ABSTRACT
It is known to all that Wnt signaling pathway plays an important role in the early development of tooth. Our previous research found that Wnt signaling pathway played crucial roles in dental development, and mutations in antagonist of Wnt signaling pathway may lead to the formation of supernumerary teeth. However, the expression pattern of Wnt signaling molecules in early development of tooth, especially genes with stage specificity, remains unclear. Hence, we applied RNA-seq analysis to determine the expression levels of wnt signal molecules at five different stages of rat first molar tooth germ. In addition, after literature review we summarized the function of Wnt signaling molecules during tooth development and the relationship between Wnt signaling molecules variation and tooth agenesis. Our research may have implications for exploring the role of Wnt signaling molecules in different stages of tooth development.
Introduction
The Wnt signaling pathway plays an important role in many processes of biological development, including embryogenesis, tissue homeostasis and wound repair. Several biological behaviors such as cell proliferation, differentiation, polarization, and apoptosis were regulated by the Wnt signal pathway. The Wnt signaling pathway is a complex system that consist of extracellular secreted glycoproteins (19 Wnt ligands), wnt receptors (10 Frizzled receptors, Frz and 2 LDL receptor related protein, Lrp5/6), cytoplasmic proteins (β-catenin, DVL, APC, AXIN and GSK-3β, etc.) and several Wnt-associated molecules (MSX1, DKK1, KREMEN1, and ANTXR1.Citation1–3 According to whether it is dependent on the activation of β-catenin, Wnt signaling pathways can be divided into the canonical signaling pathway and the noncanonical signaling pathway. The Wnt ligands, the Fzl receptors and LRP form a complex that activates the downstream canonical β-catenin pathway. Moreover, Wnt-receptor complexes are mediated and controlled by downstream degradation complexes composed of APC, AXIN and GSK-3β. The degradation complex leads to phosphorylation and ubiquitination-mediated degradation of β-catenin. Once a Wnt ligand binds to a Fzl receptor and LRP protein, the degeneration complex in cytoplasm is inhibited, leading to the accumulation of β-catenin.Citation4–6 Accumulated β-catenin translocates to the nucleus, interacts with transcription factors TCF (T-cell factor)/LEF (lymphocyte-enhancer-binding factor), and promotes the transcription of Wnt target genes.Citation7 Non-canonical Wnt signaling pathways are further divided into Wnt/planar cell polarity (PCP) signaling, Wnt/Ca2+ Pathway, Wnt RTK Pathway and Wnt Frz Pathway which are involved in the process of polarity and cell motility. Citation8
Tooth development is a dynamic process that includes the bud, cap and bell stages, root development and tooth eruption.Citation9 It is universally known that Wnt signaling pathway plays an important role in tooth development.Citation10 The Wnt signaling molecules are spatiotemporally activated in tooth development, thereby implying its essential role in the process of odontogenesis. The mutations of Wnt signaling genes may result in tooth agenesis (TA). The genetic link between TA and the Wnt pathway was first evidenced by the identification of a mutation of the Axin2 gene in an oligodontia family.Citation10 Mutation of Wnt10a may lead to odonto-onycho-dermal dysplasia, Hypohidrotic ectodermal dysplasia, Schopf-Schulz-Passarge syndrome and many non-syndromic tooth agenesis. Abnormal expression of Wnt10b is related to oligodontia, microdontia, short tooth roots, dental pulp stones, and taurodontism.Citation11–13 Mutation of Lrp6 is also associated with oligodontia, mesiodens, fusion of teeth, odontomas, microdontia, long roots, molars with unseparated roots, and taurodontism. Citation14,Citation15
Tooth development requires regulation of multiple molecules, which is the result of spatiotemporal expression of these molecules. The expression pattern of a gene or signaling are supposed to include not only the expression level but also the location at continuous stages and the study of gene expression level is an important part of learning gene expression patterns. It is known to all that Wnt signaling pathway plays an important role in tooth development. However, the expression pattern of Wnt signaling molecules in development of tooth germ are still unclear. We expect to gain a holistic understanding of the expression of the wnt signaling pathway during early tooth development and to discover genes that are specifically expressed during early tooth development. Here, we determined the expression levels of wnt signal molecules in each stage of rat first molar tooth germ development by RNA seq analysis and after literature review we summarized the function of Wnt signaling molecules during tooth development and the relationship between Wnt signaling molecules variation and tooth agenesis. Our research may have important implications for exploring the role of Wnt signaling molecules in different stages of tooth development.
Results
RNA-seq analysis
We investigated five time points (E14.5, E16.5, E18.5, P1 and P7) of rat tooth germ development. It is observed that when the embryo developed to E14.5, E16.5 and E18.5, the first molar tooth germ was in bud stage, cap stage and early bell stage respectively, while the tooth germs of 1-and 7-day-old mice were in late bell stage and eruption stage respectively. After sequencing, a total of 143 mRNA related to Wnt signal pathway were detected. Expression of Wnt signaling molecules were displayed in . Hierarchical clustering analysis revealed that the expression levels of these molecules showed significant time specificity . The results showed a high degree of similarity among the tooth germ tissues at embryonic stages, and a low degree of similarity between the embryonic and postnatal tooth germs .
Figure 1. Heatmap shows the relative expression levels of differential genes across different stages of tooth early development (E14.5–P7).
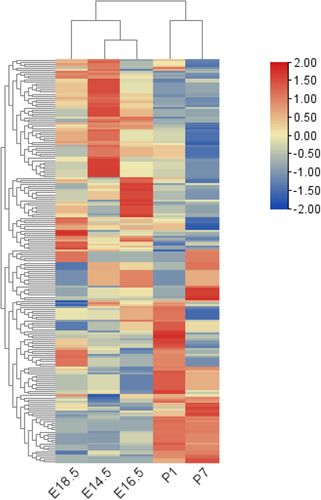
Table 1. Expression of Wnt signaling molecules during rat tooth germ development.
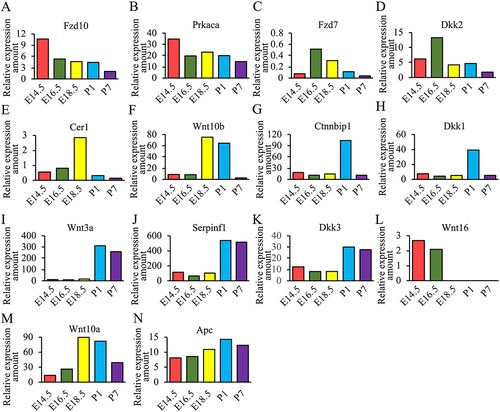
Since it is difficult to completely distinguish the molecules involved in canonical Wnt signaling pathway and the noncanonical Wnt signaling pathway, therefore we classified and integrated all genes together in . Canonical Wnt signaling pathway covered 82 different genes, and the noncanonical Wnt signaling pathway covered 81 genes. There were 20 genes with overlap between those two pathways. Besides, expression levels of genes such as Fzd10, Prkaca, Fzd7, Dkk2, Cer1, Wnt10b, Ctnnbip1, Dkk1, Wnt3a, Serpinf1, Dkk3 and Wnt16 showed obvious time specificity.
Changes in DEG mRNA levels during rat molar development
We summarized the genes with special expression levels in and used the relative expression amounts of these genes as the ordinate to draw . The mRNA expression levels of Fzd10 and Prkaca were mainly highly expressed mainly in the E14.5 stage. ThemRNAs of Fzd7 and Dkk2 were highly expressed mainly in E16.5 phase, the mRNA ofCer1 was highly expressed in E18.5 phase, while Ctnnbip1 and Dkk1 showed high expression in P1 phase. Wnt10b and Wnt10a were highly expressed in both E18.5and P1 phases. The mRNAs of Wnt3a, Serpinf1 and Dkk3 were highly expressed at the postnatal stage compared to the embryonic stage. In contrast, Wnt16 was expressed only at E14.5 and E16.5 stages. The expression of APC in the five stages of early tooth development is higher than 8.
Discussion
We investigated five time points (E14.5, E16.5, E18.5, P1 and P7) of rat tooth germ development. Considering the small size of rat tooth germs, it is difficult to separate dental papilla and dental sac, so we took the whole tooth germs for sequencing. The sequencing results include both epithelial genes and dental papilla genes. Totally 128 mRNA related to Wnt signal pathway of rat tooth germ samples from different stages of development were detected. We observed that the expression levels of these molecules showed significant time specificity, including Fzd10, Prkaca, Fzd7, Dkk2, Cer1, Ctnnbip1, Dkk1, Wnt10b, Wnt3a, Serpinf1, Dkk3, Wnt16. Wnt10b, Dkk1 and Serpinf1 have been found to be associated with tooth agenesis, but little research has been done on the relationship between the other genes mentioned above and tooth germ development. We obtained a relatively complete gene expression profile related to Wnt signaling pathway through RNA sequencing. Our study enriches the understanding of Wnt signal pathways and provides a lot of useful information for subsequent research.
Wnt signaling molecules and tooth agenesis
Gene mutation may disrupt specific signaling networks and cause a wide variety of selective tooth agenesis patterns. Tooth agenesis can either occur as an isolated condition (non-syndromic tooth agenesis) or can occur as part of a genetic syndrome (syndromic tooth agenesis). Some genes are responsible for syndromic tooth agenesis, including Robinow syndrome and Gardner syndrome. The mutations of APC may cause Gardner syndrome, besides gene mutations of Ror2, Dvl1, Dvl3 and Wnt5a are associated with Robinow syndrome. Genes responsible for tooth agenesis were displayed in . By searching for HGMD, we found that there are as many as 2037 different mutation forms in APC ().
Table 2. Wnt signaling molecules and tooth agenesis.
Apc
Adenomatous polyposis coli (APC) gene encodes a tumor suppressor protein that acts as an antagonist of the Wnt signaling pathway. It is also involved in cell migration and adhesion, transcriptional activation, and apoptosis.Citation61 Mutations in Apc may cause familial adenomatous polyposis (FAP), an autosomal dominant pre-malignant disease that usually progresses to malignancy.Citation62 Besides, other diseases such as Gardner syndrome, which is characterized by the presence of multiple intracolonic polyps and extracolonic tumors, can also be caused by Apc mutation. Our previous study found that patients with Gardner syndrome also had multiple impacted and supernumerary teeth.Citation16 We found that Apc was expressed at E14.5-P7 and there are up to 2037 different mutation forms in Apc, suggesting that Apc played an important role in all stages of early tooth development.
Wnt10b
Wingless-Type MMTVIntegration Site Family, Member 10B (Wnt10b) is a ligand of Wnt signaling pathway and has been implicated in regulation of embryogenesis. Multiple variants of Wnt10b were found by mutational analysis in patients with non-syndromic tooth agenesis.Citation11–13 Besides, mutations of Wnt10b are associated not only with oligodontia and tooth agenesis, but also with microdontia, short tooth roots, dental pulp stones, and taurodontism.Citation11 The mutations of Wnt10b decreased Wnt signaling in dental pulp stem cells, affecting the development of permanent dentition, particularly the lateral incisors.Citation13 Wnt10b mutation allele-transfected into stem cells from human exfoliated deciduous teeth resulted in decreased gene transcription, while Wnt10b protein increased with the increase of mutant alleles, indicating that Wnt10b variants have functional effects on gene regulation and reveal that these variants may affect tooth development leading to tooth agenesis.Citation63 A. Nadiri et al. observed that the relative amount of Wnt10b was maximal at E14 stage during the development of rat molar tooth germs by western blotting.Citation64 At the late bell stage, Wnt10b was mainly detected in the inner dental epithelium, indicating that Wnt10b may be involved in cell proliferation.Citation64 Unlike other studies, our study found that Wnt10b was significantly expressed at E18.5 (early bell stage) and P1 (late bell stage) stages. Therefore, the expression of Wnt10b during the tooth germ development needs to be further verified and explored.
Wnt10a
Wingless-Type MMTV Integration Site Family, Member 10A (Wnt10a) is another important ligand of Wnt signaling pathway, which appears to have specific relevance to skin, its appendages and teeth. Wnt10a is associated with both syndromic and non-syndromic tooth agenesis. Schöpf-Schulz-Passarge syndrome (SSPS) is an autosomal recessive form of ectodermal dysplasia resulting from mutations in Wnt10a.Citation65 Tooth agenesis in SSPS patients can be characterized by conical primary teeth, agenesis of permanent teeth and hypodontia.Citation24 Odonto-onycho-dermal dysplasia (OODD) is a rare autosomal recessive syndrome mainly characterized by dry hair, anodontia of permanent teeth, smooth tongue, keratoderma, and hyperhidrosis.Citation26 Recently, Wnt10a mutations were found in patients with nonsyndromic tooth agenesis as well, mainly characterized by oligodontia and agenesis of the maxillary permanent canines. Upper and lower premolars were the most affected missing teeth.Citation12,Citation21 In our research, Wnt10a was significantly expressed in E18.5 (early bell stage) and P1 (late bell stage) stages and the expression level decreased in P7 phase. Our results implied that Wnt10a played an important role in the formation of tooth hard tissue.
Dkk1
Dickkopf-related protein 1(Dkk1) is a typical antagonist of Wnt/β-catenin signaling by competing for the Wnt receptor LRP5/6. The mutation of Dkk1 may cause oligodontia and short root anomaly.Citation27,Citation66 Researchers observed that strong expression of Dkk1 was localized in preodontoblasts on the labial side of the incisors.Citation67 At postnatal day 2, Dkk1 is prominently expressed in the preodontoblasts and odontoblasts in mouse molar germs.Citation68 In Dkk1 transgenic mice, overexpression of Dkk1 in pulp and odontoblast cells delayed the maturation of dentinogenesis during post-natal development.Citation69 The dental crown begins to form in the late bell stage (P1). In this stage, peripheral cells of the dental papilla differentiate into odontoblasts that secrete dentin. Our results demonstrated that Dkk1 was significantly expressed in the P1 phase, which indicated that Dkk1 played an important role in the formation of dentin.
Serpinf1
Serpin family F member 1 (Serpinf1) encodes the 50-kDa secreted glycoprotein pigment epithelium-derived factor (PEDF), which inhibits Wnt/β-catenin signaling pathway. Mutation of Serpinf1 may cause a unique autosomal recessive disease, Osteogenesis imperfecta (OI) type VI (MIM #610968). OI is characterized by recurrent fractures, progressive skeletal deformities, and growth deficiency, blue or gray discoloration of the sclera and dentinogenesis imperfecta.Citation70 Mutations in Serpinf1 have been reported in Chinese individuals with OI.Citation46 Our findingSerpinf1 mainly high expressed during postnatal stages, indicated that Serpinf1 played an important role in the formation and mineralization of tooth hard tissues.
Other important genes
Fzd10 is one of the FZD family receptors which act through canonical Wnt signaling.Citation42 Fzd10regulates cell proliferation and mediates Wnt1-induced neurogenesis in the developing spinal cord.Citation71 Investigations have demonstrated that mutations in the gene of Prkaca result in the development of adrenocortical adenomas associated with Cushing’s syndrome.Citation72 Fzd7 plays a significant role in the regulation of multipotentiality of human pluripotent stem cells and its down-regulation accompanies differentiation and exit from the pluripotent stem cell state Dkk2 plays an important role in ovarian cancer.Citation73,Citation74 It has been well documented that Cer1 plays dual roles in neural induction and suppression of mesodermal or endodermal lineages.Citation40 We observed that the mRNA expression levels of Fzd10 and Prkaca were mainly high during bud stage (E14.5), Fzd7 and Dkk2 were mainly highly expressed during cap stage (E16.5), and Cer1 was high during early bell stage (E18.5), indicating that they may play important roles in the early stage of tooth germ development. The expression level of Ctnnnbip1 was mainly high during P1 stage (late bell stage), enlightening that Ctnnbipmay be related to the formation and mineralization of enamel and dentin. Compared to the embryonic stages, the mRNA expression levels of Wnt3a and Dkk3 were highly expressed during postnatal stages, which implied that once significant high expression of Wnt3a and Dkk3 were found, it indicated that tooth germ has developed to the stage of hard tissue formation and mineralization. By contrary, Wnt16 was expressed only during E14.5 and E16.5, suggesting that Wnt16 was one of the important markers of early development of tooth germ.
In summary, we determined the expression levels of Wnt signaling molecules at each stage of rat first molar tooth germ development by RNA-seq analysis and after literature review we summarized the relationship between Wnt signaling molecules variants and tooth agenesis. Our data provided important information about novel genes in tooth germ development. Our findings may help to elucidate the molecular mechanisms of tooth germ development and provide a theoretical basis for further studies on the expression and function of genes involved in human tooth development and regeneration.
Materials and Methods
Tissue acquisition and preparation
Specific pathogen-free (SPF)-grade Sprague Dawley (SD) rats (Department of Laboratory Animal Science, Fudan University, Shanghai, China) were used in our study. The day when the vaginal plug was formed as a reference of embryonic day 0 (E0), embryos at E14.5, E16.5 and E18.5 stages were used. One-day-old (P1) and seven-day-old (P7) rats were used as well. We separated the embryos from the uterus of pregnant female rats. Each group has 8 mouse embryos, and the sex of the embryos was randomized to both males and females. The first mandibular molar tooth germs were isolated from these embryos and rats using a zoom stereo microscope (Olympus SZ51, Tokyo, Japan) and prepared for subsequent experiments. All molars were dissected by one same researcher. The rats were euthanized with phenobarbital sodium (i.p., 50 mg/kg) anesthesia. All animal experiments were approved by the Institutional Animal Care and Use Committee of Fudan University.
RNA-seq
Total RNA was obtained from the tooth germ cells in the embryonic (E14.5, E16.5, and E18.5) and postnatal (P1 and P7) phases using TRI reagent (Invitrogen, Carlsbad, CA, USA). More than eight tooth germs were used in each phase. An RNA-seq library was constructed following the manufacturer’s instructions. Then, next-generation sequencing was performed using an Illumina NovaSeq platform (IGA, Udine, Italy). mRNAs related to wnt signal pathway were selected and hierarchical clustering analysis was performed.
Database search and literature review
We queried the Human Gene Mutation Database(HGMD®) (https://www.hgmd.cf.ac.uk/ac/index.php) for all variants of Wnt signaling genes. A comprehensive review of the PubMed database was undertaken for reports in the English language from 1January 2012 to 1 October 2022. The following search terms were used: (tooth [MeSHTerms]) AND (((Wnt Pathway [MeSH Terms]) OR Wnt Pathway, Canonical) OR Wntbeta-Catenin Signaling Pathway). Titles and abstracts were reviewed, and references and citations of relevant studies were also examined.
Declaration of conflicting interests
The authors declare no competing interests.
Acknowledgments
We would like to thank the Human Gene Mutation Database (HGMD®) for data searching.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kakugawa S, Langton PF, Zebisch M, Howell S, Chang TH, Liu Y, Feizi T, Bineva G, O’Reilly N, Snijders AP, et al. Notum deacylates Wnt proteins to suppress signalling activity. Nature. 2015;519(7542):187–12. PMID: 25731175. doi:10.1038/nature14259.
- Miller JR. The Wnts. Genome Biol. 2002;3(1):reviews3001.1. REVIEWS3001 PMID: 11806834. doi:10.1186/gb-2001-3-1-reviews3001.
- Zhang Z, Nor F, Oh M, Cucco C, Shi S, Nor JE. Wnt/β-Catenin signaling determines the vasculogenic fate of postnatal mesenchymal stem cells. Stem Cells. 2016;34(6):1576–87. PMID: 26866635. doi:10.1002/stem.2334.
- De Santis M, Di Matteo B, Chisari E, Cincinelli G, Angele P, Lattermann C, Filardo G, Vitale ND, Selmi C, Kon E. The role of wnt pathway in the pathogenesis of OA and its potential therapeutic implications in the field of regenerative medicine. Biomed Res Int. 2018; 2018: 7402947. doi: 10.1155/2018/7402947. PMID: 30410938
- van den Bosch MH, Gleissl TA, Blom AB, van den Berg WB, van Lent PL, van der Kraan PM. Wnts talking with the TGF-β superfamily: wISPers about modulation of osteoarthritis. Rheumatology (Oxford). 2016;55(9):1536–47. PMID: 26667213. doi:10.1093/rheumatology/kev402.
- Cheng J, Li M, Bai R. The Wnt signaling cascade in the pathogenesis of osteoarthritis and related promising treatment strategies. Front Physiol. 2022;13:954454. doi:10.3389/fphys.2022.954454. PMID: 36117702.
- Duchartre Y, Kim YM, Kahn M. The Wnt signaling pathway in cancer. Crit Rev Oncol Hematol. 2016;99:141–49. doi:10.1016/j.critrevonc.2015.12.005. PMID: 26775730.
- Lu X, Yang J, Zhao S, Liu S. Advances of Wnt signalling pathway in dental development and potential clinical application. Organogenesis. 2019;15(4):101–10. PMID: 31482738. doi:10.1080/15476278.2019.1656996.
- Huang XF, Chai Y. Molecular regulatory mechanism of tooth root development. Int J Oral Sci. 2012;4(4):177–81. PMID: 23222990. doi:10.1038/ijos.2012.61.
- Yu M, Wong SW, Han D, Cai T. Genetic analysis: wnt and other pathways in nonsyndromic tooth agenesis. Oral Dis. 2019;25(3):646–51. PMID: 29969831. doi:10.1111/odi.12931.
- Kantaputra PN, Hutsadaloi A, Kaewgahya M, Intachai W, German R, Koparal M, Leethanakul C, Tolun A, Ketudat Cairns JR. WNT10B mutations associated with isolated dental anomalies. Clin Genet. 2018;93(5):992–99. PMID: 29364501. doi:10.1111/cge.13218.
- Magruder S, Carter E, Williams MA, English J, Akyalcin S, Letra A. Further evidence for the role of WNT10A, WNT10B and GREM2 as candidate genes for isolated tooth agenesis. Orthod Craniofac Res. 2018;21(4):258–63. PMID: 30246922. doi:10.1111/ocr.12248.
- Yu P, Yang W, Han D, Wang X, Guo S, Li J, Li F, Zhang X, Wong SW, Bai B, et al. Mutations in WNT10B are identified in individuals with oligodontia. Am J Hum Genet. 2016;99(1):195–201. PMID: 27321946. doi:10.1016/j.ajhg.2016.05.012.
- Goto H, Kimura M, Machida J, Ota A, Nakashima M, Tsuchida N, Adachi J, Aoki Y, Tatematsu T, Takahashi K, et al. A novel LRP6 variant in a Japanese family with oligodontia. Hum Genome Var. 2021;8(1):30. PMID: 34285199. doi:10.1038/s41439-021-00162-w.
- Kantaputra PJ, Chintakanon W, Intachai K, Adisornkanj S, Pradermdutsadeeporn P, Tongsima P, Ngamphiw B, Olsen C, Tucker AS, Tucker AS, et al. Mutations in LRP6 highlight the role of WNT signaling in oral exostoses and dental anomalies. Arch Oral Biol. 2022;142:105514. PMID: 35961235. doi:10.1016/j.archoralbio.2022.105514.
- Yu F, Cai W, Jiang B, Xu L, Liu S, Zhao S. A novel mutation of adenomatous polyposis coli (APC) gene results in the formation of supernumerary teeth. J Cell Mol Med. 2018;22(1):152–62. PMID: 28782241. doi:10.1111/jcmm.13303.
- Oku T, Takayama T, Sato Y, Sato Y, Takada K, Hayashi T, Takahashi M, Kuroda M, Kato J, Niitsu Y. A case of Gardner syndrome with a mutation at codon 1556 of APC: a suggested case of genotype-phenotype correlation in dental abnormality. Eur J Gastroenterol Hepatol. 2004;16(1):101–05. PMID: 15095859. doi:10.1097/00042737-200401000-00015.
- Hartikka H, Makitie O, Mannikko M, Doria AS, Daneman A, Cole WG, Ala-Kokko L, Sochett EB. Heterozygous mutations in the LDL receptor-related protein 5 (LRP5) gene are associated with primary osteoporosis in children. J Bone Miner Res. 2005;20(5):783–89. PMID: 15824851. doi:10.1359/JBMR.050101.
- Kantaputra PN, Guven Y, Tripuwabhrut K, Adisornkanj P, Hatsadaloi A, Kaewgahya M, Olsen B, Ngamphiw C, Jatooratthawichot P, Tongsima S, et al. Mutations in LRP5 and BMP4 are associated with mesiodens, tooth agenesis, root malformation, and oral exostoses. Clin Genet. 2022;102(4):333–38. PMID: 35754005. doi:10.1111/cge.14183.
- Kantaputra PN, Kapoor S, Verma P, Kaewgahya M, Kawasaki K, Ohazama A, Ketudat Cairns JR. Al-Awadi-Raas-Rothschild syndrome with dental anomalies and a novel WNT7A mutation. Eur J Med Genet. 2017;60(12):695–700. PMID: 28917830. doi:10.1016/j.ejmg.2017.09.005.
- Parveen A, Khan SA, Mirza MU, Bashir H, Arshad F, Iqbal M, Ahmad W, Wahab A, Fiaz A, Naz S, et al. Deleterious variants in WNT10A, EDAR, and EDA causing isolated and syndromic tooth agenesis: a structural perspective from molecular dynamics simulations. Int J Mol Sci. 2019;20(21):5282. PMID: 31652981. doi:10.3390/ijms20215282.
- Guazzarotti L, Tadini G, Mancini GE, Sani I, Pisanelli S, Galderisi F, D’Auria E, Secondi R, Bottero A, Zuccotti GV. WNT10A gene is the second molecular candidate in a cohort of young Italian subjects with ectodermal derivative impairment (EDI). Clin Genet. 2018;93(3):693–98. PMID: 28976000. doi:10.1111/cge.13147.
- Martinez-Romero MC, Ballesta-Martinez MJ, Lopez-Gonzalez V, Sanchez-Soler MJ, Serrano-Anton AT, Barreda-Sanchez M, Rodriguez-Pena L, Martinez-Menchon MT, Frias-Iniesta J, Sanchez-Pedreno P, et al. EDA, EDAR, EDARADD and WNT10A allelic variants in patients with ectodermal derivative impairment in the Spanish population. Orphanet J Rare Dis. 2019;14(1):281. PMID: 31796081. doi:10.1186/s13023-019-1251-x.
- Ismail FF, McGrath J, Sinclair R. Schopf-Schulz-Passarge syndrome: a rare ectodermal dysplasia with a delayed diagnosis. Int J Dermatol. 2020;59(2):257–58. PMID: 31468502. doi:10.1111/ijd.14616.
- Hsu TC, Lee JY, Hsu MM, Chao SC. Case report of Schöpf-Schulz-Passarge syndrome resulting from a missense mutation, p.Arg104Cys, in WNT10A. J Dermatol. 2018;45(4):475–78. PMID: 29271000. doi:10.1111/1346-8138.14201.
- Yu M, Liu Y, Liu H, Wong SW, He H, Zhang X, Wang Y, Han D, Feng H. Distinct impacts of bi-allelic WNT10A mutations on the permanent and primary dentitions in odonto-onycho-dermal dysplasia. Am J Med Genet A. 2019;179(1):57–64. PMID: 30569517. doi:10.1002/ajmg.a.60682.
- Dinckan N, Du R, Petty LE, Coban-Akdemir Z, Jhangiani SN, Paine I, Baugh EH, Erdem AP, Kayserili H, Doddapaneni H, et al. Whole-Exome sequencing identifies novel variants for tooth agenesis. J Dent Res. 2018;97(1):49–59. PMID: 28813618. doi:10.1177/0022034517724149.
- Bielik P, Bonczek O, Krejci P, Zeman T, Izakovicova-Holla L, Soukalova J, Vanek J, Vojtesek B, Lochman J, Balcar VJ, et al. WNT10A variants: following the pattern of inheritance in tooth agenesis and self-reported family history of cancer. Clin Oral Investig. 2022;26(12):7045–55. PMID: 35999385. doi:10.1007/s00784-022-04664-x.
- Du R, Dinckan N, Song X, Coban-Akdemir Z, Jhangiani SN, Guven Y, Aktoren O, Kayserili H, Petty LE, Muzny DM, et al. Identification of likely pathogenic and known variants in TSPEAR, LAMB3, BCOR, and WNT10A in four Turkish families with tooth agenesis. Hum Genet. 2018;137(9):689–703. PMID: 30046887. doi:10.1007/s00439-018-1907-y.
- Grejtakova D, Gabrikova-Dojcakova D, Boronova I, Kyjovska L, Hubcejova J, Fecenkova M, Zigova M, Priganc M, Bernasovska J. WNT10A variants in relation to nonsyndromic hypodontia in eastern Slovak population. J Genet. 2018;97(5):1169–77. PMID: 30555066. doi:10.1007/s12041-018-1011-z.
- Kantaputra P, Jatooratthawichot P, Tantachamroon O, Nanekrungsan K, Intachai W, Olsen B, Tongsima S, Ngamphiw C, Cairns JRK. Novel dental anomaly–associated mutations in WNT10A protein binding sites. Int Dent J. 2022;73(1):79–86. PMID: 35537890. doi:10.1016/j.identj.2022.04.006.
- Keskin G, Karaer K, Ucar Gundogar Z. Targeted NGS(„next-generation sequencing“)-Analyse von Mutationen in Kandidatengenen für nichtsyndromale Zahnaplasie. J Orofac Orthop / Fortschritte der Kieferorthopädie. 2022;83(S1):65–74. PMID: 33725141. doi:10.1007/s00056-021-00284-4.
- Machida J, Goto H, Tatematsu T, Shibata A, Miyachi H, Takahashi K, Izumi H, Nakayama A, Shimozato K, Tokita Y. WNT10A variants isolated from Japanese patients with congenital tooth agenesis. Hum Genome Var. 2017;4(1):17047. PMID: 29367877. doi:10.1038/hgv.2017.47.
- Park H, Song JS, Shin TJ, Hyun HK, Kim YJ, Kim JW. WNT10A mutations causing oligodontia. Arch Oral Biol. 2019;103:8–11. doi:10.1016/j.archoralbio.2019.05.007. PMID: 31103801.
- Song S, Zhao R, He H, Zhang J, Feng H, Lin L. WNT10A variants are associated with non-syndromic tooth agenesis in the general population. Hum Genet. 2014;133(1):117–24. PMID: 24043634. doi:10.1007/s00439-013-1360-x.
- Xie W, Zeng B, Li P, Xu D, Yu D, Zhao W. Two novel mutations in ectodysplasin-A identified in syndromic tooth agenesis. J Coll Physicians Surg Pak. 2022;32:570–74. doi:10.29271/jcpsp.2022.05.570. PMID: 35546689.
- Zhao K, Lian M, Zou D, Huang W, Zhou W, Shen Y, Wang F, Wu Y. Novel mutations identified in patients with tooth agenesis by whole-exome sequencing. Oral Dis. 2019;25(2):523–34. PMID: 30417976. doi:10.1111/odi.13002.
- Zhou M, Zhang H, Camhi H, Seymen F, Koruyucu M, Kasimoglu Y, Kim JW, Kim-Berman H, Yuson NMR, Benke PJ, et al. Correction: analyses of oligodontia phenotypes and genetic etiologies. Int J Oral Sci. 2021;13(1):35. PMID: 34772917. doi:10.1038/s41368-021-00141-5.
- Kantaputra P, Kaewgahya M, Kantaputra W. WNT10A mutations also associated with agenesis of the maxillary permanent canines, a separate entity. Am J Med Genet A. 2014;164A(2):360–63. PMID: 24311251. doi:10.1002/ajmg.a.36280.
- Hou PS, Chuang CY, Kao CF, Chou SJ, Stone L, Ho HN, Chien CL, Kuo HC. LHX2 regulates the neural differentiation of human embryonic stem cells via transcriptional modulation of PAX6 and CER1. Nucleic Acids Res. 2013;41(16):7753–70. PMID: 23804753. doi:10.1093/nar/gkt567.
- Mishra S, Agarwalla SK, Pradhan S. Robinow syndrome: a rare diagnosis. J Clin Diagn Res. 2015;9:SD04–05. doi:10.7860/JCDR/2015/15078.6949. PMID: 26816964.
- Garcia-Morales C, Liu CH, Abu-Elmagd M, Hajihosseini MK, Wheeler GN. Frizzled-10 promotes sensory neuron development in Xenopus embryos. Dev Biol. 2009;335(1):143–55. PMID: 19716814. doi:10.1016/j.ydbio.2009.08.021.
- Arte S, Parmanen S, Pirinen S, Alaluusua S, Nieminen P, Eisenberg L. Candidate gene analysis of tooth agenesis identifies novel mutations in six genes and suggests significant role for WNT and EDA signaling and allele combinations. PLos One. 2013;8(8):e73705. PMID: 23991204. doi:10.1371/journal.pone.0073705.
- Haddaji Mastouri M, De Coster P, Zaghabani A, Jammali F, Raouahi N, Salem AB, Saad A, Coucke P, H’Mida Ben Brahim D. Genetic study of non-syndromic tooth agenesis through the screening of paired box 9, msh homeobox 1, axin 2, and Wnt family member 10A genes: a case-series. Eur J Oral Sci. 2018;126(1):24–32. PMID: 29114927. doi:10.1111/eos.12391.
- Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, Thesleff I, Pirinen S, Nieminen P. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet. 2004;74(5):1043–50. PMID: 15042511. doi:10.1086/386293.
- Zhang H, Xu Y, Yue H, Wang C, Gu J, He J, Fu W, Hu W, Zhang Z. Novel mutations of the SERPINF1 and FKBP10 genes in Chinese families with autosomal recessive osteogenesis imperfecta. Int J Mol Med. 2018;41:3662–70. doi:10.3892/ijmm.2018.3542. PMID: 29512769.
- Mostowska A, Biedziak B, Zadurska M, Dunin-Wilczynska I, Lianeri M, Jagodzinski PP. Nucleotide variants of genes encoding components of the Wnt signalling pathway and the risk of non-syndromic tooth agenesis. Clin Genet. 2013;84(5):429–40. PMID: 23167694. doi:10.1111/cge.12061.
- Chu KY, Wang YL, Chou YR, Chen JT, Wang YP, Simmer JP, Hu JC, Wang SK. Synergistic mutations of LRP6 and WNT10A in familial tooth agenesis. J Pers Med. 2021;11(11):1217. PMID: 34834569. doi:10.3390/jpm11111217.
- Huang YX, Gao CY, Zheng CY, Chen X, Yan YS, Sun YQ, Dong XY, Yang K, Zhang DL. Investigation of a novel LRP6 variant causing autosomal-dominant tooth agenesis. Front Genet. 2021;12:688241. doi:10.3389/fgene.2021.688241. PMID: 34306029.
- Ockeloen CW, Khandelwal KD, Dreesen K, Ludwig KU, Sullivan R, van Rooij I, Thonissen M, Swinnen S, Phan M, Conte F, et al. Novel mutations in LRP6 highlight the role of WNT signaling in tooth agenesis. Genet Med. 2016;18(11):1158–62. PMID: 26963285. doi:10.1038/gim.2016.10.
- Wang H, Liu Y, Zheng Y, Zhao X, Lin S, Zhang Q, Zhang X. A novel missense mutation of LRP6 identified by whole-exome sequencing in a Chinese family with non-syndromic tooth agenesis. Orthod Craniofac Res. 2021;24(2):233–40. PMID: 32844563. doi:10.1111/ocr.12424.
- Yu M, Fan Z, Wong SW, Sun K, Zhang L, Liu H, Feng H, Liu Y, Han D. Lrp6 dynamic expression in tooth development and mutations in oligodontia. J Dent Res. 2021;100(4):415–22. PMID: 33164649. doi:10.1177/0022034520970459.
- Kim SJ, Bieganski T, Sohn YB, Kozlowski K, Semenov M, Okamoto N, Kim CH, Ko AR, Ahn GH, Choi YL, et al. Identification of signal peptide domain SOST mutations in autosomal dominant craniodiaphyseal dysplasia. Hum Genet. 2011;129(5):497–502. PMID: 21221996. doi:10.1007/s00439-011-0947-3.
- Gordon CT, Vuillot A, Marlin S, Gerkes E, Henderson A, AlKindy A, Holder-Espinasse M, Park SS, Omarjee A, Sanchis-Borja M, et al. Heterogeneity of mutational mechanisms and modes of inheritance in auriculocondylar syndrome. J Med Genet. 2013;50(3):174–86. PMID: 23315542. doi:10.1136/jmedgenet-2012-101331.
- Hu R, Qiu Y, Li Y, Li J. A novel frameshift mutation of DVL1-induced Robinow syndrome: a case report and literature review. Mol Genet Genomic Med. 2022;10(3):e1886. PMID: 35137569. doi:10.1002/mgg3.1886.
- White JJ, Mazzeu JF, Hoischen A, Bayram Y, Withers M, Gezdirici A, Kimonis V, Steehouwer M, Jhangiani SN, Muzny DM, et al. DVL3 alleles resulting in a −1 frameshift of the last exon mediate autosomal-dominant robinow syndrome. Am J Hum Genet. 2016;98(3):553–61. PMID: 26924530. doi:10.1016/j.ajhg.2016.01.005.
- Roifman M, Marcelis CL, Paton T, Marshall C, Silver R, Lohr JL, Yntema HG, Venselaar H, Kayserili H, van Bon B, et al. De Novo WNT5A-associated autosomal dominant Robinow syndrome suggests specificity of genotype and phenotype. Clin Genet. 2015;87(1):34–41. PMID: 24716670. doi:10.1111/cge.12401.
- Person AD, Beiraghi S, Sieben CM, Hermanson S, Neumann AN, Robu ME, Schleiffarth JR, Billington CJ, van Bokhoven H Jr., Hoogeboom JM, et al. WNT5A mutations in patients with autosomal dominant Robinow syndrome. Dev Dyn. 2010;239:327–37. PMID: 19918918. doi:10.1002/dvdy.22156.
- Issa YA, Kamal L, Rayyan AA, Dweik D, Pierce S, Lee MK, King MC, Walsh T, Kanaan M. Mutation of KREMEN1, a modulator of Wnt signaling, is responsible for ectodermal dysplasia including oligodontia in Palestinian families. Eur J Hum Genet. 2016;24(10):1430–35. PMID: 27049303. doi:10.1038/ejhg.2016.29.
- Beck DB, Cho MT, Millan F, Yates C, Hannibal M, O’Connor B, Shinawi M, Connolly AM, Waggoner D, Halbach S, et al. A recurrent de novo CTBP1 mutation is associated with developmental delay, hypotonia, ataxia, and tooth enamel defects. Neurogenetics. 2016;17(3):173–78. PMID: 27094857. doi:10.1007/s10048-016-0482-4.
- Fang X, Svitkina TM. Adenomatous Polyposis Coli (APC) in cell migration. Eur J Cell Biol. 2022;101(3):151228. PMID: 35483122. doi:10.1016/j.ejcb.2022.151228.
- Pergolizzi M, Bizzozero L, Maione F, Maldi E, Isella C, Macagno M, Mariella E, Bardelli A, Medico E, Marchio C, et al. The neuronal protein Neuroligin 1 promotes colorectal cancer progression by modulating the APC/β-catenin pathway. J Exp Clin Cancer Res. 2022;41(1):266. PMID: 36056393. doi:10.1186/s13046-022-02465-4.
- Williams M, Zeng Y, Chiquet B, Jacob H, Kurtis Kasper F, Harrington DA, English J, Akyalcin S, Letra A. Functional characterization of ATF1, GREM2 and WNT10B variants associated with tooth agenesis. Orthod Craniofac Res. 2021;24(4):486–93. PMID: 33369218. doi:10.1111/ocr.12462.
- Nadiri A, Kuchler-Bopp S, Haikel Y, Lesot H. Immunolocalization of BMP-2/-4, FGF-4, and WNT10b in the developing mouse first lower molar. J Histochem Cytochem. 2004;52(1):103–12. PMID: 14688221. doi:10.1177/002215540405200110.
- Tziotzios C, Petrof G, Liu L, Verma A, Wedgeworth EK, Mellerio JE, McGrath JA. Clinical features and WNT 10A mutations in seven unrelated cases of Schöpf–Schulz–Passarge syndrome. Br J Dermatol. 2014;171(5):1211–14. PMID: 24902757. doi:10.1111/bjd.13158.
- Yu M, Jiang Z, Wang Y, Xi Y, Yang G. Molecular mechanisms for short root anomaly. Oral Dis. 2021;27(2):142–50. PMID: 31883171. doi:10.1111/odi.13266.
- Lavicky J, Kolouskova M, Prochazka D, Rakultsev V, Gonzalez-Lopez M, Steklikova K, Bartos M, Vijaykumar A, Kaiser J, Porizka P, et al. The development of dentin microstructure is controlled by the type of adjacent epithelium. J Bone Miner Res. 2022;37(2):323–39. PMID: 34783080. doi:10.1002/jbmr.4471.
- Fjeld K, Kettunen P, Furmanek T, Kvinnsland IH, Luukko K. Dynamic expression of Wnt signaling-related Dickkopf1, -2, and -3 mRnas in the developing mouse tooth. Dev Dyn. 2005;233(1):161–66. PMID: 15759274. doi:10.1002/dvdy.20285.
- Han XL, Liu M, Voisey A, Ren YS, Kurimoto P, Gao T, Tefera L, Dechow P, Ke HZ, Feng JQ. Post-natal effect of overexpressed DKK1 on mandibular molar formation. J Dent Res. 2011;90(11):1312–17. PMID: 21917600. doi:10.1177/0022034511421926.
- Forlino A, Marini JC. Osteogenesis imperfecta. Lancet. 2016;387(10028):1657–71. PMID: 26542481. doi:10.1016/S0140-6736(15)00728-X.
- Alrefaei AF, Munsterberg AE, Wheeler GN, Klymkowsky M. FZD10 regulates cell proliferation and mediates Wnt1 induced neurogenesis in the developing spinal cord. PLos One. 2020;15(6):e0219721. PMID: 32531778. doi:10.1371/journal.pone.0219721.
- Wu L, Xie J, Qi Y, Su T, Jiang L, Zhou W, Jiang Y, Zhang C, Zhong X, Cao Y, et al. Mutational landscape of non-functional adrenocortical adenomas. Endocr Relat Cancer. 2022;29(9):521–32. PMID: 35731037. doi:10.1530/ERC-21-0410.
- Gumber D, Do M, Suresh Kumar N, Sonavane PR, Wu CCN, Cruz LS, Grainger S, Carson D, Gaasterland T, Willert K. Selective activation of FZD7 promotes mesendodermal differentiation of human pluripotent stem cells. Elife. 2020;9: PMID: 33331818. doi:10.7554/eLife.63060.
- Fraungruber P, Kaltofen T, Heublein S, Kuhn C, Mayr D, Burges A, Mahner S, Rathert P, Jeschke U, Trillsch F. G protein-coupled estrogen receptor correlates with Dkk2 expression and has prognostic impact in ovarian cancer patients. Front Endocrinol (Lausanne). 2021;12:564002. doi:10.3389/fendo.2021.564002. PMID: 33679613.