ABSTRACT
The neuronal transcriptome changes dynamically to adapt to stimuli from the extracellular and intracellular environment. In this study, we adapted for the first time a click chemistry technique to label the newly synthesized RNA in cultured hippocampal neurons and intact larval zebrafish brain. Ethynyl uridine (EU) was incorporated into neuronal RNA in a time- and concentration-dependent manner. Newly synthesized RNA granules observed throughout the dendrites were colocalized with mRNA and rRNA markers. In zebrafish larvae, the application of EU to the swim water resulted in uptake and labeling throughout the brain. Using a GABA receptor antagonist, PTZ (pentylenetetrazol), to elevate neuronal activity, we demonstrate that newly transcribed RNA signal increased in specific regions involved in neurogenesis.
Abbreviations
| 4sU | = | 4-thiouridine |
| 4tU | = | 4-thiouracil |
| ACSF | = | Artificial cerebrospinal fluid |
| BrU | = | 5-Bromouridine |
| BrUTP | = | 5-Bromouridine-5′-triphosphate |
| DA-AP5 | = | D(−)-2-Amino-5-phosphonopentanoic acid |
| EU | = | 5-Ethynyl uridine |
| DIV | = | Days in vitro |
| DMSO | = | Dimethyl sulfoxide |
| dpf, Day post fertilization; DRB | = | D-ribofuranosylbenzimidazole |
| GABA | = | Gamma-Aminobutyric acid |
| HEPES | = | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| LUT | = | Lookup table |
| MAP2 | = | Microtubule associated protein 2 |
| mEPSC | = | Miniature excitatory postsynaptic current |
| mRNA | = | messenger RNA |
| rRNA | = | ribosomal RNA |
| RNP | = | Ribonucleoprotein |
| PABP | = | Poly(A)- Binding Protein |
| PFA | = | Paraformaldehyde |
| PIPES | = | Piperazine-N,N'-bis(2-ethanesulfonic acid) |
| PTZ | = | Pentylenetetrazol |
| TBTA | = | Tris((1-benzyl-1H-1,2,3-triazol-4-yl)methyl)amine |
| TTX | = | Tetrodotoxin |
| TUNEL | = | Terminal deoxynucleotidyl transferase dUTP nick-end labeling |
| Y10B | = | An antibody against 5.8 S rRNA |
Introduction
In the brain, selective RNA transport to subcellular compartments is a way to regulate gene expression in a spatiotemporal manner and serves to functionalize distinct domains in response to extracellular events. In neurons, RNA transport to dendrites provides a local source for new protein synthesis, which is important for some forms of synaptic plasticity and memory.Citation1,2 A recent study revealed a plethora of localized mRNAs (more than 2550) in the hippocampal neuropil, containing mainly dendritic and axonal compartments, suggesting the importance of localized RNA in local protein translation.Citation3
To visualize individual RNA species in different cellular compartments many techniques have been used including in situ hybridization,Citation4 tagging with fluorescent proteins,Citation5 molecular beaconsCitation6 or injecting fluorescently labeled RNAs into cells.Citation7 On the other hand, 4 methods have been used to label and visualize specifically newly synthesized RNA species to explore the RNA turnover, transcription and transport rates, and the dynamics of RNP (ribonucleoprotein) granules. The first method uses labeling with radioactive nucleosides and the subsequent visualization by autoradiography,Citation8 the second method relies on the delivery of BrUTP (5-Bromouridine-5’-triphosphate) or BrU (5-bromouridine) followed by immunostaining,Citation9-11 the third method on the incorporation of UTP-tagged with a fluorescent dye into RNA and further visualization via live imagingCitation12 and the fourth method on labeling with a small bioorthogonal group (i.e. alkyne, azide) coupled to a nucleoside or nucleotide followed by detection via click chemistry.Citation13,14 The first 2 methods are hindered by limited image resolution and limited access of the nucleotide analog or the antibody into the cells or tissues, respectively. The third method is powerful in terms of live imaging of RNA granules, however the nucleotide analogue needs to be injected into neurons and cannot be chemically modified for further purification. The click chemistry approach is advantageous owing to the easy penetration of the small nucleoside analogue and the tag into the cells, tissues, organs and intact animalsCitation15-17; fast detection and the amenability to high-resolution imaging.
Here, we adapted and optimized the click chemistry method to visualize the newly synthesized RNA in rat hippocampal neurons and the intact zebrafish brain. We demonstrate the time- and concentration- dependent incorporation of EU (5-ethynyl uridine) and the detection of total newly synthesized RNA signal within neuronal dendrites, and in the regions involved in the neurogenesis in the zebrafish brain. We show that RNA granules exhibit colocalization with Poly(A)-Binding Protein (PABP) and a rRNA marker (Y10B). Neuronal activity induced by KCl depolarization caused a significant decrease in the nascent RNA signal in dendrites. Moreover, we used the technique to detect newly synthesized RNA in the intact larval zebrafish brain.
Results
To initially optimize the visualization of the newly synthesized RNA in cultured hippocampal neurons, the medium was treated with EU, an alkyne-containing nucleoside, at concentrations of 0.5, 1, 5 or 10 mM for 6 hours and a click reaction was performed using an Alexa594-azide (5 μM) to label the EU-incorporated RNA with a fluorescent tag (). Imaging of the fluorescent signal in the somata and dendrites (detected by immunostaining with an anti-MAP2 antibody) of neurons revealed a dose-dependent increase in signal intensity (). The application of 0.5 mM EU resulted in labeling of nascent RNA in the nucleus and proximal dendrites but not in the distal dendrites. Labeling with 5 mM EU yielded a stronger RNA signal in the distal dendrites compared to 1 mM EU treatment. 10 mM EU labeling produced a signal comparable to that observed with 5 mM but also often resulted in the formation of precipitates after click chemistry (). In order to verify that the observed signal results from the incorporation of EU into nascently transcribed RNA, we conducted labeling experiments in the presence of transcription inhibitors (ActinomycinD, Triptolide and DRB). In contrast to the robust signal observed previously, there was only a minimal amount of background labeling evident in the vicinity of the nucleus and no dendritic signal present (). In addition, the omission of EU also resulted in a complete absence of signal ().
Figure 1. Visualization of newly synthesized RNA in cultured hippocampal neurons. (A) Illustration of the method. (B) An increase in the somatic and dendritic RNA signal is observed with increasing concentrations of EU. Fire lookup table (LUT) represents fluorescence intensity of labeled RNA (pixel intensities 0–255). Arrowheads (purple) indicate RNA granules. Scale bar for B and C, 50 μm (C) Block of transcription or absence of EU results in a severely diminished nascent RNA signal. The dendrites were detected using an anti-MAP2 antibody (green). (D) Representative straightened dendrites following 5 mM EU for the indicated treatment times. Left, proximal; right, distal. Scale bar 20 μm. LUT, Fire (pixel intensities 0–255) (E) Mean intensity of the nascent RNA fluorescence in MAP2-defined dendritic area. Per time point, 20–42 dendrites from 2 independent experiments were analyzed. ****p < 0.0001, *p = 0.0254, n.s. (non significant) p = 0.6304, *p = 0.0313, respectively. (F) Heatmap shows the RNA signal along the dendrite. LUT, Spring. (G) Electrical and synaptic properties after 5 mM EU treatment for 12 h. Upper-left, representative traces of mEPSCs from the control and EU-treated neurons; upper-right, mEPSCs amplitude in control (n = 7) and EU-treated neurons (n = 7), p = 0.8844; lower-left, mEPSC frequency in control (n = 7) and EU-treated neurons (n = 7), p = 0.6342, lower-right, resting potential in control (n = 8) and EU-treated neurons (n = 8), p = 0.9707.
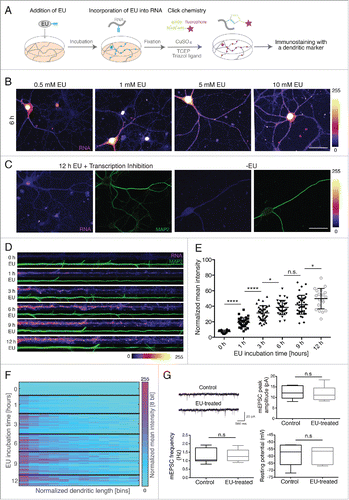
Based on the above results, we used 5 mM EU in subsequent experiments to examine the time course of EU incorporation into neuronal RNA. Labeling for as short as 10 min was sufficient to see a signal in the nucleus and after labeling for 30 min signal was evident in the somata and proximal dendrites (data not shown). 1 h of labeling resulted in the appearance of the RNA signal along the dendrites ().
To observe changes in the newly synthesized RNA signal with increasing treatment times, we quantified the RNA signal from linearized dendrites after 1, 3, 6, 9 and 12 h of EU labeling using the MAP2-immunostained dendritic area as a mask. We observed a significant increase in the mean dendritic signal after 1 and 3 h of labeling while the increase from 3 to 6 h and 9 to 12 h was less significant () cultured neurons showed some heterogeneity in RNA labeling presumably owing to variability in neuronal subtype, transcription or RNA transport rate between neurons). We also visualized the distribution of RNA signal along the dendrites using a heatmap that indicates the signal distribution in proximal and distal dendrites of examined neurons ().
To detect whether EU treatment modifies neuronal electrical or synaptic properties, whole-cell patch clamp recordings were carried out after 12 h of 5 mM EU treatment. No difference was observed between non-EU treated and EU-treated neurons in either the amplitude or frequency of mEPSCs or intrinsic properties such as resting membrane potential (), the input resistance or the AP (action potential) width (data not shown). We also tested whether EU treatment induces apoptosis using TUNEL labeling to detect DNA fragmentation. Neurons labeled with EU did not show any positive TUNEL staining (n = 9 neurons) (data not shown).
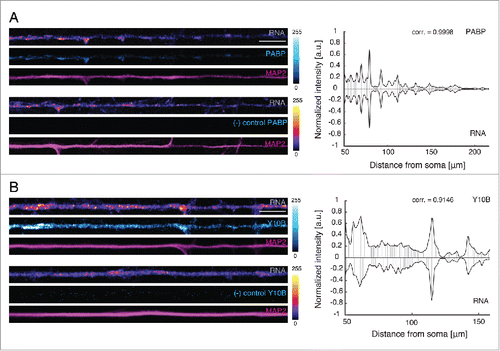
The RNA signal we detected in dendrites was often punctate in nature, suggesting that the labeled RNA was associated with ribonucleoprotein particles or granules. To address this, we labeled nascent RNA and examined the colocalization with mRNA or rRNA markers, using antibodies against the poly(A)-binding protein (PABP) or Y10B, respectively. PABP is an RNA-binding protein that regulates mRNA translationCitation18 while Y10B is an antibody specific to 5.8 rRNA.Citation19 After labeling neurons for 6 h (5 mM EU), we immunostained neurons with either an anti-PABP antibody or a Y10B antibody. We found that the PABP or Y10B signals were significantly colocalized with the nascent RNA granules (range of correlation coefficient for PABP = 0.8864–0.9998, n = 6 dendrites and 0.8735–0.9861 for Y10B, n = 7 dendrites) (), suggesting that a majority of newly synthesized RNA is associated with RNA granules that contain mRNA and rRNA.
Figure 2. PABP and Y10B colocalize with newly synthesized RNA. (A, B) Left panels: Representative dendrites showing the colocalization of the fluorescent signals from PABP or Y10B and nascent RNA. Only a defined length (104 μm) of the original straightened dendrite is shown. Antibody leave-out control (no anti-PABP) does not show any signal. Scale bars: 10 μm. LUT for PABP, Cyan Hot; LUT for RNA, Fire (pixel intensities 0–255). The dendritic marker MAP2, magenta. Right panels: Graph of the correlation of the signal from RNA granules and PABP or Y10B along the original length of the representative dendrites shown. corr., correlation coefficient.
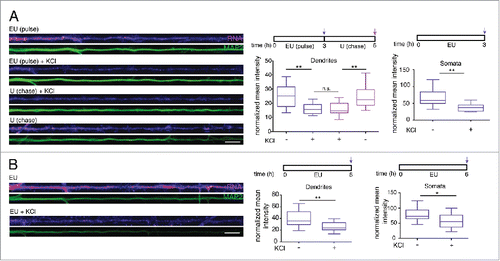
To examine whether elevated activity might alter the profile of newly transcribed RNA detected we examined the effect of depolarization. We applied EU (5 mM) together with 0 or 56 mM KCl buffer to the culture medium for 3 h or 6 h and detected a significant decrease in the newly synthesized RNA signal in both the dendrites and cell body after both 3 and 6 h (), suggesting a decay of newly synthesized RNA after depolarization. We next carried out a pulse-chase experiment to test whether the pre-existing dendritic RNA signal might decrease. Following EU labeling for 3 h, we performed a 3 h chase with or without KCl in the presence of 5 mM uridine. The delayed application of KCl during the chase period also resulted in a pronounced decrease in the previously labeled nascent RNA suggesting that depolarization results in an active degradation of the newly synthesized RNA ().
Figure 3. Depolarization of hippocampal neurons induces a decrease in detectable nascent RNA in the somata and dendrites. (A, B) Left panels: Representative dendrites showing a decrease in newly synthesized RNA signal after KCl treatment. The dendritic marker MAP2, green. Scale bars, 10 μm. Right panels: Graph showing the average intensity of RNA fluorescence in dendrites (n = 10–16) or somata (n = 9–19) in MAP2-defined dendritic area. Nascent RNA signal decreased significantly in both dendrites (top, **p = 0.0043, n.s. = 0.9999, **p = 0.0093, respectively; bottom, **p = 0.0023) and somata (top, **p = 0.0029, bottom, *p = 0.0129) following KCl treatment.
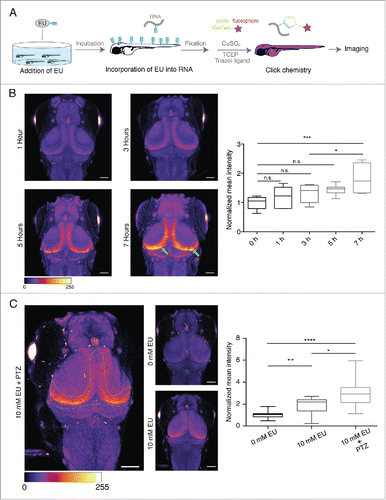
The above experiments indicate that EU can be used to label nascent RNA in cultured neurons, but do not address the potential in vivo applicability in brain. To explore this, we optimized the labeling and click reaction conditions to tag nascent RNA in the brain of intact larval zebrafish (). To visualize newly synthesized RNA in the brain we used a transgenic line, nacre HUC:GCaMPs6. HUC encodes a neuronal RNA-binding protein and its promoter is used as a panneuronal driver.Citation20,21 GCaMPs6 expression was used in order to visualize the larval brain. We incubated zebrafish larvae (4 dpf) in E2 medium supplemented with EU (10 mM) for 1, 3, 5 or 7 h to determine the rate of EU incorporation. We detected a significant increase in RNA signal after 7 h, especially in the region along the caudo-medial edge of the optic tectum, which is involved in neurogenesis (previously described by Huang et al.Citation22) (), suggesting relatively high transcription rates in differentiating neurons. To investigate whether neuronal activity induces changes in newly synthesized RNA in zebrafish brain, we explored the effect of pentylenetetrazol (PTZ; 15 mM), a GABA receptor antagonist, which induces epileptic-like seizures in zebrafish.Citation23 We incubated larvae with EU (10 mM) for 7 h and added PTZ to the medium for the last 1 h. We observed a dramatic increase in the newly synthesized RNA in the neurogenic regions, suggesting that PTZ induced an increase in the RNA synthesis ().
Figure 4. In vivo labeling of newly synthesized RNA in the larval zebrafish brain. (A) Experimental procedure. (B) Left. Representative images of the larval zebrafish head (dorsal view) showing the time-dependent increase in nascent RNA signal in specific regions (green arrows) of the tectum. LUT, Fire (pixel intensities 0–255). Right. Graph representing the time-dependent increase in the normalized mean fluorescence intensity of RNA signal for the indicated conditions. Scale bar, 25 μm. n = 7–9 larvae from 2 independent experiments. *p = 0.0221, p = 0.0001, n.s., (not significant). LUT, Fire. (C) Left. Representative images of the larval zebrafish head (dorsal view) showing PTZ-induced increase in the newly synthesized RNA signal in the indicated region of the tectum. Scale bar, 25 μm. LUT, Fire (pixel intensities 0–255). Right. Graph representing the PTZ- induced increase in the normalized mean intensity of the newly synthesized RNA signal for the indicated conditions. n = 22–24 larvae from 4 independent experiments. *p = 0.0133, **p = 0.0062, ****p < 0.0001.
Discussion
The neuronal transcriptome undergoes dynamic alterations to adapt to environmental changes through the regulation of RNA synthesis and degradation. Most studies have focused on the visualization of few RNA species at a time to detect the changes in different neuronal compartments upon activityCitation24,25 In this study, we optimized EU labeling together with click chemistry to visualize nascent RNA species in the dendrites of hippocampal neurons and zebrafish brain. Using labeling and imaging, we showed that EU was incorporated into dendritic RNA in a time- and concentration-dependent manner. A previous study reported that newly synthesized RNA signal was restricted to the cell body of hippocampal neurons after 1 h of 3H uridine labeling.Citation8 In the present study, however, we detected RNA granules in distal dendrites after just 1 h of EU labeling, indicating that the present technique may be more sensitive than radiolabeling.
It has been previously shown that mRNAs are transported to the dendrites in the form of motile RNA granules, which consist of RNA-binding proteins, mRNA, rRNA and ribosomal proteins.Citation26-29 Consistent with those findings we observed granular distribution of the newly synthesized RNA along the dendrites. These granules were colocalized with PABP and Y10B, implying that they form functional units. It is well established that RNA granules are highly dynamic structures.Citation30,31 Depolarization of neurons has been shown to reorganize granules and redistribute certain mRNAs involved in synaptic plasticity.Citation27 Park et al.Citation36 reported that a short (3–6 min) treatment with KCl led to an increase in the ratio of split to merged neuronal RNA granules, indicating a disassembly of the granule or an unmasked state of the mRNA. Here, we examine, for the first time, the effect of depolarization on de novo RNA synthesis. We demonstrated a general decrease in newly synthesized RNA in dendrites after 3 h or 6 h of KCl treatment. A decline in global protein synthesis after depolarization has also previously been observed.Citation27,32
Recent work by Hinz et al.Citation17 described the introduction of bioorthogonal groups to detect newly synthesized proteins in zebrafish larvae. In this study, we optimized the incorporation of EU into the larval zebrafish brain and visualized newly transcribed RNA in regions known to be involved in neurogenesis.Citation22 Using a GABA receptor antagonist, PTZ, we showed an increase in the newly synthesized RNA in those neurogenic regions. This increase is also consistent with an increase in the nascent larval zebrafish proteome observed after PTZ treatment.Citation17
The click chemistry method has been previously used to visualize and purify newly synthesized proteins.Citation15,16,33 Although a well-established method exists to purify the RNA after labeling with 4sU or 4tU in a cell-type specific manner,Citation34-36 this method has not been used for the visualization of labeled RNA. In the future, the techniques described can be optimized for the purification of the labeled RNA using an affinity tag so that the visualization and purification of newly transcribed RNA can be performed in parallel.
Methods
Primary hippocampal neuron culture
Sprague Dawley male and female rat pups (P0-1) were used to prepare dissociated hippocampal neurons as described previously.Citation37,38 Briefly, neurons were plated at a density of 30 × 103 cells/cm2 onto poly-D-lysine-coated glass-bottom MatTek dishes and maintained in Neurobasal A medium containing B-27 and Glutamax supplements (Invitrogen) at 37°C until they were used (DIV14-20). All experiments complied with the regulations of German animal welfare law and the Max Planck Society.
Labeling cultured neurons with EU
5-ethynyluridine (EU) was either purchased from Berry & Associates (PY 7563) or synthesized in-house as described in Jao and Salic.Citation14 EU, at the indicated concentrations, was added to 1 ml of the culture medium from a 200 mM stock in 80% DMSO. In the experiments in which EU of 0.5 mM, 1 mM, 5 mM and 10 mM was tested to find the optimal labeling concentration, for 10 mM EU only, after labeling, hippocampal neurons (DIV14) were placed back in their original conditioned medium (culture medium+secreted factors from the cultured cells) for 30 min to reduce precipitate formation caused by 10 mM EU. The neurons were washed with 1x PBS (Gibco), equilibrated to 37°C before fixation and fixed (30 min) at room temperature with 125 mM Pipes, pH 6.8, 10 mM EGTA, 1 mM magnesium chloride, 0.2% Triton X-100 and 3.7% formaldehyde in water. Fixed neurons were then washed 3 times with 1xPBS (RNase free). In experiments using transcription inhibitors, Actinomycin D (24 μM), DRB (D-ribofuranosylbenzimidazole) (100 μM) and Triptolide (1 μM) were added to the conditioned medium 1 h before labeling and were present in the medium during the EU labeling for 12 h ().
Click chemistry to detect labeled RNA and immunostaining
All the reagents and solutions used were RNAse-free. Before performing the click reaction, water and 1x PBS (pH = 7.86) (as described in Dieterich et al.Citation16) were degassed with Argon to prevent RNA degradation. The reaction mix contained 200 μM triazole ligand Tris((1-benzyl-1H-1,2,3-triazol-4-yl)methyl)amine (TBTA) (200 mM stock solution in DMSO), 500 μM TCEP (500 mM solution prepared freshly in water), 5 μM Alexa594-azide (10 mM stock solution in DMSO) and 200 μM CuSO4 (200 mM solution prepared freshly in water) in 1x PBS, pH 7.86 (modified from Dieterich et al.,Citation15 Tom Dieck et al.Citation20). The solution was vortexed vigorously after the addition of each component and then added to the cultured neurons immediately after preparation. The neurons were incubated in the click solution for 1 h 15 min at room temperature in a humidified chamber. After incubation the neurons were washed 2 times each for 10 min with 1% Tween-20, 0.5 mM EDTA in 1x PBS (pH = 7.86), followed by 2 washes each for 10 min with 1% Tween-20, 0.5 mM EDTA, 0.1% TritonX-100 in PBS (pH = 7.86) and a last wash for 10 min in PBS.
For subsequent immunostaining, neurons were treated with a blocking buffer (4% goat serum in 1× PBS) for 1 h at room temperature. Anti-MAP2 antibody (Sigma, M9942, goat anti-mouse, 1:1000) in blocking buffer was applied for 1 h at room temperature or overnight at 4°C. The neurons were washed 3x times each for 10 min with 0.1% TritonX-100 in PBS and treated with the secondary antibody (Invitrogen, A11001, Alexa 488-goat anti-mouse) for 1 h at room temperature and washed 2x each for 10 min with 0.1% TritonX-100 in PBS and a last wash for 10 min in PBS. For the co-localization experiments, PABP: anti-rabbit PABP (goat, Abcam ab21060, 1:500) and anti-mouse MAP2 (Sigma, M9942, 1:1000) as primary antibodies and anti-rabbit Alexa488 (goat, Invitrogen, A11008) and anti-mouse Alexa405 (goat, Invitrogen, A3153) as secondary antibodies were used; Y10B: anti-mouse Y10B (Abcam ab37144, 1:1000) and anti-rabbit MAP2 as primary antibodies; anti-mouse Alexa488 (Invitrogen, A11001, 1:1000) and anti-rabbit Alexa405 (goat, Invitrogen, A31556) as secondary antibodies were used. The blocking buffer included RNase inhibitor (Promega, RNasin Plus RNase inhibitor, final concentration of 0.04 U/μl) to prevent degradation of the RNA. Cells were mounted with AquaPolymount (Polysciences).
Image acquisition and processing
A Zeiss LSM780 confocal laser fluorescence microscope was used to acquire Z-stack images of neurons using a 40x/1.4-NA oil objective (Plan Apochromat 40x/1.4 oil DIC M27) with a 1024×1024 pixel resolution at 16 bit. In many experiments, the RNA signal in the nucleus was allowed to saturate in order to detect the dendritic signal in a suitable dynamic range. The images were analyzed using ImageJ. A maximum projection of the stacks was taken and, when necessary, the brightness-contrast settings were changed in the same way for all the images within an experiment. Dendrites of at least 100 μm length were linearized using the Straighten plugin (ImageJ) after conversion of the images from 16-bit to 8-bit. For the newly synthesized RNA signal analysis, a custom-written Matlab script was used to calculate the mean fluorescence intensity of the newly synthesized RNA in the dendritic area, using the MAP2 channel as a mask. The normalized mean intensity was used to evaluate the RNA signal in the dendrites ( and ). For the heatmap, the processed signal was split into an equal number of bins which defined the normalized dendritic length. Colocalization analysis between newly synthesized RNA signal and PABP or Y10B signal was performed using single planes of the dendritic segments (varied between 71–218 μm) which started 50 μm away of the soma. The colocalization analysis was conducted using the MAP2 masked intensity from each signal and correlating the local peaks of the distributions (). For the RNA signal analysis in the cell body (), the mean intensity of the signal in the somata around the nucleus was normalized to the somatic area minus nuclear area.
Electrophysiology
Primary hippocampal neurons (DIV14) were treated with 5 mM EU in the culture medium for 12 hours. Whole-cell patch clamp recordings were made from EU-treated and non-treated (control) neurons at room temperature using an Axopatch 200B amplifier. Signals were acquired at 50 KHz and Bessel filtered at 20 KHz. HEPES-buffered ACSF containing [in mM] 140 NaCl, 1.25 NaHPO4, 1 MgSO4, 3 KCl, 2 CaCl2, 1 MgCl2, 15 glucose, 10 HEPES [pH 7.4] was used as the extracellular solution. Miniature excitatory postsynaptic potentials (mEPSCs) were recorded in the presence of 1 μM TTX and 20 μM bicuculline. The patch pipette internal solution contained (in mM) 120 potassium gluconate, 20 KCl, 0.1 EGTA, 2 MgCl2, 10 HEPES, 2 ATP, 0.4 GTP (pH 7.2, ∼300 mOsm) and had resistances ranging from 4 to 7 MΩ. Cells were voltage-clamped at −70 mV and series resistance (Rs) was left uncompensated but monitored continuously. Experiments in which Rs changed more than ±3 MΩ were not included in the analysis. mEPSCs were analyzed offline using Stimfit software.Citation39 Minis were detected by template matching with a 4 pA amplitude threshold. The total recording period ranged from 10 to 15 min.
Neuronal depolarization
Hippocampal neurons (DIV20) were silenced overnight in 1 μM tetrodotoxin (TTX, Tocris) and 100 μM D(−)-2-amino-5-phosphonopentanoic acid (DA-AP5, Tocris) and subsequently treated with a medium containing 5 mM EU along with the 31% KCl buffer (described in Malik et al.Citation40 to achieve a final concentration of 56 mM KCl for 3 h or 6 h, or, 5 mM EU along with the same buffer lacking KCl. A 3 h chase was applied (using the original conditioned medium without EU) after 3 h of EU labeling in the presence of 5 mM uridine with or w/o KCl as indicated ().
Labeling of RNA in zebrafish brain
Zebrafish larvae (4 dpf, Nacre, HUC:GCaMPs6) were incubated with EU (10 mM) in E2 media (prepared as described in WesterfieldCitation41) for up to 7 hours. After incubation the larvae were washed 3 times for 5 min with E2 at room temperature, anesthetized with cold water and fixed in cold PFA-fixative (4% PFA, 4% sucrose in PBS prepared as described in Tom Dieck et al.Citation20) over night at 4°C. The permeabilization and clicking of the fluorophore onto the EU was as described by Tom Dieck et al.Citation42 with the following changes: Larvae were digested with 1 mg/ml collagenase; the click-reaction was adjusted to 4 ml and contained 0.5 ml PBS (pH 7.88), 3.5 ml PBST (pH 7.88), 0.2 mM TBTA, 0.5 mM TCEP, 5 μM Alexa 594-azide, and 0.2 mM CuSO4; the click-reaction was assembled under Argon gas. Larvae were mounted in 0.6% agarose and imaged using 10x objective and an LSM710 confocal microscope, at wavelengths 493–594 nm (GCaMP) and 599–734 nm (Alexa594-azide), excited by 488 nm and 594 nm lasers respectively. Maximum projections of the images were created using ImageJ. The fluorescent signal of GCaMPs6 was used to generate a mask to isolate the signal from labeled RNA within brain and for the identification of brain regions of interest. The neurogenic regions with high EU labeling, which overlapped with GCAMPs6 signal were selected using the Straighten plugin. The mean intensity of RNA signal was normalized to the signal from non-EU treated samples.
Statistical analysis
Statistical analysis was performed using GraphPad Prism Version 6.0b. A D'Agostino & Pearson or Shapiro-Wilk normality test (when the sample size was < 8) was used to pre-test the distribution of the data. Statistical significance was tested using a one-way Anova corrected for multiple comparisons- Tukey-Kramer method (, , ) or Kruskal-Wallis test for non-normally distributed data sets () or 2-sided unpaired t-tests (for 2 groups) (, ). Error bars in the distribution plot () represent the standard deviation of the mean. The whiskers in the box plots represent the minima and maxima of the groups and the middle line represents the median.
Disclosure of potential conflicts of interest
The authors declare no competing financial interests.
Acknowledgments
We thank I. Bartnik, N. Fuerst, A. Staab and C. Thum for the preparation of cultured hippocampal neurons. We also thank C. Glock, A.V. Traylor, M. Azcorra, I. Uyan, K. Kececi for their help in straightening the dendrites during the optimization of experiments. E.M.S. is funded by the Max Planck Society, an Advanced Investigator award from the European Research Council, DFG CRC 1080, DFG CRC 902, and the DFG Cluster of Excellence for Macromolecular Complexes.
References
- Holt CE, Schuman EM. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 2013; 80:648-57; PMID:24183017; http://dx.doi.org/10.1016/j.neuron.2013.10.036
- Trinh MA, Klann E. Translational control by eIF2alpha kinases in long-lasting synaptic plasticity and long-term memory. Neurobiol Learn Mem 2013; 105:93-9; PMID:23707798; http://dx.doi.org/10.1016/j.nlm.2013.04.013
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 2012; 74:453-66; PMID:22578497; http://dx.doi.org/10.1016/j.neuron.2012.02.036
- Burgin KE, Waxham MN, Rickling S, Westgate SA, Mobley WC, Kelly PT. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J Neurosci 1990; 10:1788-98; PMID:2162385
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell 1998; 2:437-45; PMID:9809065; http://dx.doi.org/10.1016/S1097-2765(00)80143-4
- Santangelo PJ. Molecular beacons and related probes for intracellular RNA imaging. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2010; 2:11-9; PMID:20049827; http://dx.doi.org/10.1002/wnan.52
- Wilkie GS, Davis I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell 2001; 105:209-19; PMID:11336671; http://dx.doi.org/10.1016/S0092-8674(01)00312-9
- Davis L, Banker GA, Steward O. Selective dendritic transport of RNA in hippocampal neurons in culture. Nature 1987; 330:477-9; PMID:2446139; http://dx.doi.org/10.1038/330477a0
- Halicka HD, Bedner E, Darzynkiewicz Z. Segregation of RNA and separate packaging of DNA and RNA in apoptotic bodies during apoptosis. Exp Cell Res 2000; 260:248-56; PMID:11035919; http://dx.doi.org/10.1006/excr.2000.5027
- Cmarko D, Verschure PJ, Martin TE, Dahmus ME, Krause S, Fu XD, van Driel R, Fakan S. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol Biol Cell 1999; 10:211-23; PMID:9880337; http://dx.doi.org/10.1091/mbc.10.1.211
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol 1993; 122:283-93; PMID:8320255; http://dx.doi.org/10.1083/jcb.122.2.283
- Piper M, Lee AC, van Horck FP, McNeilly H, Lu TB, Harris WA, Holt CE. Erratum to: Differential requirement of F-actin and microtubule cytoskeleton in cue-induced local protein synthesis in axonal growth cones. Neural Dev 2015; 10:16; PMID:26088085; http://dx.doi.org/10.1186/s13064-015-0043-9
- Sawant AA, Tanpure AA, Mukherjee PP, Athavale S, Kelkar A, Galande S, Srivatsan SG. A versatile toolbox for posttranscriptional chemical labeling and imaging of RNA. Nucleic Acids Res 2016; 44:e16; PMID:26384420; http://dx.doi.org/10.1093/nar/gkv903
- Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci U S A 2008; 105:15779-84; PMID:18840688; http://dx.doi.org/10.1073/pnas.0808480105
- Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci 2010; 13:897-905; PMID:20543841; http://dx.doi.org/10.1038/nn.2580
- Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc 2007; 2:532-40; PMID:17406607; http://dx.doi.org/10.1038/nprot.2007.52
- Hinz FI, Dieterich DC, Tirrell DA, Schuman EM. Non-canonical amino acid labeling in vivo to visualize and affinity purify newly synthesized proteins in larval zebrafish. ACS Chem Neurosci 2012; 3:40-9; PMID:22347535; http://dx.doi.org/10.1021/cn2000876
- Khoutorsky A, Yanagiya A, Gkogkas CG, Fabian MR, Prager-Khoutorsky M, Cao R, Gamache K, Bouthiette F, Parsyan A, Sorge RE, et al. Control of synaptic plasticity and memory via suppression of poly(A)-binding protein. Neuron 2013; 78:298-311; PMID:23622065; http://dx.doi.org/10.1016/j.neuron.2013.02.025
- Lerner EA, Lerner MR, Janeway CA, Jr, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci U S A 1981; 78:2737-41; PMID:6789322; http://dx.doi.org/10.1073/pnas.78.5.2737
- Vladimirov N, Mu Y, Kawashima T, Bennett DV, Yang CT, Looger LL, Keller PJ, Freeman J, Ahrens MB. Light-sheet functional imaging in fictively behaving zebrafish. Nat Methods 2014; 11:883-4; PMID:25068735; http://dx.doi.org/10.1038/nmeth.3040
- Park HC, Kim CH, Bae YK, Yeo SY, Kim SH, Hong SK, Shin J, Yoo KW, Hibi M, Hirano T, et al. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol 2000; 227:279-93; PMID:11071755; http://dx.doi.org/10.1006/dbio.2000.9898
- Huang HY, Liu JT, Yan HY, Tsai HJ. Arl6ip1 plays a role in proliferation during zebrafish retinogenesis. Cells Tissues Organs 2012; 196:161-74; PMID:22269635; http://dx.doi.org/10.1159/000331589
- Siebel AM, Menezes FP, da Costa Schaefer I, Petersen BD, Bonan CD. Rapamycin suppresses PTZ-induced seizures at different developmental stages of zebrafish. Pharmacol Biochem Behav 2015; 139 Pt B:163-8; PMID:26051026; http://dx.doi.org/10.1016/j.pbb.2015.05.022
- Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 1998; 21:741-51; PMID:9808461; http://dx.doi.org/10.1016/S0896-6273(00)80591-7
- Grooms SY, Noh KM, Regis R, Bassell GJ, Bryan MK, Carroll RC, Zukin RS. Activity bidirectionally regulates AMPA receptor mRNA abundance in dendrites of hippocampal neurons. J Neurosci 2006; 26:8339-51; PMID:16899729; http://dx.doi.org/10.1523/JNEUROSCI.0472-06.2006
- Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron 2001; 32:463-75; PMID:11709157; http://dx.doi.org/10.1016/S0896-6273(01)00493-7
- Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 2001; 32:683-96; PMID:11719208; http://dx.doi.org/10.1016/S0896-6273(01)00508-6
- Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci 1996; 16:7812-20; PMID:8987809
- Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sanchez-Carbente M, Servant F, Bell AW, Boismenu D, et al. Characterization of an RNA granule from developing brain. Mol Cell Proteomics 2006; 5:635-51; PMID:16352523; http://dx.doi.org/10.1074/mcp.M500255-MCP200
- Park HY, Lim H, Yoon YJ, Follenzi A, Nwokafor C, Lopez-Jones M, Meng X, Singer RH. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science 2014; 343:422-4; PMID:24458643; http://dx.doi.org/10.1126/science.1239200
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron 2006; 51:685-90; PMID:16982415; http://dx.doi.org/10.1016/j.neuron.2006.08.021
- Chotiner JK, Khorasani H, Nairn AC, O'Dell TJ, Watson JB. Adenylyl cyclase-dependent form of chemical long-term potentiation triggers translational regulation at the elongation step. Neuroscience 2003; 116:743-52; PMID:12573716; http://dx.doi.org/10.1016/S0306-4522(02)00797-2
- tom Dieck S, Kochen L, Hanus C, Heumuller M, Bartnik I, Nassim-Assir B, Merk K, Mosler T, Garg S, Bunse S, et al. Direct visualization of newly synthesized target proteins in situ. Nat Methods 2015; 12:411-4; PMID:25775042; http://dx.doi.org/10.1038/nmeth.3319
- Gay L, Miller MR, Ventura PB, Devasthali V, Vue Z, Thompson HL, Temple S, Zong H, Cleary MD, Stankunas K, et al. Mouse TU tagging: a chemical/genetic intersectional method for purifying cell type-specific nascent RNA. Genes Dev 2013; 27:98-115; PMID:23307870; http://dx.doi.org/10.1101/gad.205278.112
- Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 2008; 14:1959-72; PMID:18658122; http://dx.doi.org/10.1261/rna.1136108
- Cleary MD, Meiering CD, Jan E, Guymon R, Boothroyd JC. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat Biotechnol 2005; 23:232-7; PMID:15685165; http://dx.doi.org/10.1038/nbt1061
- Banker G, Goslin K. Culturing Nerve Cells. Second revised edition, MIT Press, Cambridge Mass., 1998, 678 pp, ISBN: 9780262024389.
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 2001; 30:489-502; PMID:11395009; http://dx.doi.org/10.1016/S0896-6273(01)00295-1
- Guzman SJ, Schlogl A, Schmidt-Hieber C. Stimfit: quantifying electrophysiological data with Python. Front Neuroinform 2014; 8:16; PMID:24600389; http://dx.doi.org/10.3389/fninf.2014.00016
- Malik AN, Vierbuchen T, Hemberg M, Rubin AA, Ling E, Couch CH, Stroud H, Spiegel I, Farh KK, Harmin DA, et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat Neurosci 2014; 17:1330-9; PMID:25195102; http://dx.doi.org/10.1038/nn.3808
- Westerfield M. The Zebrafish Book. A guide for the Laboratory use of Zebrafish (Danio rerio). University of Oregon Press, Eugene, 2000
- Tom Dieck S, Muller A, Nehring A, Hinz FI, Bartnik I, Schuman EM, Dieterich DC. Metabolic labeling with noncanonical amino acids and visualization by chemoselective fluorescent tagging. Curr Protoc Cell Biol 2012; Chapter 7:Unit7 11; PMID:22968844; http://dx.doi.org/10.1002/0471143030.cb0711s56
