ABSTRACT
Lack of genomic resources in sunn hemp (Crotalaria juncea L.), a leguminous bast fiber crop, is one of the major constraints for germplasm characterization and genetic analysis. We identified non-redundant sets of a variety of molecular markers from the de novo flower-bud transcriptome of sunn hemp. These molecular markers included 1683 simple sequence repeats, 4759 intron length polymorphism markers primarily from protein-coding genes, and 3309 single nucleotide polymorphism allele-specific markers. All of these marker resources have been integrated into a simple database for easy access. To evaluate marker efficiency and polymorphism information content of the sampled primers, a subset of SSR and intron length polymorphism markers were validated in sunn hemp germplasm accessions to obtain average PIC values of 0.26–0.32. The SSR genotyping profiles also aided in the genetic distance matrix-based clustering of sunn hemp germplasm accessions. The ability of these molecular markers to successfully amplify in cross-genera bast fiber crops further demonstrated their effectiveness in genotyping. Thus, the current study describes the identification of previously undisclosed endogenous molecular markers in sunn hemp, which have potential applications in genetic studies and breeding in sunn hemp for bast fiber improvement.
摘要
阳光大麻(Crotalaria juncea L.)是一种豆科韧皮纤维作物,缺乏基因组资源是种质鉴定和遗传分析的主要制约因素之一. 我们从太阳大麻的新花蕾转录组中鉴定了多种分子标记的非冗余组. 这些分子标记包括1683个简单序列重复、4759个主要来自蛋白质编码基因的内含子长度多态性标记和3309个单核苷酸多态性等位基因特异性标记. 所有这些标记资源都已集成到一个简单的数据库中,便于访问. 为了评估所选引物的标记效率和多态性信息含量,在阳光大麻种质资源中验证了SSR和内含子长度多态性标记的子集,以获得0.26–0.32的平均PIC值. SSR基因分型图谱也有助于基于遗传距离矩阵的向日葵种质资源聚类. 这些分子标记在跨属韧皮纤维作物中成功扩增的能力进一步证明了它们在基因分型中的有效性. 因此,本研究描述了之前未公开的向日葵内源分子标记的鉴定,这些标记在向日葵的遗传研究和育种中具有潜在的应用价值,以改善韧皮纤维.
Introduction
Sunn hemp (Crotalaria juncea L.; 2n = 2x = 16; Fabaceae), also known as Indian hemp, is a rapid-growing, nitrogen-fixing annual plant. It originated in India and is primarily grown as a green manure, pasture, and cover crop that can also suppress soil-borne nematodes (Bhandari et al. Citation2016; Hinds et al. Citation2013; Mosjidis and Wang Citation2011). It enriches organic matter by sequestering carbon in the soil and has the potential to be used as a biofuel source. Furthermore, the lignified bast fiber derived from sunn hemp stem is a significant economic product used in the manufacturing of cordage, fishing nets, ropes, twines, high-quality tissue paper, and currency papers (Cook and White Citation1996).
The late-acting gametophytic self-incompatibility system in sunn hemp prevents self-fertilization and promotes cross-pollination, resulting in a genetically heterozygous plant population (Mitra Citation1976; Rangappa Thimmaiah et al. Citation2018; Sarkar et al. Citation2015). Pollination occurs in the field when the stigmatic surface of the pistil is stimulated by the visit of specific insect pollinators such as bumblebees (Xylocopa spp.) and a giant resin bee, Megachile spp. (Bhandari et al. Citation2016; Hall and Avila Citation2016). As a highly cross-pollinated crop, estimating genetic diversity in the sunn hemp population is extremely critical for breeding applications and producing genetically pure seeds. Until now, the lack of molecular markers from the sunn hemp genome has prevented us from characterizing germplasm and exploiting them in marker-assisted breeding (Mosjidis and Wang Citation2011). However, a few attempts have been made to characterize sunn hemp germplasm using cross-species molecular markers (Satya et al. Citation2016; Wang et al. Citation2006). In addition, the genomic sequence in the public domain database is extremely limited for marker development. To date, a nucleotide search against the NCBI database using the keyword “Crotalaria” yielded only 5312 hits, with C. juncea accounting for only 201 of them. The reasons for the low genomic resources of Crotalaria in the NCBI database are its underutilized status and a lack of sequencing efforts (Mosjidis and Wang Citation2011).
Among the various molecular markers used to characterize plant genetic diversity, PCR-based genic molecular markers such as simple sequence repeat (SSR), intron-length polymorphism, and InDel markers are the most cost-effective and reliable markers for a small- to medium-budget laboratory. SSR markers are codominant in nature and are preferred in the majority of plant genotyping applications. With the advancement and popularization of next-generation sequencing (NGS) methods, genome and transcriptome sequences are being exploited to develop genic molecular markers on a large scale, even in underutilized plant species (Gujaria et al. Citation2011; Poczai et al. Citation2013; Varshney et al. Citation2007). These markers are known as functional markers because they represent nucleotide sequence variation as part of the transcribed portion of the genes. Functional markers, as opposed to random DNA-sequence derived markers, are frequently advantageous in tagging genes with adaptive phenotypic traits (Varshney et al. Citation2007). Because genomic resources and molecular markers were scarce in sunn hemp prior to this study, we used the flower-bud transcriptome assembly of C. juncea var. SUN-053 (Swastika) to identify a large number of genic molecular markers. A subset of these markers was also validated in sunn hemp and heterologous plant species for germplasm characterization and marker-assisted breeding applications.
Materials and methods
Plant samples and DNA isolations
Plant samples used in the study included 24 sunnhemp germplasm accessions (germplasm panel accessions represent collected plant lines across different agro-ecological regions of India, which are being characterized for fiber yield under a separate study), four Crotalaria wild species, two varieties each of Linum usitatissimum (flax), Hibiscus cannabinus (kenaf), Corchorus capsularis (white jute), and C. olitorious (dark jute) (species panel). A standard mini extraction protocol with cetyltrimethylammonium bromide (CTAB) buffer (Doyle and Doyle Citation1990) was used to isolate genomic DNA from the freshly harvested tender leaves of all plant accessions in liquid nitrogen. The isolated genomic DNA was purified using RNase A (20 mg/mL) followed by quality checking for an intact band on agarose gel electrophoresis and quantified in Qubit 4.0 Fluorometer (Thermo Fisher Scientific, USA) using the dsDNA HS (high sensitivity) Assay Kit (Thermo Fisher Scientific, USA). All the genomic DNA samples were diluted to 50 ng/µL and stored at −20°C in the deep freezer prior to use in the PCR reactions.
Simple sequence repeat identification from sunn hemp transcriptome assembly
A sunn hemp transcriptome assembly developed by us (Transcriptome Shotgun Assembly accession NCBI GenBank: GFBY00000000.1; Short Read Archive: SRR5043622; BioProject: PRJNA350604) was searched for simple sequence repeat (SSR) loci identification with the MISA (MIcroSAtellite) identification tool (http://pgrc.ipk-gatersleben.de/misa/). The SSR identification criteria followed dinucleotide = 8, trinucleotide = 6, tetra, penta, and hexanucleotide = 5 repeats each. Because mononucleotide repeats are abundant in the genome but are rarely used in marker studies, these repeat classes were excluded from the identification process. The maximum gap distance between the two SSRs was chosen as 100 nucleotides. Subsequently, the SSR flanking primer pairs were designed from the perfect SSR repeats using the primer3_core program (Primer3 1.0.1) (http://primer3.sourceforge.net/releases.php). The parameters used for primer designing were an amplicon size of 100–400 bp, primer annealing temperature of 55–65°C, primer length of 18–28 bp, and GC content of 45–55%. The EST-SSR primers were filtered to obtain non-redundant primers by manually removing the duplicate primer sequences followed by employing the EMBOSS primer search tool (http://emboss.sourceforge.net/apps/cvs/emboss/apps/primersearch.html) to identify and filter putative multiple primer binding sites in the sunn hemp transcriptome.
Intron length polymorphism primer identifications
The potential intron polymorphism (PIP) markers were developed from the putative introns predicted in the sunn hemp transcriptome using the PIP database’s “Develop” tool (http://ibi.zju.edu.cn/pgl/pip/index.html) (Yang et al. Citation2007) and “dicot” plant Arabidopsis as a query species. Both the EST-SSR and PIP markers were filtered further to obtain non-redundant primers using the EMBOSS primer search tool (http://emboss.sourceforge.net/apps/cvs/emboss/apps/primersearch.html), which performed a digital PCR to identify putative unique amplification products in the transcriptome sequence assembly.
Single nucleotide polymorphism loci discovery and Kompetitive allele-specific PCR primer designing
Additionally, SNPs and InDel loci were predicted from the sunn hemp transcriptome assembly using the SAMtools-mpileup/BCFtools v.1.9 (http://github.com/samtools/bcftools). For final variant loci calling, a very stringent filtering option was used to exclude variant quality scores of <40 and >100 call-depth. Kompetitive allele-specific PCR (KASP) primers were designed from the SNP flanking sequences of transcripts homologous to SI-related genes using the online Web-based Allele-Specific Primer (WASP) design tool (http://bioinfo.biotec.or.th/WASP) (Wangkumhang et al. Citation2007).
Validation of molecular markers in the sunnhemp germplasm
For marker validations with sunn hemp and other bast fiber plant DNA samples, 48 primer pairs each for EST-SSR and PIP markers were chosen at random (Table S1). Genomic DNA (50 ng) was used as a template for amplification by PCR in a Biometra TOne (Analytik Jena, Germany) thermal cycler using 94°C for 5 m, 35 cycles of 94°C for 30 s, 52–56°C for 30 s, 72°C for 30 s, and 72°C for 5 m for the EST-SSR and PIP marker profiling. The PCR amplification products were resolved on a 6% native polyacrylamide agarose gel electrophoresis (PAGE), post-electrophoresis stained with Ethidium bromide solution (0.5 μg mL−1) for 5 m, and documented using a UV transilluminator in a gel documentation system (AlphaImager HP, USA). Only the sharp bands with expected allele size were manually scored, and the primer efficiency analysis of SSR and PIP markers was conducted using PowerMarker v.3.25 (Liu and Muse Citation2005) and iMEC (Amiryousefi, Hyvönen, and Poczai Citation2018) tools, respectively. The PowerMarker generated distance matrix based on Nei’s distance measure and the neighbor joining (NJ) approach was used to generate a phylogenetic tree of 24 sunn hemp germplasm accessions.
Development of a simple database of sunn hemp molecular markers
All the sunn hemp molecular markers developed in the study were stored into a simple “Sunn hemp molecular marker database” using the Google spreadsheet for easy access. The database can be accessed using the following URL: https://docs.google.com/spreadsheets/d/1DNTYpqIHidRoPJrCgdRPt-3pWNAhbiToZwoQdOyzxQA/edit?usp=sharing.
Results and discussion
Transcriptome-wide identification of SSR loci and marker development
Despite producing commercially important bast fiber, sunn hemp has received little attention in biological research as compared to major food legumes. As a result, genetic and genomic resources such as molecular markers have been underutilized in the characterization and genetic improvement of sunn hemp germplasm. The NGS technologies now allow for the rapid generation of genomic and transcriptomic resources for the identification and development of a large number of molecular markers in any organism, including previously unknown plant species (Thudi et al. Citation2021). Transcriptome sequences of a variety of underutilized legumes were used to develop genic markers (Henry et al. Citation2012; Thakur and Randhawa Citation2018; Zhang et al. Citation2017). To date, no genomic or transcriptomic sequences from sunn hemp have been found in public domain databases for the development of molecular markers. Previously, we attempted to identify genes associated with late-acting self-incompatibility by generating a reference-quality de novo transcriptome from the flower buds of C. juncea var. SUN-053 (communicated). In this study, we analyzed the transcriptome assembly for identification of different simple sequence repeats (SSRs) and found 4064 SSR loci from 3558 contigs ().
Table 1. Summary of SSR markers designed from sunn hemp transcripts.
A small number of 437 contigs produced multiple SSR loci. Except for 243 SSR loci that were found as compound SSR types, the SSR findings yielded a high percentage (94%) of perfect SSR repeats. In terms of SSR repeat type distribution, the di-nucleotide repeat classes included 1131 (27.83%) SSR loci, while the tri-nucleotide repeats corresponded to 2485 (61.15%) loci (). There were 358 (8.8%) tetra-nucleotide repeats and a small number of penta- and hexa-nucleotide repeats found as well. In terms of SSR marker use, di- and tri-nucleotide repeat SSR markers have been found to be most effective in detecting genetic polymorphisms in a variety of plant species (He et al. Citation2009; Zhao, Prakash, and He Citation2012). Despite the lack of correlation, the abundance of tri-nucleotide SSR types was found to be very high in several plant species, including other bast fiber crops (Saha et al. Citation2017; Wang et al. Citation2014). In the sunn hemp transcriptome, we found a similar pattern, with more than 88.9% of SSR loci belonging to the di- and tri-nucleotide repeat classes. The most common repeat sequence type was found to be AG/CT repeats (20.87%), followed by AAG/CTT (15.01%) and AAT/ATT (12.87%). The AG/CT repeat motif exhibited a maximum of 39 repeating units ().
Figure 1. (a) Distribution of SSR loci types. (b) Frequency of SSR repeat motifs discovered from sunn hemp transcriptome.
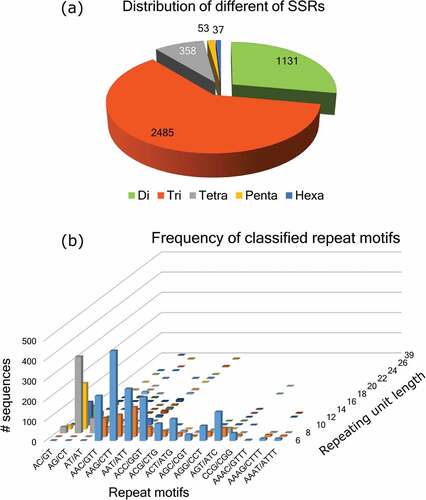
We designed PCR primers from the flanking sequences of the perfect repeats to allow amplification of the SSR loci with predicted amplicon sizes ranging from 100 to 400 bp. The total number of SSR primers designed from the perfect SSR loci was 3591, resulting in a final list of 1683 non-redundant EST-SSR primers after the filtration process (Table S2). In general, the more repeating units (≥20 nucleotide sequences), the more likely the SSR markers are to detect polymorphism, and these are referred to as “hypervariable” SSRs (Temnykh et al. Citation2001). In the sunn hemp EST-SSR primer list, 690 (41.0%) primers were designed from the hypervariable SSR loci. Thus, from the transcriptome sequence, we were able to identify a large number of EST-SSR primers that were previously undisclosed in sunn hemp.
Comparative identification of intron-length polymorphism markers
Exon-primed intron-crossing PCR (EPIC-PCR) is a functional molecular marker used primarily in animal species to assess polymorphism in intron length among a group of individuals by designing PCR primers across conserved exonic regions (Creer Citation2007). In plants, genome-wide identification and utilization of the intron length as polymorphic markers was demonstrated in rice (Wang et al. Citation2005). Subsequently, a database on potential intron polymorphism (PIP) markers in plants was developed for the purpose of designing primers by comparing EST sequences to exonic regions of model plant systems (Yang et al. Citation2007). We were able to design 4759 non-redundant PIP markers from 2053 sunn hemp transcriptome contigs using this tool and the dicotyledonous Arabidopsis thaliana intron model system as a query (Table S3). InterProScan and NCBI-BLASTx searches on the 2053 PIP marker containing transcripts yielded 8479 and 2040 hits to known protein coding sequences, respectively. This implies that the PIP markers in the sunn hemp transcriptome were derived from the functional region of the genome, which could be used as a source of genic markers for gene or trait mapping.
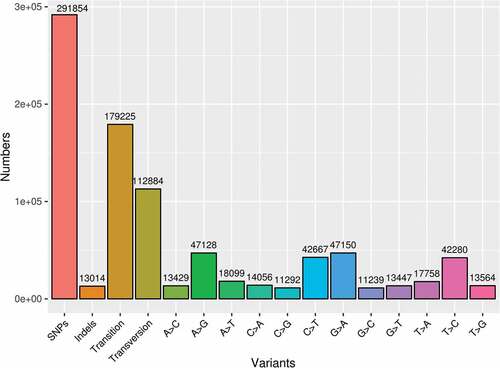
Transcriptome-level SNP marker identifications in sunn hemp and KASP primers
Among the various molecular markers that have been identified, single nucleotide polymorphism (SNP) and insertion deletion (InDel) markers are the most abundant sources of variant loci in any plant system. Although SNP markers were traditionally identified through whole-genome resequencing in a number of individuals, de novo transcriptome sequences were occasionally used to identify SNP/InDels as functional genic markers (Zhao et al. Citation2019). For example, a large number of SNP/InDel markers were identified from the pea transcriptome (Duarte et al. Citation2014) and maize RNA-sequencing data for genetic mapping and marker assisted selection (Chen et al. Citation2021). The SNP/InDel markers can be converted into PCR-based markers through designing differently labeled fluorescence primers spanning the SNP alleles known as Kompetitive allele-specific PCR (KASP) primers (Chunlin, Holme, and Anthony Citation2014). The sunn hemp transcriptome was scanned for putative SNP loci in the current study. With 291,854 SNPs and 13,014 InDels, a total of 304,868 variant alleles were predicted (). Among these variant allele types, 179225 were transition (Ts) type nucleotide substitutions, while 112,884 were transversion type (Tv) substitutions with a Ts/Tv ratio of 1.59. Since sunn hemp reproduction is characterized by a late-acting type self-incompatibility system, we focused on identifying SNP loci in 347 self-incompatibility-related homologous transcripts, which produced 3309 non-redundant KASP primers. These KASP primers were made up of three sets per locus: two primer sets that span either the wild-type or mutant alleles (which can be labeled with different fluorophores for detection) and a common primer (Table S4).
Details of the sunn hemp molecular marker database
We presented the Sunn hemp marker DB under the URL https://docs.google.com/spreadsheets/d/1DNTYpqIHidRoPJrCgdRPt-3pWNAhbiToZwoQdOyzxQA/edit#gid=0 to provide easy access to all three types of molecular markers developed from the Sunn hemp transcriptomic resource. The sunn hemp marker DB contains 1683 EST-SSR primer sets (forward and reverse primer sequences), as well as their associated SSR types, motif sequences, putative melting temperature, and probable amplicon length on cDNA. Similarly, under the PIP-primers tab, 4759 primer sets were provided in the form of a Google spreadsheet, along with primer descriptions. Finally, 3309 SNP/InDel specific KASP primers were enlisted, with three sets of primers; primers 1 and 2 specific for different SNP allele types; and primer 3 as the common reverse primer for that loci.
Validation of EST-SSR and PIP markers in sunn hemp accessions and other heterologous bast-fiber plant species
From the genic markers identified in sunn hemp, 48 each of EST-SSR and PIP markers were randomly validated in a panel of 24 sunn hemp germplasm accessions ( a-b). Additionally, these markers were also tested for their cross-species transferability in four wild Crotalaria species, two varieties each of Linum usitatissimum (flax), Hibiscus cannabinus (kenaf), Corchorus capsularis (white jute), and C. olitorious (dark jute). The details of the genetic diversity and polymorphism indices are presented in . Of the 48 EST-SSR primers evaluated, 20 were polymorphic with clear alleles (mean allele number of 2.5). The heterozygosity (He) and polymorphism information content (PIC) values for the EST-SSR markers ranged between 0.00 to 0.92 (avg. 0.32) and 0.08 to 0.52 (avg. 0.3), respectively.
Figure 3. Representative PAGE gel electrophoresis of a polymorphic (a) EST-SSR primer Cj-eSSR0219, and (b) PIP primer CjPIP01997 in germplasm panel showing polymorphic amplification profiles. (c) EST-SSR primer Cj-eSSR0722, and (d) PIP primer CjPIP01056 showing polymorphic banding profiles in a species panel. After electrophoresis, the PAGE gels were stained with Ethidium bromide solution (0.5 μg mL −1) and documented using UV transilluminator in a gel doc system.
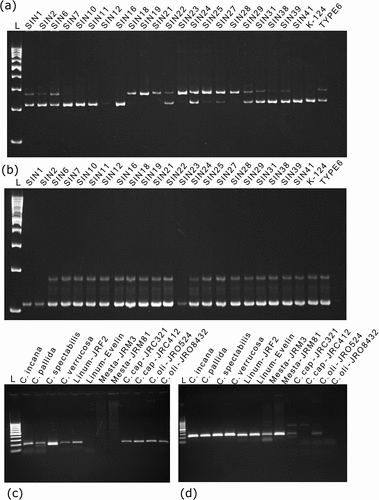
Table 2. Details of marker diversity statistics of EST-SSR markers using PowerMarker v.3.25.
Table 3. Marker diversity statistics of PIP markers using online iMEC tool.
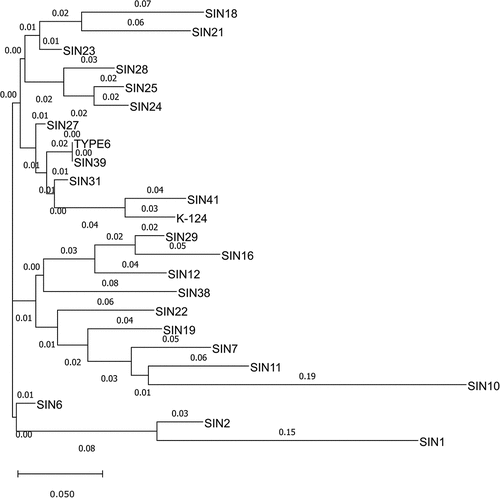
In terms of cross-species transferability of these EST-SSR markers, 14 of 48 markers (29.2%) demonstrated transferability in other plants such as flax, jute, and kenaf (). The validation of the PIP markers revealed that 26 primers were polymorphic, with PIC ranging from 0.19 to 0.31 and an average PIC of 0.26. Other marker attributes such as heterozygosity index (H), marker index (MI), and resolving power (Rp) were estimated to assess the informativeness of the PIP primer sets (). The cross-species transferability of PIP markers revealed that 23 of 48 PIP primers (47.9%) were amplifiable in heterologous plant spp. (). A clustering dendrogram () was created based on the genotyping data of 20 polymorphic EST-SSR markers in 24 sunn hemp germplasm accessions to demonstrate the utility of the EST-SSR markers in distinguishing between the sunn hemp germplasm accessions.
Figure 4. A phylogenetic tree of 24 sunn hemp germplasm using 20 polymorphic EST-SSR markers. The dendrogram was constructed using Nei’s distance measure and neighbor joining (NJ) approach and visualized using Figtree v.1.4.2. The branch length (numbers) indicated the extent of divergence from its nearest accession. The scale bar of branch length is provided below the dendrogram.
Conclusions
In the current study, the only transcriptome sequence of sunn hemp was used to report different functional/genic molecular markers for the first time in a leguminous bast-fiber crop, sunn hemp (C. juncea). Using a variety of stringent filtering criteria, we developed a non-redundant set of 1683 EST-SSR primers targeting the major SSR loci; 4759 intron-length polymorphism-specific PIP markers targeting protein-coding genes; and 3309 SNP/InDel loci-derived KASP markers targeting self-incompatibility related genes. A subset of EST-SSR markers and PIP markers has been demonstrated to be useful in characterization of genetic diversity in sunn hemp germplasm as well as cross-species applications in other heterologous bast fiber crops. Thus, the genic markers developed in the study have the potential to be used in a wide range of genetic studies, including genetic map construction and mapping of self-incompatibility genes for inter- and intra-locus association studies, as well as allelic variation studies in sunn hemp and other heterologous bast fiber crops. Although the KASP primers were designed in this study, validation of these markers in a structured sunn hemp population differing for self-incompatibility-related traits remains to be carried out. Furthermore, a simple database of sunn hemp molecular markers was developed to allow breeders and geneticists to easily access these markers.
Highlights
This is the first report on large-scale molecular marker development in Crotalaria juncea, a species with limited genomic resources.
From the only transcriptome of C. juncea, 1683 SSR, 4759 intron polymorphism, and 3309 SNP markers were developed.
A subset of 96 SSR and intron-length polymorphism markers were validated in 24 C. juncea germplasm and in other bast fiber species.
The validated molecular markers produced a satisfactory polymorphism index and efficiency in cross-species transferability.
A simple database was created for easy access of molecular markers for genetic studies on self-incompatibility and polymorphisms in C. juncea.
Ethics approval and consent to participate
No human or animal studies were performed in this research.
Acknowledgements
Authors thank Indian Council of Agriculture Research for providing with the facilities to carry out the research work under the Institutional project JB4.4.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Amiryousefi, A., J. Hyvönen, and P. Poczai. 2018. IMEC: Online marker efficiency calculator. Applications in Plant Sciences 6 (6):e01159. doi:10.1002/aps3.1159.
- Bhandari, H. R., M. K. Tripathi, B. Chaudhary, and S. K. Sarkar. 2016. Sunnhemp breeding: Challenges and prospects. Indian Journal of Agricultural Sciences 86 (11):1391–13.
- Chen, Z., D. Tang, J. Ni, P. Li, L. Wang, J. Zhou, C. Li, H. Lan, L. Li, and J. Liu. 2021. Development of genic KASP SNP markers from RNA-seq data for map-based cloning and marker-assisted selection in maize. BMC Plant Biology 21:157. doi:10.1186/S12870-021-02932-8.
- Chunlin, H., J. Holme, and J. Anthony. 2014. SNP genotyping: The KASP assay. Methods in Molecular Biology 1145:75–86. doi:10.1007/978-1-4939-0446-4_7.
- Cook, C. G., and G. A. White. 1996. Crotalaria juncea: A potential multi-purpose fiber crop. In Progress in new crops, ed. J. Janick, 389–94. Arlington, VA: ASHS Press.
- Creer, S. 2007. Choosing and using introns in molecular phylogenetics. Evolutionary Bioinformatics 3:99–108. doi:10.1177/117693430700300011.
- Doyle, J. J., and J. L. Doyle. 1990. Isolation of plant DNA from fresh tissue. Focus 12 (1):13–15.
- Duarte, J., N. Rivière, A. Baranger, G. Aubert, J. Burstin, L. Cornet, C. Lavaud, I. Lejeune-Hénaut, J.-P. Martinant, J.-P. Pichon, et al. 2014. Transcriptome sequencing for high throughput SNP development and genetic mapping in pea. BMC Genomics. 15(1):126. doi:10.1186/1471-2164-15-126.
- Gujaria, N., A. Kumar, P. Dauthal, A. Dubey, P. Hiremath, A. Bhanu Prakash, A. Farmer, M. Bhide, T. Shah, P. M. Gaur, et al. 2011. Development and use of genic molecular markers (GMMs) for construction of a transcript map of chickpea (Cicer arietinum L.). Theoretical and Applied Genetics 122 (8):1577–89. doi:10.1007/S00122-011-1556-1.
- Hall, H. G., and L. Avila. 2016. Megachile sculpturalis, the Giant Resin Bee, overcomes the blossom structure of Sunn Hemp (Crotalaria juncea) that impedes pollination. Journal of Melittology 65 (65):1–11. doi:10.17161/jom.v0i65.5887.
- Henry, R. J., M. Edwards, D. L. E. Waters, S. Gopala Krishnan, P. Bundock, T. R. Sexton, A. K. Masouleh, C. J. Nock, and J. Pattemore. 2012. Application of large-scale sequencing to marker discovery in plants. Journal of Biosciences 37 (5):829–41. doi:10.1007/s12038-012-9253-z.
- He, C., Z. L. Xia, T. A. Campbell, and G. R. Bauchan. 2009. Development and characterization of SSR markers and their use to assess genetic relationships among alfalfa germplasms. Crop Science 49 (6):2176–86. doi:10.2135/CROPSCI2007.04.0456.
- Hinds, J., K. H. Wang, S. P. Marahatta, S. L. F. Meyer, and C. R. R. Hooks. 2013. Sunn hemp cover cropping and organic fertilizer effects on the nematode community under temperate growing conditions. Journal of Nematology 45 (4):265–71.
- Liu, K., and S. V. Muse. 2005. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 21 (9):2128–29. doi:10.1093/bioinformatics/bti282.
- Mitra, G. C. 1976. Male sterility in sunn hemp (Crotalaria juncea L.). Current Science 45:873–77.
- Mosjidis, J. A., and M. L. Wang. 2011. Crotalaria. In Wild crop relatives: Genomic and breeding resources, 63–69. Berlin, Heidelberg: Springer. doi:10.1007/978-3-642-21102-7_3.
- Poczai, P., I. Varga, M. Laos, A. S. Cseh, N. Bell, J. P. T. Valkonen, and J. Hyvönen. 2013. Advances in plant gene-targeted and functional markers: A review. Plant Methods 9:6. doi:10.1186/1746-4811-9-6.
- Rangappa Thimmaiah, M., S. B. Choudhary, H. K. Sharma, A. Anil Kumar, H. Bhandari, J. Mitra, and P. G. Karmakar. 2018. Late-acting self-incompatibility: A barrier to self-fertilization in sunnhemp (Crotalaria juncea L.). Euphytica 214:19. doi:10.1007/s10681-017-2096-9.
- Saha, D., R. S. Rana, S. Chakraborty, S. Datta, A. Anil Kumar, A. K. Chakraborty, and P. G. Karmakar. 2017. Development of a set of SSR markers for genetic polymorphism detection and interspecific hybrid jute breeding. The Crop Journal 5 (5):416–29. doi:10.1016/J.CJ.2017.02.006.
- Sarkar, S. K., S. K. Hazra, H. S. Sen, P. G. Karmakar, and M. K. Tripathi. 2015. Sunnhemp in India, 1–140. Barrackpore: ICAR-Central Research Institute for Jute and Allied Fibres.
- Satya, P., R. Banerjee, M. Karan, E. Mukhopadhyay, B. Chaudhary, A. Bera, R. T. Maruthi, and S. K. Sarkar. 2016. Insight into genetic relation and diversity of cultivated and semi-domesticated under-utilized Crotalaria species gained using Start Codon Targeted (SCoT) markers. Biochemical Systematics and Ecology 66:24–32. doi:10.1016/j.bse.2016.02.032.
- Temnykh, S., G. DeClerck, A. Lukashova, L. Lipovich, S. Cartinhour, and S. McCouch. 2001. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): Frequency, length variation, transposon associations, and genetic marker potential. Genome Research 11 (8):1441–52. doi:10.1101/GR.184001.
- Thakur, O., and G. S. Randhawa. 2018. Identification and characterization of SSR, SNP and InDel molecular markers from RNA-seq data of guar (Cyamopsis tetragonoloba, L. Taub.) roots. BMC Genomics 19:951. doi:10.1186/s12864-018-5205-9.
- Thudi, M., R. Palakurthi, J. C. Schnable, A. Chitikineni, S. Dreisigacker, E. Mace, R. K. Srivastava, C. Tara Satyavathi, D. Odeny, V. K. Tiwari, et al. 2021. Genomic resources in plant breeding for sustainable agriculture. Journal of Plant Physiology 257:153351. doi:10.1016/J.JPLPH.2020.153351.
- Varshney, R. K., M. Thudi, R. K. Aggarwal, and A. Börner. 2007. Genic molecular markers in plants: Development and applications. In Genomics-assisted crop improvement, 13–29. Dordrecht: Springer. doi:10.1007/978-1-4020-6295-7_2.
- Wangkumhang, P., K. Chaichoompu, C. Ngamphiw, U. Ruangrit, J. Chanprasert, A. Assawamakin, and S. Tongsima. 2007. WASP: A web-based allele-specific PCR assay designing tool for detecting SNPs and mutations. BMC Genomics 8 (1):275. doi:10.1186/1471-2164-8-275.
- Wang, M. L., J. A. Mosjidis, J. B. Morris, R. E. Dean, T. M. Jenkins, and G. A. Pederson. 2006. Genetic diversity of Crotalaria germplasm assessed through phylogenetic analysis of EST-SSR markers. Genome 49 (6):707–15. doi:10.1139/G06-027.
- Wang, Z., G. Yu, B. Shi, X. Wang, H. Qiang, and H. Gao. 2014. Development and characterization of simple sequence repeat (SSR) markers based on RNA-sequencing of Medicago sativa and in silico mapping onto the M. truncatula genome. PLoS One 9 (3):e92029. doi:10.1371/JOURNAL.PONE.0092029.
- Wang, X., X. Zhao, J. Zhu, and W. Wu. 2005. Genome-wide investigation of intron length polymorphisms and their potential as molecular markers in rice (Oryza sativa L.). DNA Research 12 (6):417–27. doi:10.1093/DNARES/DSI019.
- Yang, L., G. Jin, X. Zhao, Y. Zheng, Z. Xu, and W. Wu. 2007. PIP: A database of potential intron polymorphism markers. Bioinformatics 23 (16):2174–77. doi:10.1093/bioinformatics/btm296.
- Zhang, Z., X. Min, Z. Wang, Y. Wang, Z. Liu, and W. Liu. 2017. Genome-wide development and utilization of novel intron-length polymorphic (ILP) markers in Medicago sativa. Molecular Breeding 37:87. doi:10.1007/s11032-017-0659-z.
- Zhao, Y., C. S. Prakash, and G. He. 2012. Characterization and compilation of polymorphic simple sequence repeat (SSR) markers of peanut from public database. BMC Research Notes 5:362. doi:10.1186/1756-0500-5-362.
- Zhao, Y., K. Wang, W. L. Wang, T. T. Yin, W. Q. Dong, and C. J. Xu. 2019. A high-throughput SNP discovery strategy for RNA-seq data. BMC Genomics 20 (1):160. doi:10.1186/S12864-019-5533-4.