ABSTRACT
Forkhead box P3 (FOXP3) is the essential transcription factor for the function of regulatory T-cell (Treg). However, the gene mutation of FOXP3 in patients with chronic obstructive pulmonary disease (COPD) at different stages has not been reported. We aim to investigate four single nucleotide polymorphisms (SNPs) and the mRNA expression of FOXP3 in smokers with normal lung function and smokers with COPD at different stages. FOXP3 mRNA expression and SNPs in FOXP3 were assessed in nonsmokers with normal lung function (N), smokers with normal lung function (S), smokers with COPD in the Global Initiative for Chronic Obstructive Lung Disease (GOLD) 1 or 2 grade (COPD 1–2), and smokers with COPD in GOLD 3 or 4 grade (COPD 3–4). In peripheral blood sample, FOXP3 mRNA was assessed using real-time quantitative PCR and SNPs were analyzed by TaqMan PCR. FOXP3 mRNA level in peripheral blood sample was decreased when COPD was aggravated. The frequency of FOXP3 rs5902434 genotype del/del and allele del are lower in COPD 1–2 and COPD 3–4 than that in N or S. The rs5902434 genotype del/del and allele del were, respectively, associated with decreased risk of COPD and lung function decline. The rs5902434 genotypic distribution was correlated with FOXP3 mRNA level. In conclusion, both FOXP3 rs5902434 genotypes and alleles were differently distributed in COPD patients and smokers with normal lung function. The distribution of del/del genotype was associated with systemic expression of FOXP3 mRNA. More research is needed to explore the role of FOXP3 gene polymorphism in immunoinflammation of COPD.
Background
Regulatory T-cells (Tregs) have significant anti-inflammatory and immunomodulatory effects Citation(1, 2). In chronic obstructive pulmonary disease (COPD), Tregs were shown to be differently expressed in large airways, small airways, lung parenchyma, and peripheral blood in COPD patients and in animal model Citation(3–7). Since intracellular expression of forkhead box P3 (FOXP3) is an essential transcription factor for the induction, development, and activity of human Tregs; the expression of FOXP3 in COPD patients may reflect the activity of Tregs. Our previous study has found a decrease in expression of FOXP3 in lung tissues of COPD patients compared with nonsmokers Citation(5). In bronchoalveolar lavage fluid (BALF), the expression of FOXP3 also was found to be decreased in smokers than in nonsmokers Citation(8). Similarly, a recent study found a decreased expression of FOXP3 mRNA in peripheral blood of patients with COPD at different stages Citation(9). Thus, abnormal FOXP3 expression is associated with COPD.
Based on this, we wonder if the abnormality of Tregs activity is the result of COPD development, or is an upstream factor determined by individual factors. Therefore, we analyzed gene polymorphism of Tregs in COPD pathogenesis in this study. It is widely accepted that the overwhelming majority of COPD is caused by environmental exposures. This exposure is primarily via cigarette smoke—about 85–90% of cases of this disorder are due to cigarette consumption Citation(10). Only 10–15% of smokers develop COPD Citation(11), suggesting an important role for genetic susceptibility. On the other hand, some studies indicated that smoking cessation failed to reverse the chronic airway inflammation, which would be involved into irregulation of immune response Citation(12–14). Therefore, the gene polymorphism involved in immunoinflammatory response would provide novel insights into COPD pathogenesis. In the present study, we tested single nucleotide polymorphism (SNP) of Tregs in COPD patients to investigate the basic role of Tregs in COPD pathogenesis.
FOXP3 gene is located on chromosome Xp11.23. FOXP3 SNPs, such as rs5902434 and rs3761548, have been shown to be associated with autoimmune or inflammatory diseases, such as Crohn's disease, systemic lupus erythematosus, and allergic rhinitis Citation(15–18). However, the association between FOXP3 SNPs and COPD has not been reported. In the present study, we analyzed FOXP3 rs2280883, rs3761548, rs3761549, and rs5902434 SNPs in COPD patients at different stages to investigate the origin of Tregs abnormality in COPD pathogenesis. At the same time, we tested the expression of FOXP3 mRNA in peripheral blood of patients with COPD at different stages to explore whether there is any relationship between FOXP3 SNPs and Tregs in patients with COPD at different stages.
Materials and methods
Study population
COPD diagnosis was made by using FEV1/forced vital capacity (FVC) lower limits of normal (LLN) and Global Lungs Initiative predicting equations Citation(19) in the First Affiliated Hospital of Guangxi Medical University (Guangxi, China). Furthermore, the stage of COPD was evaluated according to the definition of the Global Initiative for Chronic Obstructive Lung Disease (GOLD) Citation(20). For comparison, nonsmokers with normal lung function and current smokers with normal lung function were selected as control subjects. The control subjects had no history of cardiovascular disease, hypertension, diabetes, cancer, allergic diseases and other immune-related diseases, tuberculosis, bronchiectasis, pulmonary fibrosis, and other lung diseases other than COPD, who were recruited from a pool of volunteers participating physical examination in our hospital. Information on demographic characteristics, including age and smoking habits, was obtained from personal interviews administered by trained personnel. The study was approved by the local ethics committee (the First Affiliated Hospital of Guangxi Medical University ethical committee, Guangxi, China) and conformed to the Declaration of Helsinki; written informed consent was obtained from each subject.
Selection of SNPs in FOXP3 gene region
We selected SNPs in the FOXP3 gene region from chromosome X with a minor allele frequency (MAF) >5% available in the international HapMap database (http://hapmap.ncbi.nlm.nih.gov/index.html.en) and Ensembl SNP databases (http://www.ensembl.org/index.html). The choice of this MAF was based on the factors including the sample size, the number of SNPs to be genotyped, and the power of this study to detect the risk allele (80%,α = 0.05). Moreover, SNPs in FOXP3 gene region were selected based on the evidence that they were involved in inflammatory or autoimmune diseases Citation(15–18, 21, 22). Therefore, rs2280883, rs3761548, rs3761549, and rs5902434 with MAF >5% in Asian population were selected in our study.
Quantitative real-time PCR
Total RNA was extracted from peripheral blood using the TRIzol reagent (Invitrogen, Carlsberg, CA, USA) according to the manufacturer's instructions and stored at −80°C until use. The concentration and purity of RNA were estimated using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). RNA purity was calculated from the ratio of absorbance at 260 and 280 nm. The mean RNA purity (A260/A280) was evaluated as 1.9 ± 0.0. The cDNA was synthesized with RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, Canada).
A quantitative real-time polymerase chain reaction (PCR) was performed with a ABI StepOnePlus Real-Time PCR System (Applied Biosystems, CA, US) and the SYBR® Premix Ex Taq™ (TaKaRa) in accordance with the manufacturer's protocol. Since β-actin is the most common marker in peripheral blood mononuclear cells in human according to the lowest percent coefficient of variation and standard deviation and the highest correlation coefficient compared to other housekeeping genes, including GAPDH and beta-2 microglobulin Citation(23), we chose only β-actin to be reference. The following were the primers used: human Foxp3 F, 5′-CTGGCAAATGGTGTCTGCAAGT-3′ and R, 5′-CTGCCCTTCTCATCCAGAAGATG-3′; β-actin F, 5′-ACACTGTGCCCATCTACG-3′ and R, 5′-TGTCACGCACGATTTCC-3′. Each set of experiments was repeated three times. The 25 μl PCR reaction mixtures (with 12.5 μl SYBR green, 5 × 10−6 μl M forward and 5 × 10−6 μl M reverse primers, and 2 μl cDNA) underwent the following reactions: 30 s at 95°C, 5 s at 90°C, and then 40 cycles of 30 s at 60°C. Spectral data were captured by using StepOne Software v2.1. Gene expression was normalized to the expression of β-actin.
Genotype method
Whole blood samples (2 ml) from the 378 subjects were collected in EDTA tubes. Genomic DNA was extracted using a DNA Purification Kit (Quick-gDNA™ Blood MicroPrep, Zymo Research, Irvine, CA, US). Genotyping of the FOXP3 rs2280883, rs3761548, rs3761549, and rs5902434 SNPs were performed using TaqMan assays (ABI StepOnePlus Real-Time PCR System; Applied Biosystems, CA, US). Genotypes were scored automatically using the StepOne Software v2.1. The quality and potential misclassification of the genotyping results were assessed by evaluating 10% of duplicate DNA samples that were randomly selected from the whole samples. Their replicates were 100% concordant. The genotyping was performed by technicians blind to the groups of subjects.
Statistical method
The difference in general characteristics between the multiple groups was tested using one-way analysis of variance (ANOVA) followed by the Student–Newman–Keuls test or the Games–Howell test. Hardy–Weinberg equilibrium and linkage disequilibrium (LD) test was estimated using the Haploview program (version 4.2, Broad Institute of Harvard and MIT, Cambridge, MA, USA) Citation(24). The difference in the genotype and allele distributions between COPD patients and controls were tested for significance by using standard Chi-square test or Fisher exact test. Correlation between FOXP3 mRNA levels and lung function or SNPs distribution was calculated using Spearman's rank method. Multiple logistic regression analysis was performed with adjustment for age, smoking history, and body mass index (BMI) to obtain the odds ratio (OR) for the risk of COPD or lung function decline at 95% confidence interval (CI). The results were presented as ORs and 95% CIs. p-values < 0.05 were considered to be statistically significant. Group data are expressed as mean ± std. deviation (SD). All statistical analyses were performed using SPSS 17.0 (IBM SPSS Inc., Chicago, IL, USA).
Results
Characteristics of the study participants
shows the general characteristics and lung function of the subjects included in the study on FOXP3 SNPs. Of the 378 subjects who met the inclusion criteria and were genotyped, 140 met GOLD 3 and 4 stage of COPD definition criteria (COPD 3–4 group), 67 were in GOLD 1 and 2 stage of COPD (COPD 1–2 group), 100 were unaffected smoking controls (S group), and 71 were nonsmoking controls (N group). All subjects were selected from November 1st, 2011 to October 31th, 2013, and were males. Age, smoking history, and BMI were different between N group, S group, and COPD patients, but their effect on the distribution of SNP in all subjects was analyzed by multiple logistic regressions. The lung functions in COPD patients were lower than that in N group and S group, and were declined as the severity of COPD developed.
Table 1. Subject characteristics in the study on FOXP3 rs5902434 SNP.
shows the general characteristics and lung function of the subjects in the study on the expression of FOXP3 mRNA. All 80 subjects were selected from November 1st, 2011 to January 31th, 2012, and were subdivided into four groups: 20 subjects were nonsmokers with normal lung function (N group), 20 subjects were current smokers with normal lung function (S group), 20 subjects were smokers with COPD at GOLD 1 and 2 stage (COPD 1–2 group), and 20 subjects were smokers with COPD at GOLD 3 and 4 stage (COPD 3–4 group). No differences with age and BMI were observed in all four groups. Smoking history was not different between S group and COPD groups. Meanwhile, the lung function in COPD patients was worse than S and N groups.
Table 2. Subject characteristics in the study on the expression of FOXP3 mRNA.
Expression of FOXP3 mRNA in peripheral blood
FOXP3 mRNA expression levels were significantly lower in COPD 3–4 group (0.498 ± 0.235) and in COPD 1–2 group (0.931 ± 0.237) than in unaffected smoking controls (1.215 ± 0.258) and nonsmokers (1.452 ± 0.254). Furthermore, Foxp3 mRNA expression level in COPD 3–4 group was significantly decreased compared with that in COPD 1–2 group. (ANOVA p < 0.001, all p-values were <0.05 when every two groups were compared between the four groups) ().
Figure 1. The expression of FOXP3 mRNA in peripheral blood. The expressions of FOXP3 mRNA in COPD 3–4 group and in COPD 1–2 group were significantly decreased compared with smokers with normal lung function and nonsmokers. N: nonsmokers with normal lung function. S, smokers with normal lung function; COPD 1–2 group, COPD patients at 1–2 stage; COPD 3–4 group, COPD patients at 3–4 stage. *p < 0.05.
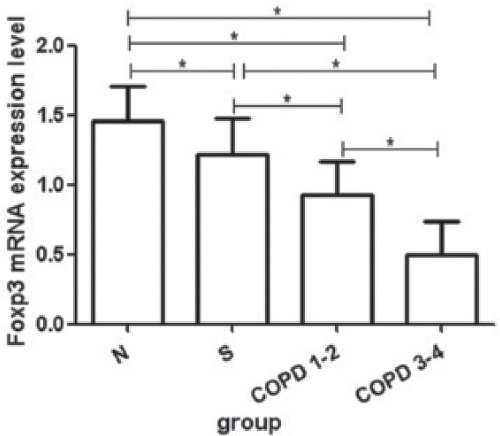
Figure 2. Correlation. A: The FOXP3 mRNA expression level was negatively correlated with smoking history. B: The expression of Foxp3 mRNA in peripheral blood was positively correlated with FEV1%pred. FEV1%pred: forced expiratory volume in 1 s % predicted.
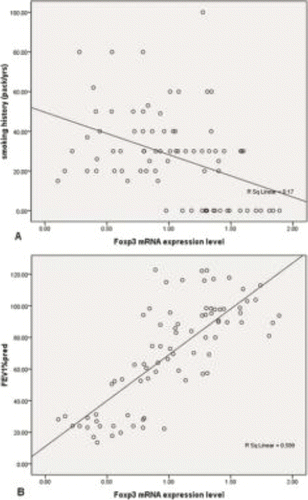
The correlation analysis showed that the FOXP3 mRNA expression level was negatively correlated with smoking history (R = −0.424,p < 0.001), and positively correlated with forced expiratory volume in 1 s % predicted (FEV1%pred) (R = 0.759, p < 0.001) (). However, no significant correlation between FOXP3 mRNA level and age was observed.
Genotypic and allelic distribution of FOXP3 SNPs
All FOXP3 SNPs, namely rs2280883, rs3761548, rs3761549, and rs5902434, were consistent with Hardy–Weinberg equilibrium for COPD patients and control groups with a p-value from 0.079 to 0.213. These four SNPs were under a low linkage disequilibrium (LD) in all subjects (). The genotypic frequency of four SNPs in COPD patients and controls is shown in . Among four SNPs, only rs5902434 SNP was observed to differently distribute among all groups (p = 0.008) (, ). The genotypic frequency of del/del, del/ATT, and ATT/ATT was 59.15%, 35.21%, and 5.63% in N group; 58.00%, 34.00%, and 8.00% in S group; 41.79%, 44.78%, and 13.43% in COPD 1–2 group; and 37.14%, 46.43%, and 16.43% in COPD 3–4 group, respectively. Furthermore, the rs5902434 SNP genotype was differently distributed in COPD 3–4 group (p = 0.004) compared with S group. However, there was no significant difference in the genotypic frequency of rs5902434 SNP between N group and S group, and between COPD 1–2 group and COPD 3–4 group, respectively. Therefore, genotypic frequencies of FOXP3 rs5902434 SNP were differently distributed between non-COPD subjects and COPD patients. However, no difference was observed among COPD patients in different grades.
Figure 3. Schematic overview of human FOXP3 gene and linkage disequilibrium analysis in COPD cases and controls (Haploview 4.20 version). The standard (D/LOD) was represented by red color and r2 was presented as a number.
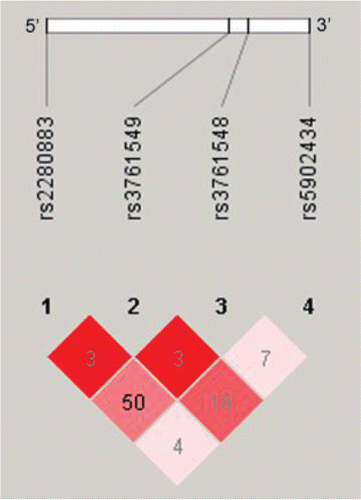
Table 3. Genotypic frequency of rs2280883, rs3761548, rs3761549, and rs5902434.
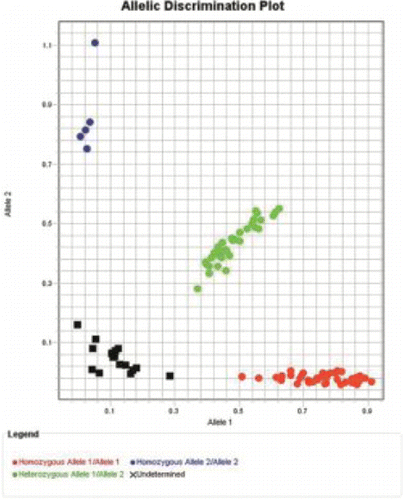
Figure 4. The genotyping scatterplot of FOXP3 SNP rs5902434. SNP: single nucleotide polymorphism. Red: −/−, green: -/ATT, blue: ATT/ATT, black: negative control.
The allelic distribution of these four SNPs in COPD patients and controls is shown in . Similar to the genotypic distribution of these four SNPs, only the rs5902434 SNP was observed to be differently expressed among all groups (p < 0.001) that the frequency of del and ATT was 76.76% and 23.24% in N group; 75.00% and 25.00% in S group; 64.18% and 35.82% in COPD 1–2 group; and 60.36% and 39.64% in COPD 3–4 group, respectively. Moreover, the rs5902434 SNP allele was differently distributed in COPD 1–2 group (p = 0.033) and COPD 3–4 group (p = 0.001), respectively, compared with S group. However, there was no significant difference in the allelic frequencies of rs5902434 SNP between N group and S group, and between COPD 1–2 group and COPD 3–4 group, respectively. Therefore, the alleles were differently distributed between non-COPD subjects and COPD patients.
Table 4. Allelic frequency of rs2280883, rs3761548, rs3761549, and rs5902434.
Association between SNPs and risk of COPD and lung function
We investigated the association between all four SNPs and the risk of COPD with multiple logistic regression model after adjusting for smoking history, age, and BMI (). Only rs5902434 genotype del/del was associated with a decreased risk of COPD (COPD 1–2: OR = 0.284, 95%CI 0.076–0.956; COPD 3–4: OR = 0.195, 95%CI 0.060–0.635). Moreover, rs5902434 allele del was associated with a decreased risk of COPD 3–4 (OR = 0.270, 95%CI 0.086–0.848).
Table 5. The association between FOXP3 SNPs genotypes and risk of COPD.
Furthermore, we explored the association between rs5902434 and lung function using multiple logistic regression after adjusting for smoking history, age, and BMI. The genotype del/del was associated with protection from forced expiratory volume in 1 s % predicted (FEV1%pred) less than 80% (FEV1%pred = 30–49: OR = 0.194 95%CI 0.080–0.473; FEV1%1pred = 50–79: OR = 0.333, 95%CI 0.121–0.912), and from FEV1/FVC less than 70% (FEV1/FVC<50%: OR = 0.236, 95%CI 0.101–0.550). In addition, the allele del was associated with protection of lung function decline (FEV1%pred = 30–49: OR = 0.268, 95%CI = 0.115–0.623; FEV1/FVC<50%: OR = 0.318, 95%CI 0.141–0.716; FEV1/FVC = 50–69%: OR = 0.377, 95%CI 0.148–0.962).
Correlation between FOXP3 mRNA level and rs5902434
The correlation between FOXP3 mRNA level and genotype frequencies of FOXP3 rs5902434 SNP was analyzed by Spearman's rank method in 80 subjects included in FOXP3 mRNA level investigation. The correlation analysis showed FOXP3 mRNA level was correlated with genotypic distribution of FOXP3 rs5902434 SNP (R = −0.326,p = 0.003). This finding suggested that the genotype del/del was associated with higher expression of FOXP3 mRNA.
Discussion
Our present study found the expression level of FOXP3 mRNA was decreased in COPD patients, especially in patients at GOLD 3 and 4, compared with smokers and nonsmokers, which was correlated with lung function. SNP analysis showed that genotypic and allelic frequencies of FOXP3 rs5902434 genotype del/del and allele del are lower in COPD patients than that in nonsmokers or smokers with normal lung function. We further found that rs5902434 genotype del/del and allele del were, respectively, associated with decreased risk of COPD and lung function decline. Both rs5902434 genotypic distribution and lung function were correlated with FOXP3 mRNA level. Our results suggest that the abnormal expression of FOXP3 exists in COPD patients, which aggravates as the lung function declines. FOXP3 rs5902434 genotype del/del and allele del may be associated with decreased risk of COPD and lung function decline in Chinese males, probably by altering Foxp3 expression.
The results of the present study showed that the FOXP3 expression in system was decreased as the severity of COPD aggravates, which confirmed previous finding by Wang et al. Citation(9). In COPD patients, expression of FOXP3 decreased in small airways Citation(25). Moreover, our previous study has showed a decrease expression of FOXP3 in lung tissues of COPD patients compared with normal smokers and nonsmokers Citation(5). These findings suggested the possible role of Tregs in the respiratory inflammation of COPD patients. The result of the present study is consistent with FOXP3 expression in small airway and lung parenchyma. Furthermore, the FOXP3 level was significantly lower in COPD patients in GOLD 3 or 4 grade than that in GOLD 1 or 2 grade, suggesting that inflammation involved in FOXP3 in system may be similar to that in lung parenchyma, which aggravated as COPD developed. Moreover, the impaired function of FOXP3+ cells was found in COPD patients Citation(26), and similar finding was observed in the rats with cigarette smoke-induced airway inflammation Citation(27), which confirmed the role of FOXP3 in inflammation of COPD.
Based on the findings of FOXP3 expression in lung parenchyma and in system, we further investigated the origin of FOXP3 abnormality in COPD patients in view of gene polymorphism. Individual susceptibility to COPD has been shown to vary with the presence of SNPs in several genes of critical inflammatory cytokines Citation(28–30). The rs5902434 SNP was evaluated in relation to psoriasis in a Han Chinese population by Gao et al. Citation(31) and no association was found, but the evaluation of rs5902434 SNP with respect to unexplained recurrent spontaneous abortion by Wu et al. Citation(32) showed that this polymorphism of the FOXP3 gene may confer an important susceptibility in Chinese population. This study found that the genotypic frequency of the rs5902434 SNP between COPD patients and smokers with normal lung function was different, while this genotypic frequency between nonsmokers and smokers with normal lung function, or between COPD 1–2 group and COPD 3–4 group, was not significantly different. The present study further found that the genotype del/del was associated with a decreased risk of COPD. The analysis of association between the genotype del/del and lung function confirmed this result, as the genotype del/del may be a protective factor for the decline of lung function. A similar finding was shown in the analysis of rs5902434 allele del. The allele del was associated with a decreased risk of COPD, in particular severe or very severe COPD. However, we cannot find a high LD among all four SNPs. Since some introns could encode functional RNAs after splicing to noncoding RNAs Citation(33), our result suggested a potential role of noncoding RNAs in immunoinflammation of COPD, which needs to be explored by further study.
Although our study observed a different distribution of rs5902434 and FOXP3 mRNA expression among COPD patients and non-COPD patients, our studies cannot directly prove a causal relationship between rs5902434 and FOXP3 mRNA levels. The precise relationship between SNPs and FOXP3 expression or even Tregs' function, and its possible mechanism need further investigation in in vivo and in vitro study. Our findings in the present study may provide an insight into immunoinflammation of COPD pathogenesis from genetic polymorphisms.
We acknowledge that the diagnostic criteria of COPD according to GOLD might lead to over-diagnosis in some older subjects. Since the FEV/FVC ratio declines with age in healthy persons Citation(34), the fixed 0.7 threshold for the FEV1/FVC ratio may limit diagnostic accuracy in the older people. Thus, in our study, we made a diagnosis of COPD by using FEV1/FVC LLN, and then evaluated the stage of COPD according to GOLD.
Conclusion
In conclusion, both FOXP3 rs5902434 genotypes and alleles were differently distributed in COPD patients and smokers with normal lung function. The distribution of del/del genotype was associated with systemic expression of FOXP3 mRNA. More research is needed to explore the role of FOXP3 gene polymorphism in immunoinflammation of COPD.
Declaration of interest
The content of this report does not necessarily reflect the position or policy of the Chinese Government. No conflict of interest, financial or otherwise, is declared by the authors.
Funding
This study was funded by the National Natural Science Foundation of China [81160010] and [81260013].
References
- Bluestone JA, Tang Q. How do CD4+CD25+ regulatory T cells control autoimmunity? Curr Opin Immunol 2005; 17:638–642.
- Jiang H, Chess L. Regulation of immune responses by T cells. N Engl J Med 2006; 354:1166–1176.
- Hou J, Sun Y, Hao Y, Zhuo J, Liu X, Bai P, et al. Imbalance between subpopulations of regulatory T cells in COPD. Thorax 2013; 68(12):1131–1139.
- Isajevs S, Taivans I, Strazda G, Kopeika U, Bukouskis M, Gordjusina V, et al. Decreased FOXP3 expression in small airways of smokers with COPD. Eur Respir J 2009; 33(1):61–67.
- Chu S, Zhong X, Zhang J, Lao Q, He Z, Bai J. The expression of Foxp3 and ROR gamma t in lung tissues from normal smokers and chronic obstructive pulmonary disease patients. Int Immunopharmacol 2011; 11(11):1780–1788.
- Bai J, Qiu SL, Zhong XN, Huang QP, He ZY, Zhang JQ, et al. Erythromycin enhances CD4+Foxp3+ regulatory T-cell responses in a rat model of smoke-induced lung inflammation. Mediators Inflamm 2012; 2012:410232.
- Domagała-Kulawik J, Hoser G, Dąbrowska M, Safianowska A, Chazan R. CD4+/CD25+ cells in systemic inflammation in COPD. Scand J Immunol 2011; 73(1):59–65.
- Roos-Engstrand E, Pourazar J, Behndig AF, Bucht A, Blomberg A. Expansion of CD4+CD25+ helper T cells without regulatory function in smoking and COPD. Respir Res 2011; 12:74.
- Wang H, Ying H, Wang S, Gu X, Weng Y, Peng W, et al. Imbalance of peripheral blood Th17 and Treg responses in patients with chronic obstructive pulmonary disease. Clin Respir J 2015; 9(3):330–341.
- Sørheim I, Johannesse A, Gulsvik A, Bakke PS, Silverman EK, DeMeo DL. Gender differences in COPD: are women more susceptible to smoking effects than men? Thorax 2010; 65:480–485.
- Løkke A, Lange P, Scharling H, Fabricius P, Vestbo J. Developing COPD: a 25-year follow up study of the general population. Thorax 2006; 61:935–939.
- Gamble E, Grootendorst DC, Hattotuwa K, O'Shaughnessy T, Ram FS, Qiu Y, et al. Airway mucosal inflammation in COPD is similar in smokers and ex-smokers: a pooled analysis. Eur Respir J 2007; 30 (3):467–471.
- Willemse BW, ten Hacken NH, Rutgers B, Lesman-Leegte IG, Postma DS, Timens W. Effect of 1-year smoking cessation on airway inflammation in COPD and asymptomatic smokers. Eur Respir J 2005; 26 (5):835–845.
- Turato G, Di Stefano A, Maestrelli P, Mapp CE, Ruggieri MP, Roggeri A, et al. Effect of smoking cessation on airway inflammation in chronic bronchitis. Am J Respir Crit Care Med 1995; 152 (4 Pt 1):1262–1267.
- Park O, Grishina I, Leung PS, Gershwin ME, Prindiville T. Analysis of the Foxp3/scurfin gene in Crohn's disease. Ann N Y Acad Sci 2005; 1051:218–228.
- Lin YC, Lee JH, Wu AS, Tsai CY, Yu HH, Wang LC, et al. Association of single-nucleotide polymorphisms in FOXP3 gene with systemic lupus erythematosus susceptibility: a case-control study. Lupus 2011; 20(2):137–143.
- Lan Y, Tang XS, Qin J, Wu J, Qin JM. Association of transcription factor FOXP3 gene polymorphism with genetic susceptibility to systematic lupus erythematosus in Guangxi Zhuang population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2010; 27(4):433–436.
- Zhang L, Zhang Y, Desrosiers M, Wang C, Zhao Y, Han D. Genetic association study of FOXP3 polymorphisms in allergic rhinitis in a Chinese population. Hum Immunol 2009; 70(11):930–934.
- Quanjer PH, Stanojevic S, Cole TJ, Baur X, Hall GL, Culver BH, et al. Multi-ethnic reference values for spirometry for the 3-95-yr age range: The global lung function 2012 equations. Eur Respir J. 2012;40(6):1324–1343. Available at: http://www.ers-education.org/guidelines/global-lung-function-initiative
- Rabe KF, Hurd S, Anzueto A. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 2007; 176(6):532–555.
- Fodor E, Garaczi E, Polyánka H, Koreck A, Kemény L, Széll M. The rs3761548 polymorphism of FOXP3 is a protective genetic factor against allergic rhinitis in the Hungarian female population. Hum Immunol 2011; 72(10):926–929.
- Bossowski 1, Borysewicz-Sańczyk H, Wawrusiewicz-Kurylonek N, Zasim A, Szalecki M, Wikiera B, et al. Analysis of chosen polymorphisms in FoxP3 gene in children and adolescents with autoimmune thyroid diseases. Autoimmunity 2014; 47(6):395–400.
- Sudchada P, Oo-puthinan S, Kerdpin O, Saelim N. ABCB1 gene expression in peripheral blood mononuclear cells in healthy Thai males and females. Genet Mol Res 2010; 9(2):1177–1185.
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21(2):263e5.
- Isajevs S1, Taivans I, Strazda G, Kopeika U, Bukovskis M, Gordjusina V, et al. Decreased FOXP3 expression in small airways of smokers with COPD. Eur Respir J 2009; 33(1):61–67.
- Tan DB, Fernandez S, Price P, French MA, Thompson PJ, Moodley YP. Impaired function of regulatory T-cells in patients with chronic obstructive pulmonary disease (COPD). Immunobiology 2014; 219(12):975–979.
- Meng JJ, Zhong XN, Bai J, He ZY, Zhang JQ, Huang QP. Changes of CD(4)(+)Foxp3(+) regulatory T cells and CD(4)(+)IL-17(+)T cells in cigarette smoke-exposed rats. Zhonghua Jie He He Hu Xi Za Zhi 2012; 35(1):55–60.
- He JQ, Foreman MG, Shumansky K, Zhang X, Akhabir L, Sin DD, et al. Associations of IL6 polymorphisms with lung function decline and COPD. Thorax 2009; 64(8):698–704.
- Trajkov D, Mirkovska-Stojkovikj J, Petlichkovski A, Strezova A, Efinska-Mladenovska O, Sandevska E, et al. Association of cytokine gene polymorphisms with chronic obstructive pulmonary disease in Macedonians. Iran J Allergy Asthma Immunol 2009; 8(1):31–42.
- Ito M, Hanaoka M, Droma Y, Hatayama O, Sato E, Katsuyama Y, et al. The association of transforming growth factor beta 1 gene polymorphisms with the emphysema phenotype of COPD in Japanese. Intern Med 2008; 47(15):1387–1394.
- Gao L, Li K, Li F, Li H, Liu L, Wang L, et al. Polymorphisms in the FOXP3 gene in Han Chinese psoriasis patients. J Dermatol Sci 2010; 57(1):51–56.
- Wu Z, You Z, Zhang C, Li Z, Su X, Zhang X, et al. Association between functional polymorphisms of Foxp3 gene and the occurrence of unexplained recurrent spontaneous abortion in a Chinese Han population. Clin Dev Immunol 2012; 2012:896458.
- Rearick D, Prakash A, McSweeny A, Shepard SS, Fedorova L, Fedorov A. Critical association of ncRNA with introns. Nucleic Acids Res 2011; 39(6):2357–2366.
- Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med 1999; 159(1):179–187.
