ABSTRACT
In a recent publication, we reported a unique interaction between a protein encoded by the giant myovirus phiKZ and the Pseudomonas aeruginosa RNA degradosome. Crystallography, site-directed mutagenesis and interactomics approaches revealed this ‘degradosome interacting protein’ or Dip, to adopt an ‘open-claw’ dimeric structure that presents acidic patches on its outer surface which hijack 2 conserved RNA binding sites on the scaffold domain of the RNase E component of the RNA degradosome. This interaction prevents substrate RNAs from being bound and degraded by the RNA degradosome during the virus infection cycle. In this commentary, we provide a perspective into the biological role of Dip, its structural analysis and its mysterious evolutionary origin, and we suggest some therapeutic and biotechnological applications of this distinctive viral protein.
Introduction
The relationship between bacteria and the viruses that prey upon them is complex and ever evolving. Although studied intensively, discoveries continue to be made of different strategies employed by both virus and host to aid or evade infection, respectively. Bacteriophages have evolved multiple and varied mechanisms to efficiently infect their bacterial hosts. An ubiquitous strategy utilized by phages during infection is the production of proteins that modulate or redirect the functionality of specific host proteins.Citation1,2 From a bacteriophage perspective, these interactions are often crucial to evade the multitude of bacterial defense mechanisms or to alter the host metabolism in order to ensure an efficient infection cycle. The discovery of phage effector proteins that target regulatory hubs of the host bacterium could open new doors toward drug discovery and design.Citation3 The best studied example of such a hub is the RNA polymerase which is targeted by several phages at different interaction sites, influencing transcription by a wide range of mechanisms.Citation4 Another key regulatory hub is the RNA degradosome, a multiprotein complex responsible for RNA turnover and posttranscriptional gene regulation in bacteria. A general model for the Escherichia coli RNA degradosome has been described, with a core complex comprising the hydrolytic endonuclease RNase E, a phosphorolytic exoribonuclease, PNPase, the ATP dependent helicase RhlB, and a glycolytic enzyme, enolase.Citation5 However, the exact makeup and variability of the complex in important bacterial pathogens like Pseudomonas aeruginosa was not previously characterized.
Intracellular levels of any RNA are balanced by both synthesis and degradation, and must be well synchronized with cellular processes. As such, the degradation rate of individual RNAs is an important aspect of the control of gene expression. In bacteria, mRNA has a half-life of only 2 to 3 minutes, which allows the cell to quickly adapt to alterations in the environment and govern stress responses.Citation6,7 Therefore, we reasoned that an important regulatory hub such as the RNA degradosome would be a potential candidate for targeting by phage effector proteins, thereby disrupting this level of cellular control. The identification of such phage proteins had previously been limited to 2 examples, a phosphorylation-based inhibitor (Protein kinase 0.7, phage T7) that selectively stabilizes phage transcripts and an RNA degradosome activator from coliphage T4 (Srd), which has been found to destabilize host mRNAs.Citation8,9 In our recent publication,Citation10 a phage effector protein was identified, encoded by the giant Pseudomonas phage phiKZ, able to specifically target the RNA degradosome of P. aeruginosa. This ‘degradosome interacting protein’ (Dip) was shown to act by inhibiting the activity of the host ribonuclease RNase E. Additionally, the methods used to identify Dip also shed further light on the protein composition of the P. aeruginosa RNA degradosome. In this point-of-view commentary, we expand on this interaction and reflect on the impact of identifying this inhibitor of RNase E and understanding the mechanism of Dip.
The composition of the P. aeruginosa RNA degradosome
The strategy to identify phage-encoded proteins that interact with bacterial host proteins was based on the pull down of bacterial proteins (and/or complexes) during the early phase of a phage infection cycle.Citation11 By performing affinity purifications on P. aeruginosa cells containing a StrepII-tagged RNase E, Dip was identified in interaction with the RNA degradosome during phiKZ-infection.Citation10 Moreover, this and other pull-downs using 6 unrelated Pseudomonas phages provided information on the composition of the P. aeruginosa degradosome itself for the first time ( and Table S1). The exoribonuclease PNPase co-purified with RNase E following infection with all used phages and was present in the pull-down experiment using heterologously expressed Dip and wild type Pseudomonas cell lysate. In this Dip-based pull down the RNA helicase DeaD was detected as well. Remarkably, during infection with the different phages, one to 3 different DEAD-box RNA helicases (RhlB, RhlE and DeaD) were co-purified with RNase E. Moreover, protein chaperone DnaK, which has previously been identified in complex with the RNA degradosome in E. coli,Citation12,13 was co-purified during some phage infections. These findings indicate that the composition of the RNA degradosome may vary in response to different phage infections and might suggest that some phages possess more indirect mechanisms to affect the RNA degradosome as well.
Enolase could not be identified in any of the pull-down experiments, suggesting that this canonical component of the E. coli RNA degradosome does not form part of the P. aeruginosa complex, even though enolase is predicted to be present in the P. aeruginosa cytoplasm.Citation14 Finally, given the presence of ATP synthase and NADH quinone oxidoreductase (NuoD) and in these experiments, it is tempting to speculate that the list of metabolic enzymes capable of binding to the RNA degradosome in different bacterial organisms can be expanded. However, whether these proteins are genuine components of the degradosome assembly in P. aeruginosa remains to be established.
The functional role of Dip during phage infection
Having identified the RNA degradosome as a target of Dip, the question arose as to the functional consequences of this interaction. The role of Dip could be inferred from its in vitro inhibition of RNase E mediated cleavage of RNA substrates. This inhibitory effect was found on substrates of both bacterial and viral origin, indicating a lack of any specificity toward RNA substrates. Additionally, we found that the Dip protein reaches detectable levels in P. aeruginosa 9 minutes post infection, which was in agreement with a previously published RNA-seq analysis of phage phiKZ-infected Pseudomonas cells.10,15 Since the protein remains present in the cells during the remaining infection cycle, a time-regulated mechanism by which the phage subverts the role of the RNA degradosome in transcript degradation and processing is suggested. It can be speculated that a delay between initial infection and Dip production allows for RNase E mediated degradation of host RNAs prior to the inhibition of this enzyme. In addition, the stabilization of the viral RNA during the middle and later stages of the phage infection cycle is consistent with the fivefold increase in cellular RNA levels during late infection stages.Citation15 In contrast, coliphage T4 uses a different strategy, since it over-activates the host RNase E with Srd, increasing degradation of host RNA during early infection stages.Citation9 The importance of Dip for efficient infection of P. aeruginosa by phiKZ remains to be established, but it is apparent from comparative genomics analyses that this protein does not share sequence homology to proteins of other (closely) related phage. In addition, the unique fold of this protein raises the question of its evolutionary origin and could support the observations that phiKZ forms a distinct branch of the Myoviridae family.Citation16
Toward a structure-based interaction model for Dip and the RNA degradosome
Dip forms a dimer that prevents RNA from being bound and degraded by the RNA degradosome (). Crystallography, site-directed mutagenesis and interactomics approaches revealed the novel structure of Dip (PDB ID 5FT0 and 5FT1) and identified the RNase E interaction site as the outer surface of the Dip dimer. Dip is able to hijack the RNA binding sites of the RNase E scaffold domain via extensive, acidic patches on its outer surface. A double mutation within the acidic surface patch was shown to abolish the interaction with RNase E in vitro (by electrophoretic mobility assays) and retained the wild type phenotype of P. aeruginosa when overexpressed in vivo. This suggests that multiple amino acids in the acidic surface patch contribute to the global interaction between Dip and the RNase E scaffold domain. Two bacterial regulatory factors, RraA and RraB have been found to control RNase E activity in a similar way in E. coli. Both of these regulators bind the C-terminal domain of RNase E, but only RraA does so by occluding the RNA binding sites.Citation17 However, there is no structural similarity between RraA and Dip.
Figure 1. Displacement of RNA from the RNase E scaffold domain by Dip. (A) global composition of the P. aeruginosa RNA degradosome as observed in pull down assays. In the absence of Dip, RNA (dotted line) is bound by the RNase E scaffold domain and degraded by the catalytic domain. (B) When Dip is present, the RNase E scaffold domain is hijacked and RNA is no longer degraded.
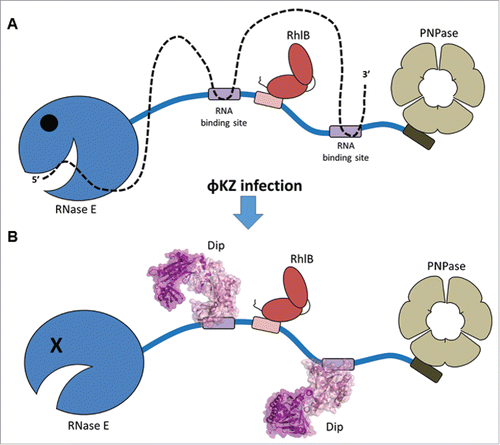
With the current crystallographic data, it is tempting to speculate that Dip is capable of assembling into a higher order structure when bound to RNase E. Such an oligomer of Dip may mimic an RNA duplex strand to misguide the P. aeruginosa RNA degradosome. This ‘nucleic acid-mimicking’ strategy would not be unprecedented, as it has been shown to occur in other phages. For example, the Ocr protein of coliphage T7 mimics B-form DNA to hijack bacterial restriction enzymes and thereby protects T7 genomic DNA.Citation18 Current efforts are concentrating on structural approaches to elucidate the interaction model.
Potential therapeutic applications to treat bacterial infections inspired by Dip
The overuse of antibiotics has led to an ever increasing number of multidrug resistant bacteria since 2000.Citation19 Due to its remarkable capacity to withstand antibiotics, P. aeruginosa has joined the ranks of these ‘superbugs’.Citation20,21 Since phiKZ uses Dip to inhibit the RNA degradosome in a direct and efficient way, it is tempting to apply the targeting of the degradosome in a similar manner as part of a new antibacterial strategy. Even though Dip decreases the growth rate of P. aeruginosa and E. coli, it does not kill these bacteria. Therefore, it should be evaluated whether the RNA binding segments of the RNA degradosome would be a good antibacterial target and if the action of Dip on these specific RNA binding sites can be mimicked by small molecules. Given that previous successful efforts have been made to identify compounds to target the catalytic domain of RNase E in E. coli and Mycobacterium tuberculosis,Citation22 it may be worthwhile to develop Dip-based small molecule inhibitors against the scaffold domain of RNase E to complement the catalytic domain inhibitors.
Although it remains to be seen whether Dip-based molecules can be effective inhibitors, the real strength of Dip may lay in its broad interaction range rather than in its toxicity.Citation10 It has been shown that in addition to being able to inhibit P. aeruginosa RNase E, Dip can inhibit the E. coli degradosome as well. Moreover, interactions with the RNA degradosome of the distantly related Caulobacter crescentus were detected in vivo. Therefore, Dip based inhibitors could be tested against a series of pathogens, in isolation and in conjunction with the small molecule inhibitors against the catalytic domain.
In addition, it is conceivable that heterologous expression of Dip might even improve the infection of a series of designer phages for species like P. aeruginosa, E. coli and C. crescentus, by protecting phage mRNA and increasing expression efficiency during the infection cycle. The ability to generate designer phage with boosted virulence may be particularly appealing to the field of phage therapy, which currently uses natural phage to treat, among others, P. aeruginosa infections in severe burn wounds.Citation23,24
Biotechnological applications of Dip
Since Dip can interact with the RNase E of several bacterial species and can inhibit RNA degradation without killing the bacterial cell, several possible biotechnological applications can be envisaged. Dip may have potential applications in improving recombinant protein expression in bacteria by stabilizing the mRNA of the recombinant protein in vivo, by co-expression of Dip (or addition of a small-molecule inhibitor). Also, Dip could be used as a protein additive in total RNA extraction kits to help stabilize RNA. In both examples, the dose and timing of Dip or small molecule application would need to be optimized to obtain a maximal yield, since RNase E plays important roles in RNA processing as well as degradation.Citation26
In addition, there is great research interest in identifying specific enzymes responsible for the degradation or processing of RNA targets, e.g. when testing stress responses of bacteria. A specific example is that of the degradation of stable RNAs, rRNA and tRNA, upon starvation. Although RNase E is involved in the maturation of rRNA and tRNA, the contribution of the degradosome in the degradation of these stable RNAs remains underexplored.Citation27,28,29 Heterologously expressed Dip or Dip-based small molecule inhibitors could be employed to specifically lower the activity of the RNA degradosome, and thus facilitate such experiments. Presently, such experiments are typically performed using a strain of bacteria with a temperature sensitive RNase E gene product, since knock outs are non-viable. Raising the bacterial culture to a non-permissive growth temperature for the RNase E mutant strain will effectively result in inhibition of RNase E activity, however the change in temperature could also result in multiple undesired and unrelated heat shock responses in the bacterial cell. Although RraA inhibits the degradosome activity via the same mechanism as Dip, It was shown that RNase E has a higher affinity for RNA than for RraA.Citation30 Dip on the other hand, is able to displace bound RNA from RNase E and is therefore believed to have a higher affinity toward its target than RraA.
Finally, Dip or Dip-based small molecule inhibitors could potentially be used in conjunction with CRISPR-Cas editing or RNA interference applications. Both the RNA interference and CRISPR-Cas mechanisms use short RNA fragments in association with a ribonuclease (complex) to target RNA or DNA substrates in the cell. In the case of CRISPR-Cas for example, one may want to use a CRISPR array to knock out several genes at once. The corresponding pre-crRNA will be long and could be subject to degradation by the bacterial RNase E. In antisense RNA experiments, on the other hand, the specificity of the antisense transcript increases with its length. However, long antisense RNAs are unstable and might be subject to degradation by ribonucleases. Therefore Dip or Dip-based small molecule inhibitors might improve the performance of CRISPR-Cas and long antisense RNAs in vivo to knock down targeted gene expression.
In conclusion, these data indicate that molecular phage-bacteria interactions continue to reveal novel mechanisms of metabolism regulation as well as unique protein structures which can inspire application-driven biotechnological developments.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary_Table_1.docx
Download MS Word (35.4 KB)Funding
AV was doctoral fellow supported by the ‘Fonds voor Wetenschappelijk Onderzoek’ (FWO, Belgium). SH, KB and BFL are supported by the Wellcome Trust. This research was further supported by Grant G.0599.11 from the FWO, the SBO-project 100042 of the IWT (‘Agentschap voor Innovatie door Wetenschap en Technologie in Vlaanderen), the KULeuven GOA “Phage Biosystems," the JPN project R-3986 of the Herculesstichting and the grants CREA/09/017 and IDO/10/012 from the KU Leuven Research Fund.
References
- Wagemans J & Lavigne R. In Phages and their hosts: a web of interactions - applications to drug design. In Hyman P & Abedon ST, editors. Bacteriophages in health and disease Ashland (USA): Cabi; 2012. p.119-33.
- Häuser R, Blasche S, Dokland T, Haggård-Ljungquist E, von Brunn A, Salas M, Casjens S, Molineux I & Uetz P. Bacteriophage protein–protein interactions. Protein–protein interactions. Adv Virus Res 2012; 83:219-98; PMID:22748812; http://dx.doi.org/10.1016/B978-0-12-394438-2.00006-2
- Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, Callejo M, Ferretti V, Ha N, Kwan T, et al. Antimicrobial drug discovery through bacteriophage genomics. Nat Biotechnol 2004; 22(2):185-91; PMID:14716317; http://dx.doi.org/10.1038/nbt932
- Yang H, Ma Y, Wang Y, Yang H, Shen W, Chen X. Transcription regulation mechanisms of bacteriophages: recent advances and future prospects. Bioengineered 2014; 5(5):300-4; PMID:25482231; http://dx.doi.org/10.4161/bioe.32110
- Bandyra KJ, Bouvier M, Carpousis AJ, Luisi BF. The social fabric of the RNA degradosome. Biochimica et Biophysica Acta 2013; 1829(6–7):514-22; PMID:23459248; http://dx.doi.org/10.1016/j.bbagrm.2013.02.011
- Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res 2006; 34 (2): 659-66; PMID:16452296; http://dx.doi.org/10.1093/nar/gkj472
- Bandyra KJ, Luisi BF. Licensing and due process in the turnover of bacterial RNA. RNA Biol 2013; 10(4):627-35; PMID:23580162; http://dx.doi.org/10.4161/rna.24393
- Marchand I, Nicholson AW, Dreyfus M. Bacteriophage T7 protein kinase phosphorylates RNase E and stabilizes mRNAs synthesized by T7 RNA polymerase. Mol Microbiol 2001; 42:767-76; PMID:11722741; http://dx.doi.org/10.1046/j.1365-2958.2001.02668.x
- Qi D, Alawneh AM, Yonesaki T & Otsuka Y. Rapid degradation of host mRNAs by stimulation of RNase E activity by Srd of bacteriophage T4. Genetics 2015; 201(3):977-87; PMID:26323881; http://dx.doi.org/10.1534/genetics.115.180364
- Van den Bossche A, Hardwick SW, Ceyssens P-J, Hendrix H, Voet M, Dendooven T, Bandyra KJ, De Maeyer M, Aertsen A, Noben J-P, et al. Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome. eLife 2016; 5:e16413; PMID:27447594; http://dx.doi.org/10.7554/eLife.16413
- Van den Bossche A, Ceyssens P-J, De Smet J, Hendrix H, Bellon H, Leimer N, Wagemans J, Delattre A-S, Cenens W, Aersten A, et al. Systematic identification of hypothetical bacteriophage proteins targeting key protein complexes of pseudomonas aeruginosa. J Proteome Res 2014; 13(10):4446-56; PMID:25185497; http://dx.doi.org/10.1021/pr500796n
- Górna MW, Carpousis AJ, Luisi BF. From conformational chaos to robust regulation: the structure and function of the multi-enzyme RNA degradosome. Q Rev Biophys 2012; 45(2):105-45; PMID:22169164; http://dx.doi.org/10.1017/S003358351100014X
- Miczak A, Kaberdin VR, Wei CL, Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proceedings of the National Academy of Sciences of the United States of America 1996; 93(9):3865-3869.
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000; 406(6799):959-64; PMID:10984043; http://dx.doi.org/406(6799):959-64
- Ceyssens P-J, Minakhin L, Van den Bossche A, Yakunina M, Klimuk E, Blasdel B, De Smet J, Noben JP, Bläsi U, Severinov K, et al. Development of giant bacteriophage φKZ is independent of the host transcription apparatus. Hutt-Fletcher L, ed. Journal of Virology 2014; 88(18):10501-10; PMID:24965474; http://dx.doi.org/10.1128/JVI.01347-14
- Krylov V, Pleteneva E, Bourkaltseva M, Shaburova O, Volckaert G, Sykilinda N, Kurochkina L & Mesyanzhinov V. Myoviridae bacteriophages of Pseudomonas aeruginosa: a long and complex evolutionary pathway. Research in Microbiology 2003; 154(4):269-275. doi: 10.1016/S0923-2508(03)00070-6
- Gao J, Lee K, Zhao M, Qiu J, Zhan X, Saxena A, Moore CJ, Cohen SN, Georgiou G. Differential modulation of E. coli mRNA abundance by inhibitory proteins that alter the composition of the degradosome. Molecularl 2006; 61(2):394-406; PMID:16771842; http://dx.doi.org/10.1111/j.1365-2958.2006.05246.x
- Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM & Dryden DTF. Structure of Ocr from Bacteriophage T7, a protein that mimics B-form DNA. Molecular Cell 2002; 9(1):187-194. http://dx.doi.org/10.1016/S1097-2765(02)00435-5
- Bassetti M, Merelli M, Temperoni C, Astilean A. New antibiotics for bad bugs: where are we? Ann Clin Microbiol Antimicrob 2013; 12:22; PMID:23984642; http://dx.doi.org/10.1186/1476-0711-12-22
- Breidenstein EB, de la Fuente-Núñez C, Hancock RE. Pseudomonas aeruginosa: all roads lead to resistance. Trends in Microbiology 2011; 19(8):419-26. doi: 10.1016/j.tim.2011.04.005
- Courvalin P. Predictable and unpredictable evolution of antibiotic resistance. Journal of internal medicine 2008; 264(1):4-16. doi: 10.1111/j.1365-2796.2008.01940.x
- Kime L, Vincent HA, Gendoo DMA, Jourdan SS, Fishwick CWG, Callaghan AJ & McDowall KJ. The first small-molecule inhibitors of members of the ribonuclease E family. Scientific Reports 2015; 5:8028. doi: 10.1038/srep08028
- Rose T, Verbeken G, Vos DD, Merabishvili M, Vaneechoutte M, Lavigne R, Jennes S, Zizi M, Pirnay J-P. Experimental phage therapy of burn wound infection: difficult first steps. Int J Burns Trauma 2014 Oct 26; 4(2):66-73; PMID:25356373
- Brüssow H. What is needed for phage therapy to become a reality in Western medicine? Virology 2012; 434(2):138-42. doi: 10.1016/j.virol.2012.09.015
- Endersen L, O'Mahony J, Hill C, Ross RP, McAuliffe O, Coffey A. Phage therapy in the food industry. Annual review of food science and technology 2014; 5:327-49. doi: 10.1146/annurev-food-030713-092415
- Mackie GA. RNase E: at the interface of bacterial RNA processing and decay. Nature reviews. Microbiology 2013; 11(1):45-57. doi: 10.1038/nrmicro2930
- Maiväli Ü, Paier A, Tenson T. When stable RNA becomes unstable: the degradation of ribosomes in bacteria and beyond. Biol Chem 2013; 394(7):845-55; PMID:23241849; http://dx.doi.org/10.1515/hsz-2013-0133
- Deutscher MP. Degradation of stable RNA in Bacteria. The Journal of Biological Chemistry 2003; 278:45041-45044. doi: 10.1074/jbc.R300031200
- Sulthana S, Basturea GN & Deutscher MP. Elucidation of pathways of ribosomal RNA degradation: an essential role for RNase E. RNA 2016; 22(8):1163-71. Doi: 10.1261/rna.056275.116
- Górna MW, Pietras Z, Tsai YC, Callaghan AJ, Hernández H, Robinson CV, Luisi BF. The regulatory protein RraA modulates RNA-binding and helicase activities of the E. coli RNA degradosome. RNA 2010; 16(3):553-62. doi: 10.1261/rna.1858010
