 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
A recent study revealed that poly(A)-binding protein (PABP) bound to poly(A) RNA exhibits a sharply bent configuration at the linker region between RNA-recognition motif 2 (RRM2) and RRM3, whereas free PABP exhibits a highly flexible linear configuration. However, the physiological role of the bent structure of mRNA-bound PABP remains unknown. We investigated a role of the bent structure of PABP by constructing a PABP variant that fails to form the poly(A)-dependent bent structure but maintains its poly(A)-binding activity. We found that the bent structure of PABP/poly(A) complex is required for PABP's efficient interaction with eIF4G and eIF4G/eIF4E complex. Moreover, the mutant PABP had compromised translation activation function and failed to augment the formation of 80S translation initiation complex in an in vitro translation system. These results suggest that the bent conformation of PABP, which is induced by the interaction with 3′ poly(A) tail, mediates poly(A)-dependent translation by facilitating the interaction with eIF4G and the eIF4G/eIF4E complex. The preferential binding of the eIF4G/eIF4E complex to the bent PABP/poly(A) complex seems to be a mechanism discriminating the mRNA-bound PABPs participating in translation from the idling mRNA-unbound PABPs.
Introduction
The start codon of eukaryotic mRNAs is not directly recognized by the translation-competent preinitiation complex (43S), which is composed of a 40S ribosomal subunit, eIF1, eIF1A, eIF3, eIF2–GTP–Met-tRNAMet, and probably eIF5.Citation1 Instead, the 43S complex is transferred to the start codon after binding to the 7-methyl guanosine cap structure (m7G) at the 5′ end in the presence or absence of poly(A) at the 3′ end of an mRNA molecule.Citation2,3 The poly(A)-mediated stimulation of translation occurs through concerted actions of translation factors associated with the 3′ poly(A) tail and the 5′ cap structure. The poly(A)-binding protein (PABP) binds to the 3′ poly(A) tail and then interacts with eIF4G to bring the 3′ end of an mRNA close to its 5′ cap structure. Sequential interactions of poly(A)-bound PABP – eIF4G – eIF4E – 5′ cap circularize the mRNA, which apparently facilitates the translation of 5′-capped and 3′- poly(A)-tailed mRNAs.Citation4 Curiously, the poly(A) tail augments translation even in the absence of the 5′ cap structure and the cap-binding protein eIF4E.Citation3,5-8 Moreover, although PABP has been well documented to play a pivotal role in poly(A)-dependent translation, the mechanistic basis of its involvement in poly(A)-dependent translation has not been fully elucidated.Citation9
PABP contains four RNA-recognition motifs (RRMs) responsible for poly(A) binding, plus a hydrophobic C-terminal domain (PABC) that has no RNA-binding activity. The RRMs and PABC all participate in interactions with various proteins that contribute to translational activation and modulation. A recent single-molecule study showed that PABP undergoes a drastic conformational change upon binding to poly(A) RNA, acquiring a sharp bend in the RRM2-3 linker region, a slight bend in the RRM3-4 linker region, and maintaining a straight conformation of RRM1-2 to yield a highly stable bent structure in which RRM1 is located close to RRM4.Citation10 A structural study also revealed that RRM1-2-3 undergo conformational changes when this fragment of PABP binds to poly(A) RNA.Citation11 These studies further showed that the conformation of poly(A)-bound PABP is allosterically altered by the binding of PABP-interacting protein 2 (Paip2) to RRM2-3 or eIF4G to RRM2.Citation10,11 However, it was not known how the bent conformation of 3′ poly(A)-bound PABP might participate in poly(A)-dependent translation.
To study how the bent conformation of full-length PABP contributes to translation, we constructed a mutant PABP in which linker 2 (that between RRM2 and RRM3) was replaced with linker 1 (that between RRM1 and RRM2), such that the mutant retained a strong poly(A) RNA-binding affinity similar to that of wild-type (WT) PABP, but exhibited a linear alignment of RRMs instead of the bent conformation upon poly(A) binding. Single-molecule Förster Resonance Energy Transfer (smFRET) analysis of WT and mutant PABP confirmed that the conformational change of the linker 2 region is critical to the overall conformational change of PABP upon poly(A) binding. Importantly, the mutant PABP showed decreased translation activation and eIF4G interaction in the presence of poly(A), relative to WT PABP. The results suggest that the specific conformation of PABP on the 3′ poly(A) tail of an mRNA augments poly(A)-dependent translation by facilitating interactions with eIF4G. We also found that poly(A)-bound PABP interacts more strongly with the eIF4G/eIF4E complex than with monomeric eIF4G. These preferential interactions are likely to represent a mechanism through which the cellular translational machinery distinguishes mRNA-bound PABP (i.e., that participating in translation) from idling mRNA-unbound PABP, and discriminates the eIF4G/eIF4E complex, which interacts with the 5′ cap to circularize an mRNA, from monomeric eIF4G, which lacks cap-binding ability.
Results
The conformations and poly(A) RNA-binding affinities of WT and mutant PABPs
RRM1-2 of human PABP is linearly aligned along poly(A), but RRM2-3 bound to poly(A) RNA has a sharply bent conformation.Citation10 To investigate the role of the bent conformation of PABP, we replaced linker 2 (RKEREAELGARAKE; between RRM2 and RRM3) with linker 1 (DPSLRKSGV; between RRM1 and RRM2) to generate a mutant PABP (PABP 2–3 mt) that lacks poly(A)-dependent bending but maintains its poly(A)-binding activity ().
Figure 1. Generation of a PABP mutant that fails to exhibit poly(A)-dependent bending. (A) Schematic diagrams of WT PABP and the generated mutant PABP (PABP 2–3 mt). The linker region between RRM2 and RRM3 (linker 2) was replaced with that between RRM1 and RRM2 (linker 1). (B) Histograms of the FRET values obtained from poly(A)25 containing a Dy547-Dy649 FRET pair, as assessed in the presence of truncated PABPs corresponding to RRM2-3 (left) and RRM2-3 mt (right) of PABP and PABP 2–3 mt, respectively. The FRET value was 0.41 ± 0.09 in the absence of proteins (Unbound); it increased to 0.57 ± 0.15 in the presence of RRM2-3 (left, Bound), but was only 0.20 ± 0.16 in the presence of RRM2-3 mt (right, Bound). (C) Histograms of the FRET values obtained from poly(A)25 in the presence of WT PABP or PABP 2–3 mt. The FRET increased to 0.81 ± 0.13 upon binding of WT PABP (left, Bound), but decreased to 0.00 ± 0.07 upon binding of PABP 2–3 mt (right, Bound).
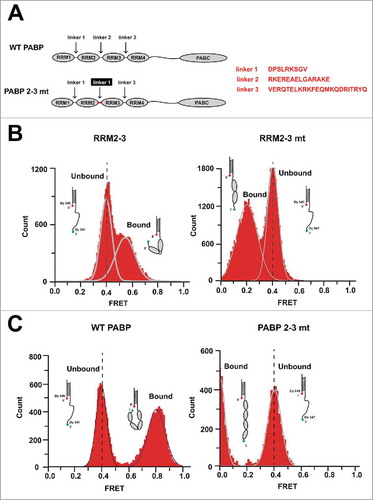
We used smFRET imaging to determine the configuration of the mutant PABP on poly(A) RNA, as previously described.Citation10 Briefly, a partial RNA-DNA duplex with a 25-nucleotide poly(A) RNA tail was immobilized on a surface that had been passivated with PEG/PEG-biotin using a PEG-biotin/streptavidin/biotin-3′ DNA oligo interaction. The poly(A)25 template contains a donor Dy 547 at the 3′ end of the RNA oligo and an acceptor Dy 649 at the 5′ end of the DNA oligo, enabling smFRET. The FRET efficiency of the RNA-DNA hybrid was 0.41 ± 0.09 in the absence of PABP (). The addition of the WT RRM2-3 fragment increased the FRET value of the poly(A) RNA to 0.57 ± 0.15 (N = 108; and Fig. S2A). In contrast, the addition of RRM2-3 mt decreased the FRET value of the RNA-DNA hybrid to 0.21 ± 0.16 (N = 80; and Fig. S2B). We used a similar strategy to confirm the configuration of full-length WT PABP and full-length mutant PABP (PABP 2–3 mt). The FRET efficiency of the RNA-DNA hybrid was 0.81 ± 0.13 (N = 160; and Fig. S2C) in the presence of WT PABP, suggesting that, consistent with the previous report,Citation10 RRM1 is located near RRM4 in poly(A)-bound WT PABP. In contrast, the addition of PABP 2–3 mt sharply decreased the FRET signal to 0.00 ± 0.07 (N = 43; ), indicating that the RRMs of the mutant are aligned linearly along the poly(A) RNA. We measured the fluorescence of a donor-acceptor pair to distinguish the zero FRET state (0.00 ± 0.07) from the fluorescence bleaching of the acceptor (Fig. S2D). Taken together, these results strongly indicate that linker 2 is critical for the ability of PABP to undertake the bent conformation upon binding to poly(A) RNA. Interestingly, the poly(A)25-binding affinities of WT PABP and PABP 2–3 mt were nearly identical, with estimated Kd values of ∼5.9 nM and ∼6.9 nM, respectively (). This Kd value for WT PABP is consistent with previous reports,Citation12-14 and our findings indicate that the conformational change introduced into the mutant PABP did not affect its association with the poly(A) tail.
Figure 2. WT PABP and PABP 2–3 mt have the same affinity for poly(A) RNA. (A and B) Electrophoretic mobility shift assays were performed to measure the affinities of WT PABP and PABP 2–3 mt for poly(A)25. A constant amount (20 fmol) of [32P]-oligo(A)25 RNA was incubated with 0, 0.04, 0.2, 1, 2, 10, 20, 40, 100, 500, 1000, 3000, 5000, or 7000 fmol of WT PABP or PABP 2–3 mt. PABP-bound poly(A)25 [PABP-poly(A) complex] and free poly(A)25 RNA were resolved by native gel electrophoresis, and the band intensities were quantified. The apparent Kd values for WT PABP and PABP 2–3 mt were estimated to be ∼5.9 nM and ∼6.9 nM, respectively.
![Figure 2. WT PABP and PABP 2–3 mt have the same affinity for poly(A) RNA. (A and B) Electrophoretic mobility shift assays were performed to measure the affinities of WT PABP and PABP 2–3 mt for poly(A)25. A constant amount (20 fmol) of [32P]-oligo(A)25 RNA was incubated with 0, 0.04, 0.2, 1, 2, 10, 20, 40, 100, 500, 1000, 3000, 5000, or 7000 fmol of WT PABP or PABP 2–3 mt. PABP-bound poly(A)25 [PABP-poly(A) complex] and free poly(A)25 RNA were resolved by native gel electrophoresis, and the band intensities were quantified. The apparent Kd values for WT PABP and PABP 2–3 mt were estimated to be ∼5.9 nM and ∼6.9 nM, respectively.](/cms/asset/29732b03-cd31-43ec-8fc8-6e8ea7c49ab5/krnb_a_1280224_f0002_b.jpg)
The bent conformation of poly(A)-bound PABP enhances translation
To test whether and how the bent conformation of poly(A) RNA-bound PABP affects translation, we performed in vitro translation reactions using PABP-depleted HeLa lysates supplemented with purified full-length WT PABP or PABP 2–3 mt (). Endogenous PABP was depleted from translation-competent HeLa lysates using GST-Paip2-conjugated glutathione Sepharose beads, as previously described.Citation15 GST-conjugated beads incubated with HeLa lysates served as a control lysate.
Figure 3. Translation-enhancing activities of WT PABP and PABP 2–3 mt. (A) Depletion of PABP from HeLa lysates was confirmed by Western blotting. Western blot analyses were performed with antibodies against eIF4GI (upper panel), PABP (middle panel) and GAPDH (lower panel) (B) In vitro translation reactions were performed with a reporter mRNA, which contains the 5′ cap, firefly luciferase gene, and poly(A)120 tail, in HeLa lysates that were either PABP-depleted (lanes 4–6) or mock-depleted (lanes 1–3). The HeLa lysates were supplemented with control buffer (lanes 1 and 4), WT PABP (10 g/ml, lanes 2 and 5), or PABP 2–3 mt (10
g/ml, lanes 3 and 6). Translation efficiencies observed at specific reaction conditions were normalized to those in PABP-depleted lysate (lane 4) which were set to 1. The individual data points of three independent experiments are depicted. The mean values and standard deviations are shown by thick and thin bars, respectively. Statistical significance is calculated by unpaired t-test: *P < 0.05.
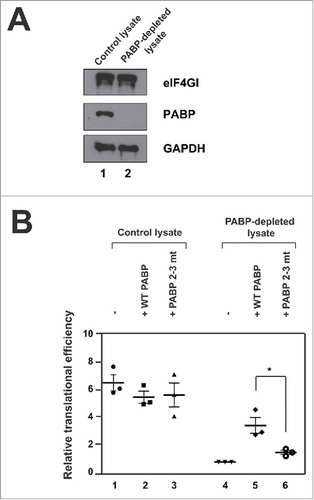
PABP was almost completely removed from HeLa lysates treated with GST-Paip2-conjugated glutathione Sepharose beads and other initiation factor, eIF4G, was not affected by the depletion of PABP (). In vitro translation reactions were performed with a 5′-capped and 3′-poly(A)120-tailed RNA. Our results revealed that translation was decreased by 6-fold upon PABP depletion (). The addition of WT PABP (10 μg/ml: the amount of endogenous PABP in the original lysate) to the PABP-depleted lysate enhanced translation by 3.5-fold (), whereas the addition of PABP 2–3 mt (10 μg/ml) augmented translation to only by 1.6-fold (). These results clearly show that the bent conformation of 3′ poly(A)-bound PABP is required for this protein to fully function as a translational activator.
The bent conformation of poly(A)-bound PABP facilitates the formation of 80S ribosomal complex
To study whether the conformation of PABP affects the initiation step of translation, we used a sucrose density gradient analysis to monitor 80S ribosomal complex formation (). Briefly, a radiolabeled 5′-capped and 3′-poly(A)120-tailed RNA was incubated in control or PABP-depleted lysates in the presence of cycloheximide (20 mM) which blocks the elongation reaction of the 80S ribosomal complex. The extent of 80S ribosomal complex formation was significantly decreased in PABP-depleted lysate (open squares in ). When WT PABP was supplemented with PABP-depleted lysate (WT PABP), the level of 80S complex formation was dramatically increased (open triangles in ), but not when PABP 2–3 mt was added (symbol x in ). These data strongly suggest that the bent configuration of PABP augments translation initiation by facilitating the formation of 80S ribosomal complex.
Figure 4. The bent structure of poly(A)-bound PABP facilitates the formation of 80S ribosomal complex. Sucrose density gradient analyses were performed on mock-depleted HeLa lysates (Control lysate), PABP-depleted lysates supplemented with translation buffer (buffer), WT PABP (WT PABP), or PABP 2–3 mutant (PABP 2–3 mt). In vitro translation reactions were executed as described in the legend to except that cycloheximide (20 mM final) was added in the reaction mixtures. The amounts of 80S complex on [32P]-labeled reporter RNAs were monitored by sucrose density gradient analyses after incubation of HeLa lysates at 30for 15min.
![Figure 4. The bent structure of poly(A)-bound PABP facilitates the formation of 80S ribosomal complex. Sucrose density gradient analyses were performed on mock-depleted HeLa lysates (Control lysate), PABP-depleted lysates supplemented with translation buffer (buffer), WT PABP (WT PABP), or PABP 2–3 mutant (PABP 2–3 mt). In vitro translation reactions were executed as described in the legend to Fig. 3 except that cycloheximide (20 mM final) was added in the reaction mixtures. The amounts of 80S complex on [32P]-labeled reporter RNAs were monitored by sucrose density gradient analyses after incubation of HeLa lysates at 30°C for 15min.](/cms/asset/2a16d48c-8317-4408-8290-2bc021614882/krnb_a_1280224_f0004_b.jpg)
Interaction between eIF4G and the poly(A)25-PABP complex
To assess whether and how the bent conformation of 3′ poly(A)-bound PABP affects translation, we investigated the binding affinities of poly(A)-bound WT PABP or PABP 2–3 mt with eIF4G, which is known to interact with RRM2 of PABP.Citation11 Because it is technically difficult to purify full-length mammalian eIF4G, we generated a truncated human eIF4GI containing the PABP- and eIF4E-binding sites (designated as eIF4G-N and corresponding to amino acids 42–653 of eIF4GI; Fig. S3A). We confirmed that purified eIF4G-N could interact with both eIF4E and PABP (Fig. S3B and S3C). When equal amounts of truncated PABPs (RRM2-3 or RRM2-3 mt) were precipitated with poly(A) RNA (approximately 100-nt long) conjugated to Sepharose beads (lower panel), the interaction between RRM2-3 mt and eIF4G-N was ∼70% lower than that between RRM2-3 and eIF4G-N (upper panel in ). This indicates that the bent conformation of the RRM2-3 region plays an important role in eIF4G binding, possibly by exposing the eIF4G-binding site in RRM2 upon the binding of PABP to poly(A) RNA (see below).
Figure 5. Interactions of eIF4G-N with WT PABP or PABP 2–3 mt. (A) The interaction between purified eIF4G-N and poly(A)-bound RRM2-3 (lanes 5 and 7) or RRM2-3 mt (lanes 6 and 8). Poly(A) pull-down experiments were performed by incubating purified eIF4G-N with poly(A) beads pre-incubated with RRM2-3 (lane 7) or RRM2-3 mt (lane 8). Western blotting was performed with anti-eIF4GI (upper panel) and anti-His (to detect PABP; lower panel). (B) The interactions of poly(A)-bound WT PABP (lanes 5 and 7) or PABP 2–3 mt (lanes 6 and 8) with purified eIF4G-N and eIF4E. Poly(A) pull-down experiments were performed with eIF4E, eIF4G-N, and WT PABP or PABP 2–3 mt. Western blotting was performed with anti-eIF4E (upper panel), anti-eIF4GI (middle panel), or anti-Flag (to detect PABP; lower panel). (C) The interaction of eIF4G-N with WT PABP or PABP 2–3 mt in the presence or absence of poly(A)25 RNA. Recombinant WT PABP or PABP 2–3 mt proteins were incubated with eIF4G/eIF4E complexes, and Flag immunoprecipitations were used to pull down eIF4G in the presence (lanes 7 and 8) or absence (lanes 5 and 6) of poly(A)25 RNA. Lanes 1–3 depict input controls loaded with 20% of the proteins used in the reactions. Western blot analyses were performed with anti-eIF4E (upper panel), anti-eIF4GI (middle panel) and anti-Flag (to detect PABP; lower panel).
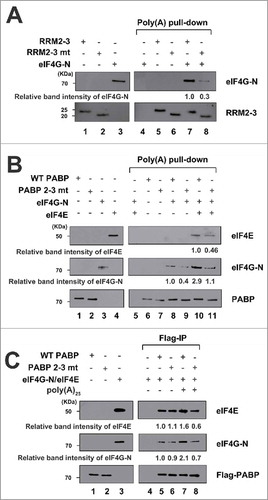
Similarly, the interaction between poly(A)-bound WT PABP (full length) and eIF4G-N was impaired by ∼60% in the mutant (middle panel in ). The addition of eIF4E increased the binding affinity of eIF4G-N to WT PABP by about 3-fold (compare lane 10 with 8 in ). However, the interaction between poly(A)-bound WT PABP (full length) and eIF4G-N was impaired by ∼60% in the mutant even in the presence of eIF4E (middle panel in ), similarly to the results obtained in the absence of eIF4E (middle panel in ). Co-precipitation of eIF4E, which is precipitated in association with eIF4G-N, was also 55% lower for the mutant relative to WT PABP (upper panel in ).
Flag-immunoprecipitation was used to assess the binding affinity of eIF4G-N/eIF4E complexes to Flag-tagged PABPs (WT PABP or PABP 2–3 mt) in the presence or absence of poly(A)25 RNA (). In the absence of poly(A) RNA, the binding affinities of WT PABP and PABP 2–3 mt to the eIF4G/eIF4E were nearly identical (upper and middle panels in ). Surprisingly, however, the presence of poly(A) RNA dramatically increased the interaction between eIF4E-bound eIF4G-N and WT PABP, but decreased that between eIF4E-bound eIF4G-N and PABP 2–3 mt (upper and middle panels in ). The ability of poly(A) RNA to enhance the interaction between PABP and eIF4G is consistent with a previous report,Citation11 but this is the first study to show that this effect was abolished when the bending of PABP was genetically abrogated. Taken together, our results suggest that the bent conformation of PABP, which is induced by poly(A) binding, strongly augments the interaction of PABP with eIF4G/eIF4E.
Discussion
Our previous single-molecule study suggested that the poly(A)-binding-induced bent conformation of PABP might regulate translation through an interaction between RRM2-3 and one or more translation initiation factors.Citation10 The linker 2 between RRM2 and RRM3 is well conserved among human cytoplasmic PABP isoforms (PABPC1, PABPC3 and PABPC4). Moreover, the linker 2 of PABPC1 is well conserved in human, frog, fish and fruit fly, but less well conserved in yeast.
To investigate whether the bent configuration of poly(A)-bound PABP is required for the ability of this protein to enhance translation, we replaced linker 2 with linker 1 to generate a mutant PABP (PABP 2–3 mt). PABP 2–3 mt showed the same binding affinity to poly(A) RNA as WT PABP, but exhibited a linear alignment of RRM2 and RRM3 upon poly(A) binding ( and ). The structure of RRM1-2 with linkers 1 and 2 was previously reported by Safaee et al., (2012) using X-ray crystallography. The report showed that both linker 1 and linker 2 regions form α-helices (RRM1-α3 and RRM2-α3, respectively) with or without poly(A) binding. These α-helices and the β sheets of each RRM domain form clamp-like structures to hold the RNA. The authors suggested that Arg94 in the first helix (RRM1-α3) and Arg179 in the second helix (RRM2-α3) contribute to poly(A) binding (Safaee et al., 2012). Arg94 contributes to poly(A) binding by forming hydrogen bonds with 2′ OH of ribose moieties. On the other hand, Arg179 contributes to poly(A) binding by forming hydrogen bonds with the backbone phosphate oxygen. The report indicates that the huge configuration difference (linear and sharply bent configurations) of linkers 1 and 2 bound to poly(A) is attributed not only to the length difference of linkers but also to the different modes of interaction between amino acids and nucleotides.
The addition of PABP to PABP-depleted HeLa lysate restored translation of a reporter mRNA but that of PABP 2–3 mt only partially augmented translation, and the sucrose density gradient analysis indicated that the initiation step of translation was hampered by the mutation in PABP ( and ). Regarding the molecular basis of the activity differences between the WT and mutant PABPs, we speculated that the bent conformation of RRM2-3 may play an important role in the interaction with eIF4G. Previous studies showed that assembly of the PABP-eIF4G complex with a poly(A) tail enhances the recruitment of ribosomes to mRNAs in yeasts, mammals, and plants.Citation9,16,17 The N terminus of eIF4G interacts with the RRM2 domain of PABP, triggering conformational changes in both PABP and eIF4G, and the binding of poly(A) RNA increases the affinity of eIF4G to PABP.Citation11 As expected, we observed that poly(A) RNA augmented the binding of PABP to the eIF4G/eIF4E complex (compare lane 5 with 7 in ). It should be noted that poly(A) RNA-bound WT PABP had a 2- to 3-fold higher binding affinity to eIF4G (compare lane 8 with 9 in ) and the eIF4G/eIF4E complex (compare lane 10 with 11 in ) compared to poly(A) RNA-bound PABP 2–3 mt. This suggests that the bent structure of PABP, which is induced by the binding of poly(A), augments the interaction between PABP and eIF4G. Notably, the amino acid residues of PABP RRM2 that are responsible for interacting with eIF4G are located at a distance from linker 2 and the RNA-binding regions of PABP.Citation11 Therefore, we speculate that the poly(A)-binding-induced bending of PABP exposes the eIF4G-binding site in RRM2, which in turn allows eIF4G to physically interact with PABP. Consistent with this, the association of poly(A) RNA to the PABP 2–3 mutant, which straightens domains 2 and 3 (), reduced the binding affinity of eIF4G/eIF4E to PABP (compare lanes 6 and 8 in ).
The induced exposure of the eIF4G-binding site on PABP by an allosteric interaction of poly(A) RNA enables the translational machinery to ensure that eIF4G and the eIF4G/eIF4E complex preferentially interact with mRNA-bound PABPs rather than mRNA-unbound PABPs. Given that the level of PABP is about ∼6-fold higher than that of eIF4G in cells,Citation12,18 an interaction between eIF4G and mRNA-unbound PABP would potentially sequester eIF4G and prevent it from executing its functions in translation. We also found that eIF4E augments the binding of eIF4G to poly(A)-bound PABP (). Mechanistically, this ensures that an eIF4F complex (composed of eIF4G, eIF4E, and eIF4A), which facilitates cap-dependent translation through an interaction between eIF4E and the 5′ cap, is recruited to the mRNA-bound PABP rather than monomeric eIF4G that cannot bind to the 5′ cap. This would be a critical preference, since cells contain ∼2.5-fold more eIF4G than eIF4E.Citation18 In this study, we provided evidences that a conformational change of PABP induced by poly(A) tail-binding plays an important role in the communication between a PABP on the 3′ tail of an mRNA and an eIF4F on the 5′ cap structure of the mRNA which is required for enhancing translation initiation. We also propose the molecular mechanism underlying the balanced interactions among translation factors eIF4E, eIF4G, and PABP which exist seemingly unbalanced amounts in cells.
Materials and methods
Plasmids
A plasmid expressing a mutant PABP that failed to undergo bending upon poly(A) binding (pQE31-PABP 2–3 mt-Flag) was generated by site-directed mutagenesis: linker 2 was replaced with linker 1 using specific primers: (forward, 5′-TCCTAAATGATCGCAAAGTATTTGTTGGACGATTTAAGTCTGATCCATCACTTCGCAAAAGTGGAGTATTCACCAATGTTTACATCAAGAATTTTGGAGAAGACATGGA-3TT and reverse, 5′-TCCATGTCTTCTCCAAAATTCTTGATGTAAACATTGGTGAATACTCCACTTTTGCGAAGTGATGGATCAGACTTAAATCGTCCAACAAATACTTTGCGATCATTTAGGA-3′). The construction of pQE31-PABP-Flag was described in a previous report.Citation10 pET28a-RRM2-3 was constructed by PCR amplification of full-length human PABP using specific primers (forward, 5′ -GCTAGCGGAGTAGGCAACATATTCATTAAAAAT-3′; and reverse, 5′ -GGATCCTTATTTCTGAGCTCGACCAACATAAAT-3′). pET28a-RRM2-3 mt was generated by site-directed mutagenesis of the RRM2-3 fragment amplified as described above. The PCR products were treated with NheI-BamHI and ligated into NheI-BamHI-treated pET-28a. pET28a-eIF4GN was generated by treating pcDNA 3.1-eIF4GI with HindIII-Klenow-XhoI and ligating the obtained fragments with NdeI-Klenow-XhoI-treated pET-28a. pGEX-eIF4E was constructed by ligating HindIII-Klenow-NotI-treated eIF4E into EcoRI-Klenow-NotI-treated pGEX-4T-3.
Protein expression and purification
For purification of His-PABP-Flag, His-PABP 2–3 mt-Flag, His-RRM2-3, His-RRM2-3 mt, and His-eIF4G-N, E. coli M15 cells were transformed with pQE-PABP-Flag or pQE-PABP 2–3 mt-Flag, and E. coli Bl21 cells were transformed with pET-RRM2-3, pET-RRM2-3 mt, or pET-eIF4G-N. Proteins were induced with 1 mM isopropyl--D-thiogalactopyranoside (IPTG) at 16
for 16 h. The cells were harvested and lysed in lysis buffer [50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 1% Triton X-100, 5 mM
-mercaptoethanol, 20 mM imidazole, 10% glycerol], and the His-tagged recombinant proteins were purified with Ni-NTA beads (Qiagen). For purification of GST-eIF4E, E. coli Bl21 cells were transformed with pGEX-eIF4E and lysed with lysis buffer [50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 1% Triton X-100, 5 mM
-mercaptoethanol, 1 mM reduced glutathione, 10% glycerol], and proteins were purified using glutathione-Sepharose 4B beads (GE Healthcare). The purity, concentration and stability (melting temperature: Tm) of purified WT PABP and PABP 2–3 mt were similar as shown in Fig. S1.
Single-molecule FRET and data analysis
smFRET experiments were performed as previously described.Citation10 Equal amounts (10 nM) of RRM2-3, RRM2-3 mt, WT PABP, or PABP 2–3 mt were infused into the flow chamber, which was coated with Dy 547-labeled poly(A)25 RNA. smFRET signals were collected, and data analysis was performed as previously described.Citation10
Electrophoretic mobility shift assay (EMSA)
Poly(A)25 RNA was subjected to 5′ 32P labeling and incubated with increasing amounts of WT PABP or PABP 2–3 mt. PABP-bound poly(A)25 [PABP-poly(A) complex] and free poly(A)25 RNA were separated by native gel electrophoresis, and quantification was performed.
Preparation of PABP-depleted lysates and in vitro translation
Translation-competent HeLa cell lysates were prepared as described previously.Citation19 Lysates were incubated with GST-Paip2-conjugated GSH beads to deplete the endogenous PABP.Citation15 In vitro-transcribed RNAs (final concentration, 5 nM) were translated in 12.5 μl of PABP-depleted or control lysates 35% (v/v) in the presence of WT PABP or PABP 2–3 mt (10g/ml). In vitro translation reactions and luciferase activity measurements were performed as previously described.Citation20
Sucrose density gradient analyses
Sucrose density gradient analyses of HeLa cell lysates were performed as described previouslyCitation19 with minor modifications. In vitro translation reaction mixtures (50 l) composed of HeLa cell lysates were incubated with 20 mM cycloheximide (CHX) at 30
for 15min. In the presence of cycloheximide, the 80S ribosomal complexes are stalled on [32P]-labeled mRNAs containing poly(A)120. The reaction samples were loaded onto 5–20% sucrose gradients with sucrose buffer [50 mM HEPES (pH 7.4), 100 mM KOAc, 100 mM NH4Cl, 10 mM MgCl2, 2 mM DTT and 0.1 mM EDTA] and centrifuged at 30000rpm in a SW41Ti rotor (Beckman) at 4
for 3h. Fractions were collected using a Brandel gradient density fractionator and an Econo UV monitor (Bio-Rad), and the radioactivity on each fraction was measured by using a scintillation counter.
Poly(A) pull-down assay
Freeze-dried Poly(A)-Sepharose 4B beads (GE Healthcare) were swollen for 15 min in 0.1 M NaCl (pH 7.5) and washed with binding buffer [40 mM HEPES-KOH (pH 7.5), 100 mM KCl, 1 mM EDTA, 10 mM -glycerophosphate, 10 mM NaF, 2 mM Na3VO4, 0.1% NP-40, 1 mM PMSF]. The beads were equilibrated with five bed volumes of starting buffer and incubated with WT or mutant PABPs and eIF4G-N at 4
for 1.5 h. The beads were washed three times, and the proteins were eluted, resolved by SDS-PAGE, and detected by Western blot analysis.
Immunoprecipitation
Flag-antibody conjugated beads (anti-Flag M2 affinity gel; Sigma) were washed in binding buffer [40 mM HEPES-KOH (pH 7.5), 100 mM KCl, 1 mM EDTA, 10 mM -glycerophosphate, 10 mM NaF, 2 mM Na3VO4, 0.1% NP-40, 1 mM PMSF], and equal amounts of beads (10
) were incubated with His-WT PABP-Flag or His-PABP 2–3 mt-Flag in the presence or absence of 20 nM poly(A)25 RNA in 700
l of binding buffer at 4
for 30 min. His-eIF4G-N/GST-eIF4E complexes were added, and the mixtures were incubated at 4
for 30 min. The beads were washed three times with binding buffer, and the proteins were eluted, resolved by SDS-PAGE, and detected by Western blot analysis.
Thermal shift assay
WT PABP or PABP 2–3 mt (1 μ each) were incubated in 50
l of reaction buffer [10 mM Sodium Phosphate (pH 7.0), 100 mM NaCl and 5 x SYPRO orange dye (Sigma-Aldrich)]. Samples were incubated at 25
for 2 min, and the temperature was gradually increased from 25
to 95
at the rate of 2
/min by using a real-time PCR machine (Bio-Rad iQ5). Emitted fluorescence intensity was measured by a real-time PCR machine (Bio-Rad iQ5).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Figures_1-3.docx
Download MS Word (860.9 KB)Funding
This study was supported by a grant from the Bio R&D Program (No. 2012M3A9A9054974) through NRF grant funded by the Korean government (MSIP).
References
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010; 11(2):113-27; PMID:20094052; http://dx.doi.org/10.1038/nrm2838
- Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol 1990; 10(7):3441-55; PMID:1972543; http://dx.doi.org/10.1128/MCB.10.7.3441
- Tarun SZ, Jr, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci U S A 1997; 94(17):9046-51; PMID:9256432; http://dx.doi.org/10.1073/pnas.94.17.9046
- Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 1998; 2(1):135-40; PMID:9702200; http://dx.doi.org/10.1016/S1097-2765(00)80122-7
- Borman AM, Michel YM, Kean KM. Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5′-end. Nucleic Acids Res 2000; 28(21):4068-75; PMID:11058101; http://dx.doi.org/10.1093/nar/28.21.4068
- Preiss T, Hentze MW. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature 1998; 392(6675):516-20; PMID:9548259; http://dx.doi.org/10.1038/33192
- Yanagiya A, Suyama E, Adachi H, Svitkin YV, Aza-Blanc P, Imataka H, Mikami S, Martineau Y, Ronai ZA, Sonenberg N. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol Cell 2012; 46(6):847-58; PMID:22578813; http://dx.doi.org/10.1016/j.molcel.2012.04.004
- Preiss T, Muckenthaler M, Hentze MW. Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA 1998; 4(11):1321-31; PMID:9814754; http://dx.doi.org/10.1017/S1355838298980669
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 2005; 19(1):104-13; PMID:15630022; http://dx.doi.org/10.1101/gad.1262905
- Lee SH, Oh J, Park J, Paek KY, Rho S, Jang SK, Lee JB. Poly(A) RNA and Paip2 act as allosteric regulators of poly(A)-binding protein. Nucleic Acids Res 2014; 42(4):2697-707; PMID:24293655; http://dx.doi.org/10.1093/nar/gkt1170
- Safaee N, Kozlov G, Noronha AM, Xie J, Wilds CJ, Gehring K. Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol Cell 2012; 48(3):375-86; PMID:23041282; http://dx.doi.org/10.1016/j.molcel.2012.09.001
- Gorlach M, Burd CG, Dreyfuss G. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp Cell Res 1994; 211(2):400-7; PMID:7908267; http://dx.doi.org/10.1006/excr.1994.1104
- Kuhn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol 1996; 256(1):20-30; PMID:8609610; http://dx.doi.org/10.1006/jmbi.1996.0065
- Deardorff JA, Sachs AB. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J Mol Biol 1997; 269(1):67-81; PMID:9193001; http://dx.doi.org/10.1006/jmbi.1997.1013
- Svitkin YV, Yanagiya A, Karetnikov AE, Alain T, Fabian MR, Khoutorsky A, Perreault S, Topisirovic I, Sonenberg N. Control of translation and miRNA-dependent repression by a novel poly(A) binding protein, hnRNP-Q. PLoS Biol 2013; 11(5):e1001564; PMID:23700384; http://dx.doi.org/10.1371/journal.pbio.1001564
- Simon AE, Miller WA. 3′ cap-independent translation enhancers of plant viruses. Annual Rev Microbiol 2013; 67:21-42; PMID:23682606; http://dx.doi.org/10.1146/annurev-micro-092412-155609
- Tarun SZ, Jr, Sachs AB. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol Cell Biol 1997; 17(12):6876-86; PMID:9372919; http://dx.doi.org/10.1128/MCB.17.12.6876
- Duncan R, Milburn SC, Hershey JW. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem 1987; 262(1):380-8; PMID:3793730
- Park SM, Paek KY, Hong KY, Jang CJ, Cho S, Park JH, Kim JH, Jan E, Jang SK. Translation-competent 48S complex formation on HCV IRES requires the RNA-binding protein NSAP1. Nucleic Acids Res 2011; 39(17):7791-802; PMID:21715376; http://dx.doi.org/10.1093/nar/gkr509
- Paek KY, Hong KY, Ryu I, Park SM, Keum SJ, Kwon OS, Jang SK. Translation initiation mediated by RNA looping. Proc Natl Acad Sci U S A 2015; 112(4):1041-6; PMID:25583496; http://dx.doi.org/10.1073/pnas.1416883112
