COMMITTEE (CEI PI 48_2021-v2). All three ES cell lines were established from different blastocysts that from different couples. Animal experiments were performed in accordance with the guidelines stated in the International Guiding Principles of Biomedical Research Involving Animals, developed by the Council for International Organizations of Medical Sciences (CIOMS), and were approved by Ethical Committee (CEIyBA) (PROEX 106.7/20).
Cell culture
hESCs and hiPSCs were maintained on matrigel (Corning)-coated (1:100) plates with mTeSR1 (STEMCELL Technologies). Cells were generally passaged by EDTA (0.5 mM) every 4 days at a ratio of 1:6. BJ human foreskin fibroblasts were purchased from the American Type Culture Collection (ATCC) (population doubling times 23) and grown in Dulbecco´s modified Eagle´s medium (DMEM) supplied with 10% of fetal bovine serum (FBS) and 1% (vol/vol) penicillin-streptomycin (Gibco). Cells were grown at 37°C with 5% CO2 and regularly tested for mycoplasma using the Myco Alert Mycoplasma Detection Kit (Lonza).
Generation of TERT and TERC knock-out BJ fibroblasts
To knockout TERT and TERC in BJ fibroblasts, cells were transiently transfected with plasmid pSpCas9(BB)-2A-GFP (PX458) with different sgRNAs. For TERT, one sgRNA was desigened to target exon 2 to induce frameshift mutations, while for TERC, two sgRNAs were designed to delete the whole gene. Single cell was sorted by FACS and plated into 96-well plates for monoclonal cell expansion. Once the cells grow out, genomic DNA was isolated using a DNeasy kit (69504, QIAGEN) and different locus fragments were amplified using Phusion High-Fidelity DNA Polymerase (F53OL, Thermo Fisher). Sanger sequencing was used to identify the correct clones. Sequence of sgRNAs are listed in Supplementary Table 1.
Generation of hiPSCs
Reprogramming of BJ fibroblasts was done as previously described [Citation28]. In brief, retroviral supernatants were produced by transfecting HEK-293T cells (5 × 106 cells per 100-mm-diameter dish) with the ecotropic packaging plasmid pCL-Ampho (4 μg) together with one of the following retroviral constructs (4 μg): pMXs-hKlf4, pMXs-hSox2, pMXs-hOct4 and pMXs-hc-Myc (Addgene #13370, 13367, 17217, 17220, respectively). BJ fibroblasts (WT, TERT-/- and TERC-/-) had been seeded the previous day (1 × 105 cells per well of 6-well plate) and received 0.5 ml of each of the corresponding retroviral supernatants. This procedure was repeated every 12 h for 2 days (a total of four rounds of infections). The day after infection was completed, media was replaced by human fibroblast media, and kept for a further 2 days. At day 8, cells were trypsinized and reseeded on feeder plate (CF-1 MEF, 1 × 106 cells per 100-mm-diameter dish). At day 9, medium was changed to hES cell medium. At day 20, colonies with ES-like morphology became visible under the microscope. Colonies were picked after 3 weeks and expanded on feeder fibroblasts using standard procedures or adapted to matrigel-coated plates with mTeSR1.
Generation of TERT knockdown hESCs
Lentivirus armed with short hairpin RNA for TERT (shTERT, SHCLNG-NM_003219, SIGMA ALDRICH) were packaged and transduced into hESCs. Cells were selected by puromycin (0.5 ug/ml) for 2 days after transduction.
Telomerase activity by TRAP assay
To perform the TRAP analysis, cell lysates were prepared with S-100 lysis buffer as previously described [Citation45]. Protein concentration of the lysates was determined by Bradford assay (Bio-Rad). Cell extracts (50 ug) were incubated with telomeric TS primers for a 60 min initial extension step at 30°C in vitro. The extended reaction was subjected to PCR amplification (25 cycle of 30 s at 94°C, 30 s at 59°C, 30 s at 72°C) with the TS primer labeled with 32 P ɣ-ATP, the 36-bp internal standard control (TSNT) and reverse primers (NT and ACX) for amplification of internal standard control (IC) and telomerase products. The PCR reactions were resolved by 8% polyacrylamide, 7 M urea gel electrophoresis, and the gel was exposed to a phosphor-imager and scanned by a Typhoon scanner. Primer sequences are listed in Supplementary Table 1.
Telomere restriction fragment (TRF) analysis
Cells were harvested and embedded in agarose plugs following digestion with MboI (R0147, New England Biolabs). Telomeres were separated by gel electrophoresis in 0.5 X TBE buffer at 14°C using a CHEF DR-II pulsed-field apparatus (Bio-Rad) for 12 h at a 5 V/cm constant voltage and 5 s constant pulse time. The gel was transferred to nylon membrane (Hybond-XL, GE Healthcare) and probed with a 32P-labeled telomeric probe (TTAGGG)n (a kind gift from T. de Lange).
Quantitative fluorescence in situ hybridization (Q-FISH) analysis
Paraffin-embedded tissue sections were deparaffinized and fixed with 4% formaldehyde, permeabilized with 0.5% Triton X-100 for 2 h. To perform Q-FISH on metaphase, ESCs or iPSCs were incubated with 0.1 μg/ml colcemide for 3–4 h. After hypotonic swelling in 0.03 M sodium citrate for 20–30 min at 37°C, cells were fixed in methanol:acetic acid (3:1) and spread onto clean slides. Both tissue sections and metaphase spreads were digested with pepsin/HCl and a second fixation with 4% formaldehyde. Slides were dehydrated with increasing concentrations of EtOH (70%, 90%, 100%) and incubated with the telomeric C-rich probe (CCCTAA)3-Cy3 (Panagene, F1002) at 85°C for 3 min followed by 2 h at room temperature in a wet chamber. The slides were extensively washed with 50% formamide and 0.08% TBS-Tween 20. Images of tissue sections were captured by confocal microscopy with a laser-scanning microscope (Leica TSC SP5) using a Plan Apo 63Å-1.40 NA oil immersion objective (HCX). Images of metaphase spreads were captured using microscope Leica DM6B using a 100× oil objective. Telomere length was analyzed using Leica Application Suite X Software.
Immunohistochemistry (IHC) and immunofluorescence (IF) analysis
Immunohistochemistry (IHC) was performed on de-paraffined tissue sections with antigen retrieval by 10 mM sodium citrate (pH 6.5) cooked under pressure for 2 min. The staining was performed with primary antibodies against phosphor-Histone H2AX (Ser139) (05–636, Millipore), p21-WAF1/Cip1 (SX118) (M7202, DAKO) and Ki67 (IR626, Dako). Images were analyzed by ZEISS ZEN Microscope Software v2.3 and quantified by ImageJ.
For Immunofluorescence (IF), ESCs and iPSCs were plated in Poly-L-lysine-coated coverslips, fixed 10 min in 4% buffered formaldehyde, following permeabilization with 0.2% PBS-Triton for 10 min and blocked with 5% fetal bovine serum in PBS for 1 h. The staining was performed with primary antibodies, anti-53BP1 (1:500; Novus Biologicals, NB100–304), anti-PML (1:500; Santa Cruz, 966). Secondary antibodies conjugated with 488-Alexa or 555-Alexa were used to detect the binding of the primary antibodies. When indicated, a Q-FISH was performed on IF stained slides fixed with 4% formaldehyde for 20 minutes.
Chromosome orientation fluorescence in situ hybridization (CO-FISH)
Sub-confluent ESCs were incubated with 5’-bromo-2’-deoxyuridine (BrdU; Sigma) at the concentration of 10 mM for 12–14 h. Colcemide was added at a concentration of 0.1 μg/ml during the last 3–4 hours, and metaphase spreads were prepared for telomere Q-FISH. Chromosome slides were treated with RNaseA, fixed with 4% formaldehyde, then stained with Hoechst 33,258 (0.5 mg/ml) for 15 min and exposed to 365 nm UV light for 40 min. The BrdU-substituted DNA was digested with Exonuclease III (Takara). PNA-FISH was performed with telomeric C-rich probe (CCCTAA)3-Cy3 (Panagene, F1002) and G-rich probe (TTAGGG)3-Alexa488 (Panagene, F1008). Images were captured using microscope Leica DM6B using a 100× oil objective.
Chromatin immune precipitation (ChIP) assay and telomere dot-blot
ChIP assay was performed as previously described [Citation15] with the following antibodies: 3 ug of anti-H3K9me3 (ab8898, abcam), 3 ug of anti-H4K20me3 (ab9053, abcam), 2 ug of anti-TRF2 (NB110–57130, Novus Biologicals), 5 ug of anti-HP-1ɣ (05–690, Sigma) and IgG (sc-2025, Santa Cruz Biotechnology). Samples were transferred to a Hybond-N+ membrane (Amersham) on a dot blot, and hybridized with the same telomeric probe (TTAGGG)n used for TRF. The signal was quantified with the ImageJ software. The amount of telomeric DNA in each ChIP was normalized to total telomeric DNA signal.
Western blot analysis
Cells were lysed by Laemmli Sample Buffer. Lysates were boiled at 100°C for 5–10 min and separated in 4–12% SDS-PAGE gels (NuPAGE Invitrogen). After transferring to nitrocellulose membranes (Amersham Protan), blots were blocked and incubated with the indicated primary antibodies. Antibody bindings were detected by incubating with IRDye®-Conjugated secondary antibody (LI-COR). The primary antibodies used were anti-TRF1 (1:1000; BED5, Bio-Rad), anti-TERT (1:1000; ab320320, abcam), anti-NANOG (1:1000; Cell Signaling, 4903), anti-OCT4 (1:1000; Cell Signaling, 2750), anti-SOX2 (1:1000; Cell Signaling, 3579), and anti-β-Action (1:500; Santa Cruz 47,778).
Real-time qPCR
Total RNA from cells was extracted with the RNeasy Mini kit (74106, QIAGEN) and reverse transcribed using the iSCRIPT cDNA synthesis kit (1708891, BIO-RAD). Quantitative real-time PCR was performed with the QuantStudio 6 Flex (Applied Biosystens, Life Technologies) using Go-Taq Green Master Mix (M7123, Promega) according to the manufacturer’s protocol. Primer sequences can be found in Supplementary Table 1.
Teratoma formation assay
For xenograft experiments, 6-week athymic nude females were purchased from Harlan (Foxn1nu/nu). Mice were maintained at the Spanish National Cancer Centre (CNIO) in accordance with the recommendations of the Federation of European Laboratory Animal Science Associations (FELASA) under specific pathogen-free conditions. Along with those guidelines, mice were monitored in a daily or weekly basis and they sacrificed in CO2 chambers when the human end-point was considered.
hiPSCs were harvested by EDTA (0.5 mM) and centrifuged. Cell pellets were re-suspended in the mixture of mTeSR1 and Matrigel in a 1:1 ratio. Foxn1nu/nu mice were subcutaneously injected with 2 × 106 cells to dorsal flank of the mice. Tumors were measured every other days. Tumor volume was determined by the following equation: V = a*b2, a and b are length and width respectively. At the human end-point, tumors were dissected, weighted, and fixed with PBS containing 4% paraformaldehyde. Paraffin-embedded tissue was sliced and stained with hematoxylin and eosin.
Karyotype analysis
Metaphase spreads were prepared for the hiPSCs. Chromosomal G-band analyses were performed at Cytogenetics Unit in Centro Nacional de Investigaciones Oncológicas (CNIO), Spain.
RNA dot-blot
Fifteen ug of total RNA (in 1 mM EDTA, 7% formaldehyde, 6 × SSC – Volume 100 µl) was denatured in a thermocycler at 60°C for 30 min. Denaturated RNA was dot-blotted onto 6 × SSC-soaked Hybond-N+ membrane (Amersham) and then UV cross-linked. Hybridization was performed with the telomeric probe (TTAGGG)n. Dot-blot was normalized using 18S probes and quantified using ImageJ software.
Author contributions
B.Y.M. designed experiments, performed most of the experiments, data analysis and wrote the manuscript. P.M. designed experiments, data curation, methodology and wrote the manuscript. R.S.V. designed experiments and methodology. M.A.B. secured funding, conceived the project, designed experiments, and wrote the manuscript.
Supplementary figures.pdf
Download PDF (3 MB)Acknowledgements
We thank to Histopathology and Cytogenetics units at CNIO for histology and karyotype analysis, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
All relevant data are within the manuscript and its Supporting Information files.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2023.2285551.
Additional information
Funding
References
- Chan SW, Blackburn EH. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene. 2002;21(4):553–563. PubMed PMID: 11850780. doi: 10.1038/sj.onc.1205082
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19(18):2100–2110. PubMed PMID: 16166375. doi: 10.1101/gad.1346005
- Lingner J, Cooper JP, Cech TR. Telomerase and DNA end replication: no longer a lagging strand problem? Science. 1995;269(5230):1533–1534. PubMed PMID: 7545310. doi: 10.1126/science.7545310
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43(2 Pt 1):405–413. PubMed PMID: 3907856. doi: 10.1016/0092-8674(85)90170-9
- Dunham MA, Neumann AA, Fasching CL, et al. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26(4):447–450. PubMed PMID: 11101843. doi: 10.1038/82586
- Blasco MA. Telomere length, stem cells and aging. Nat Chem Biol. 2007;3(10):640–649. PubMed PMID: 17876321. doi: 10.1038/nchembio.2007.38
- Armanios M, Greider CW. Telomerase and cancer stem cells. Cold Spring Harb Symp Quant Biol. 2005;70:205–208. PubMed PMID: 16869755. doi: 10.1101/sqb.2005.70.030
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345(6274):458–460. PubMed PMID: 2342578. doi: 10.1038/345458a0
- Fumagalli M, Rossiello F, Clerici M, et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol. 2012;14(4):355–365. PubMed PMID: 22426077; PubMed Central PMCID: PMCPMC3717580. doi: 10.1038/ncb2466
- Martinez P, Blasco MA. Telomere-driven diseases and telomere-targeting therapies. J Cell Bio. 2017;216(4):875–887. PubMed PMID: 28254828; PubMed Central PMCID: PMCPMC5379954. doi: 10.1083/jcb.201610111
- Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012;13(10):693–704. PubMed PMID: 22965356; PubMed Central PMCID: PMCPMC3548426. doi: 10.1038/nrg3246
- Wong JM, Collins K. Telomere maintenance and disease. Lancet. 2003;362(9388):983–988. PubMed PMID: 14511933. doi: 10.1016/S0140-6736(03)14369-3
- Tomas-Loba A, Flores I, Fernandez-Marcos PJ, et al. Telomerase reverse transcriptase delays aging in cancer-resistant mice. Cell. 2008;135(4):609–622. PubMed PMID: 19013273. doi: 10.1016/j.cell.2008.09.034
- Bernardes de Jesus B, Vera E, Schneeberger K, et al. Telomerase gene therapy in adult and old mice delays aging and increases longevity without increasing cancer. EMBO Mol Med. 2012;4(8):691–704. PubMed PMID: 22585399; PubMed Central PMCID: PMCPMC3494070. doi: 10.1002/emmm.201200245
- Garcia-Cao M, O’Sullivan R, Peters AH, et al. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet. 2004;36(1):94–99. PubMed PMID: 14702045. doi: 10.1038/ng1278
- Benetti R, Gonzalo S, Jaco I, et al. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Bio. 2007;178(6):925–936. PubMed PMID: 17846168; PubMed Central PMCID: PMCPMC2064618. doi: 10.1083/jcb.200703081
- Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8(4):299–309. PubMed PMID: 17363977. doi: 10.1038/nrg2047
- Gonzalo S, Jaco I, Fraga MF, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8(4):416–424. PubMed PMID: 16565708. doi: 10.1038/ncb1386
- Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10(2):228–236. PubMed PMID: 18157120. doi: 10.1038/ncb1685
- Azzalin CM, Reichenbach P, Khoriauli L, et al. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318(5851):798–801. PubMed PMID: 17916692. doi: 10.1126/science.1147182
- Montero JJ, Lopez-Silanes I, Megias D, et al. TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat Commun. 2018;9(1):1548. PubMed PMID: 29670078; PubMed Central PMCID: PMCPMC5906467. doi: 10.1038/s41467-018-03916-3
- Liu L, Bailey SM, Okuka M, et al. Telomere lengthening early in development. Nat Cell Biol. 2007;9(12):1436–1441. PubMed PMID: 17982445. doi: 10.1038/ncb1664
- Schaetzlein S, Lucas-Hahn A, Lemme E, et al. Telomere length is reset during early mammalian embryogenesis. Proc Natl Acad Sci U S A. 2004;101(21):8034–8038. PubMed PMID: 15148368; PubMed Central PMCID: PMCPMC419552. doi: 10.1073/pnas.0402400101
- Varela E, Schneider RP, Ortega S, et al. Different telomere-length dynamics at the inner cell mass versus established embryonic stem (ES) cells. Proc Natl Acad Sci U S A. 2011;108(37):15207–15212. PubMed PMID: 21873233; PubMed Central PMCID: PMCPMC3174656. doi: 10.1073/pnas.1105414108
- Varela E, Munoz-Lorente MA, Tejera AM, et al. Generation of mice with longer and better preserved telomeres in the absence of genetic manipulations. Nat Commun. 2016;7:11739. PubMed PMID: 27252083; PubMed Central PMCID: PMCPMC4895768. doi: 10.1038/ncomms11739
- Marion RM, Strati K, Li H, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4(2):141–154. PubMed PMID: 19200803. doi: 10.1016/j.stem.2008.12.010
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. PubMed PMID: 16904174. doi: 10.1016/j.cell.2006.07.024
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. PubMed PMID: 18035408. doi: 10.1016/j.cell.2007.11.019
- Marion RM, Strati K, Li H, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460(7259):1149–1153. PubMed PMID: 19668189; PubMed Central PMCID: PMCPMC3624089. doi: 10.1038/nature08287
- Huang J, Wang F, Okuka M, et al. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 2011;21(5):779–792. PubMed PMID: 21283131; PubMed Central PMCID: PMCPMC3203670. doi: 10.1038/cr.2011.16
- Munoz-Lorente MA, Cano-Martin AC, Blasco MA. Mice with hyper-long telomeres show less metabolic aging and longer lifespans. Nat Commun. 2019;10(1):4723. PubMed PMID: 31624261; PubMed Central PMCID: PMCPMC6797762. doi: 10.1038/s41467-019-12664-x
- Yehezkel S, Rebibo-Sabbah A, Segev Y, et al. Reprogramming of telomeric regions during the generation of human induced pluripotent stem cells and subsequent differentiation into fibroblast-like derivatives. Epigenetics. 2011;6(1):63–75. PubMed PMID: 20861676; PubMed Central PMCID: PMCPMC3052915. doi: 10.4161/epi.6.1.13390
- Liu L. Linking Telomere Regulation to Stem Cell Pluripotency. Trends Genet. 2017;33(1):16–33. PubMed PMID: 27889084. doi: 10.1016/j.tig.2016.10.007
- Schneider RP, Garrobo I, Foronda M, et al. TRF1 is a stem cell marker and is essential for the generation of induced pluripotent stem cells. Nat Commun. 2013;4:1946. doi: 10.1038/ncomms2946. PubMed PMID: 23735977.
- Marion RM, Montero JJ, Lopez de Silanes I, et al. TERRA regulate the transcriptional landscape of pluripotent cells through TRF1-dependent recruitment of PRC2. Elife. 2019;8. PubMed PMID: 31426913; PubMed Central PMCID: PMCPMC6701927. doi: 10.7554/eLife.44656
- Zeng S, Liu L, Sun Y, et al. Telomerase-mediated telomere elongation from human blastocysts to embryonic stem cells. J Cell Sci. 2014;127(Pt 4):752–762. PubMed PMID: 24338368. doi: 10.1242/jcs.131433
- Draper JS, Smith K, Gokhale P, et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22(1):53–54. PubMed PMID: 14661028. doi: 10.1038/nbt922
- I AK, Andrews PW, Anyfantis G, et al. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol. 2011;29(12):1132–1144. PubMed PMID: 22119741; PubMed Central PMCID: PMCPMC3454460. doi:10.1038/nbt.2051
- Mender I, Gryaznov S, Dikmen ZG, et al. Induction of telomere dysfunction mediated by the telomerase substrate precursor 6-thio-2’-deoxyguanosine. Cancer Discov. 2015;5(1):82–95. PubMed PMID: 25516420; PubMed Central PMCID: PMCPMC4293221. doi: 10.1158/2159-8290.CD-14-0609
- Winkler T, Hong SG, Decker JE, et al. Defective telomere elongation and hematopoiesis from telomerase-mutant aplastic anemia iPscs. J Clin Invest. 2013;123(5):1952–1963. PubMed PMID: 23585473; PubMed Central PMCID: PMCPMC3635755. doi: 10.1172/JCI67146
- Batista LF, Pech MF, Zhong FL, et al. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature. 2011;474(7351):399–402. PubMed PMID: 21602826; PubMed Central PMCID: PMCPMC3155806. doi: 10.1038/nature10084
- Wang H, Zhang K, Liu Y, et al. Telomere heterogeneity linked to metabolism and pluripotency state revealed by simultaneous analysis of telomere length and RNA-seq in the same human embryonic stem cell. BMC Biol. 2017;15(1):114. PubMed PMID: 29216888; PubMed Central PMCID: PMCPMC5721592. doi: 10.1186/s12915-017-0453-8
- Liu CC, Ma DL, Yan TD, et al. Distinct responses of stem cells to telomere uncapping-A potential strategy to improve the safety of cell therapy. Stem Cells. 2016;34(10):2471–2484. PubMed PMID: 27299710. doi: 10.1002/stem.2431
- Li H, Collado M, Villasante A, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460(7259):1136–1139. PubMed PMID: 19668188; PubMed Central PMCID: PMCPMC3578184. doi: 10.1038/nature08290
- Blasco MA, Lee HW, Hande MP, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91(1):25–34. PubMed PMID: 9335332. doi: 10.1016/s0092-8674(01)80006-4
