ABSTRACT
Base 37 in tRNA, 3′-adjacent to the anticodon, is occupied by a purine base that is thought to stabilize codon recognition by stacking interactions on the first Watson-Crick base pair. If the first codon position forms an A.U or U.A base pair, the purine is likely further modified in all domains of life. One of the first base modifications found in tRNA is N6-isopentenyl adenosine (i6A) present in a fraction of tRNAs in bacteria and eukaryotes, which can be further modified to 2-methyl-thio-N6-isopentenyladenosine (ms2i6A) in a subset of tRNAs. Homologous tRNA isopentenyl transferase enzymes have been identified in bacteria (MiaA), yeast (Mod5, Tit1), roundworm (GRO-1), and mammals (TRIT1). In eukaryotes, isopentenylation of cytoplasmic and mitochondrial tRNAs is mediated by products of the same gene. Accordingly, a patient with homozygous mutations in TRIT1 has mitochondrial disease. The role of i6A in a subset of tRNAs in gene expression has been linked with translational fidelity, speed of translation, skewed gene expression, and non-sense suppression. This review will not cover the action of i6A as a cytokinin in plants or the potential function of Mod5 as a prion in yeast.
Modification of purines at position 37 in tRNA
Among the over 100 base modifications occurring in tRNA, many target the anticodon stem loop, in particular base 34 reading the wobble base and the dangling base 37 3′-adjacent to the anticodon.Citation1,2 Base modifications present at base 37 include mostly bulky additions to adenosine, i.e. N6-isopentenyladenosine (i6A), 2-methyl-thio i6A (ms2i6A), 6-hydroxy ms2i6A (ms2io6A), N6-threonylcarbamoyladenosine (t6A), ms2t6A, and modification of guanosine, to N1-methylguanosine (m1G) or wybutosine (yW) (). The i6A modified base was one of the first hypermodified bases identified and has been found in bacteria and eukaryotes, but not in archeae.Citation3,4 Codon-anticodon interactions are considered weaker in the case of A.U base pairs than in case of G.C base pairs.Citation5 It has been argued that A.U base pairs on the first codon position (base 36 of tRNA) need a modified base 37 which would stabilize the Watson-Crick base pair by base stacking. In addition, the modification prevents illicit hydrogen bonding between U33 and A37 that would otherwise disturb the anticodon loop.Citation6,7 Codons starting with A are coding for Thr (ACN), Ser (AGY), Arg (AGR), Lys (AAR), Asn (AAY), Met (AUG), and Ile (AUY and AUA). Invariably, as for example in tRNALys, U36 is followed by t6A37.Citation2,8,9 Codons starting with U are coding for Ser (UCN), Cys (UGY), Trp (UGG), Phe (UUY), Leu (UUR), and Tyr (UAY). Accordingly, these codons are read by tRNAs having an A at position 36 that is usually followed by a modified base at position 37.Citation2 Modification of G37 to yW in tRNAPhe (UUU) is a classical example and remarkable, because yW37 is exclusively found in tRNAPhe.Citation10 Most other tRNAs reading codons starting with U and bearing a sequence of A36-A37-A38 are isopentenylated, except e.g. mt-tRNALeu (UUR).Citation8 While the AAA motif thus appears necessary, it is not sufficient to signal isopentenylation of A36 in tRNA.Citation11,12 Predictions on the presence and identity of A37 modifications from sequence alone are thus not possible at the moment. Therefore, a systematic study on bovine mitochondrial tRNA modification has been conducted using mass spectrometry that has identified all base 37 modifications.Citation8 Unfortunately, data on mammalian cytoplasmic tRNAs is still incomplete with respect to analyzed tRNAs, and has not always conclusively demonstrated the identity of the modified base.Citation11 The matter is further complicated by potential species differences. We have tried to summarize available data on tRNAs carrying i6A and ms2i6A in E. coli (), the yeasts S. cerevisiae, S. pombe (), and mammals (). We included tRNAs that are isopentenylated in one of the species, but not in others, to highlight species differences. Data from tRNAdb was integrated.Citation13 In tRNA, all available data suggests that i6A and its derivatives are exclusively found at position 37.
Figure 1. Canonical tRNA structure with base 37 modifications. Canonical tRNA structure and 5 individual anticodon loops of tRNAs that carry different base 37 modifications. Where appropriate, base 32 and 34 modifications are indicated as well. Structural formulae indicate the modified nucleosides at position 37. Respective drawings of base modifications have been copied from www.modomics.genesilico.pl with permission.Citation14,15 Cm 2′O-methylcytosine; m1G 1-Methylguanosine; m3C 3-Methylcytosine, τm5U 5-taurinomethyluridine, Gm 2′O-methyl Guanosine, mcm5U 5-Methoxycarbonylmethyluridine, mcm5s2U 5-Methoxycarbonylmethyl-2-thiouridine, ms2i6A 2-Methylthio-N6-isopentenyladenosine, ms2t6A 2-Methylthio-N6-threonylcarbamoyladenosine, yW wybutosine.
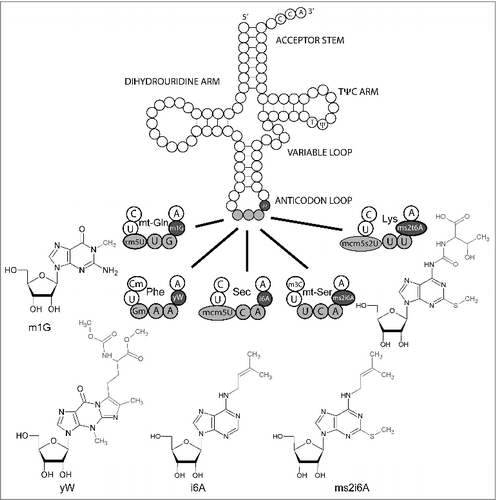
Table 1. E. coli tRNAs carrying i6A37 or ms2i6A37.
Table 2. Yeast tRNAs carrying i6A37 and tRNAs that carry i6A in other species.
Table 3. Mammalian tRNAs carrying i6A37 or ms2i6A37 and tRNAs that carry i6A in other species.
Isopentenyladenosine was among the first modified bases identified in tRNA, and was localized next to the anticodon in 2 Ser isoacceptors from yeast that were among the first tRNA sequenced.Citation16,17 The biochemical pathway leading to i6A or ms2i6A formation in E. coli was initially discovered with the trpX−/miaA mutant lacking ms2i6A in tRNATrp18-22 Subsequently, miaB was identified as the gene required for formation of ms2i6A, and miaE was found to catalyze ms2io6A in S. typhimurium.Citation3 Isopentenyladenosine is thus further modified to ms2i6A by MiaB and can be hydroxylated by MiaE to ms2io6A in Salmonella. MiaB is the founding member of a family of radical SAM enzymes involved in tRNA modification.Citation23 When tRNA from mod5–1 mutants in S. cerevisiae was found deficient in i6A,Citation24 the first eukaryotic isopentenyl transferase gene was identified. Cloning revealed that miaA and mod5 are homologous genes,Citation21,25,26 and homology cloning led to the identification of TRIT1 in humans,Citation27 suggesting that all tRNA isopentenyl transferases are homologous genes in all domains of life. The biochemistry of isopentenyl transferases will be shortly discussed toward the end of the article. In mammals, thiomethylation reactions are performed by the radical SAM enzymes CDK5RAP1 and CDKAL1 acting on i6A37 and t6A37, respectively.Citation28,29 While i6A and ms2i6A have been reported in miRNA and poly A+ RNA from HeLa cells,Citation28 their potential role in these RNAs and in other species is not known.
Delineating the biologic functions of i6A and ms2i6A
E.coli (miaA) and S. typhymurium (miaA1)
Escherichia coli tRNAs decoding codons starting with U contain i6A or ms2i6A at position 37, i.e., Phe UUY, Leu UUR, Cys UGY, Trp UGG, Tyr UAY, and Ser UCNCitation30 (). The product of the selC gene in E. coli is a bacterial tRNA[Ser]Sec that reads UGA codons as selenocysteine (Sec). This tRNA carries i6A37 like its mammalian counterpartCitation31 (see below). In ribosome binding assays it was shown that tRNAs lacking their natural ms2i6A or i6A modification had a reduced affinity to their cognate codons.Citation32 Similarly, the function of non-isopentenylated tRNATrp is apparently impaired, as a mutant that was defective in attenuation of the trp operon, trpX− lacks isopentenylation of tRNATrp.Citation22 The trp operon is not solely transcriptionally regulated through a repressor/operator system, but also by a process called attenuation. In this regulatory process, termination of trp operon transcription is regulated by translation of a short translational reading frame that contains 2 consecutive UGG Trp codons. If Trp-tRNATrp is plentiful in the cell, a ribosome swiftly passes through the open reading frame and disrupts by its presence a potential stem loop in the newly synthesized mRNA. Formation of an alternative stem loop induces termination of transcription. If Trp is scarce, translation through this open reading frame is slow, the terminator stem loop does not form, but another, and transcription of the trp operon proceeds. As trpX−/miaA− cells lack isopentenylation of tRNATrp (and others) and are deficient in attenuation, the hypomodified tRNATrp is apparently less efficient in translation than the fully modified species. In fact, it was this publication in which the gene name miaA was coined as the first gene related to deficiency in ms2i6A.Citation22 Iron restriction likewise impaired attenuation of the phe and trp operons in E. coli,Citation33 probably because the MiaB enzyme contains a [4Fe-4S] cluster needed for methylthiolation of i6A-modified A37.Citation3,23 The ms2i6A tRNA modification appears to be harnessed as a regulatory mechanism to induce genes involved in iron accumulation in bacteria. To slow bacterial growth, bacteria-infected hosts will reduce iron available in body fluids by increasing iron-chelating protein. Pathogenic bacteria in turn will make the Fe3+-chelating compound enterochelin (enterobactin) and express a membrane receptor for the Fe3+-enterochelin complex to maintain their iron supply and growth.Citation34-36 Enterochelin biosynthesis is constitutively enhanced in miaA S. typhimurium. Cysteine stress and anaerobic growth likewise enhanced enterochelin along with decreased formation of ms2io6A.Citation37 Whether only the modification of tRNAs for aromatic amino acids or also tRNASer play a role in this process remained unclear.Citation37
Lack of tRNA isopentenylation reduced growth rates in E. coli only marginally when grown in full media.Citation22,38 Salmonella typhimurium miaA mutants show slower growth rates and cellular protein production on several substrates.Citation39 The metabolic effects of impaired tRNA isopentenylation have been reviewed in great detail.Citation1,3 While modification of A37 was not demonstrated to influence tRNA charging reactions, it was shown to make decoding of UAG codons by suppressor tRNAs within a lacI-lacZ reporter gene more independent of codon context in E. coli and S. typhimurium.Citation40 The same paper also showed that near-cognate misreading of certain codons is reduced when the base 37 modification is not present.Citation40 Likewise, in an in vitro system of polyU translation with ms2i6A-dependent tRNAPhe, decoding was more accurate in the absence of the modification,Citation41 possibly because near cognate decoding was less favored. The role of base 37 modifications of tRNAPheGAA and tRNATyrQUA was dissected in S. typhimurium miaA, miaB, and miaE mutants. P-site slippage (+1 frame-shifting) was found increased 3–9-fold by tRNAs lacking the ms2-modification, while lack of i6A had no greater effect than lack of the thiomethylation alone.Citation42 In contrast, of tRNAs normally carrying m1G37, only tRNAArgCCG caused a 3-fold increase in P-site slippage. Lack of modification of G37 thus seems less critical than lack of modification of A37 with respect to +1 frame-shifting.Citation42
A miaA mutation in the pathogenic bacterium Shigella flexneri decreased i6A modification of tRNAs, decreased expression of several virulence genes, but did not affect their mRNA levels suggesting that the effect was at the level of translation.Citation43 Complementation with a functional miaA gene demonstrated that the effect on virulence gene expression depended on tRNA isopentenylation.
Skewed gene expression that may be modulated by the use of rare codons or codons depending on specific modifications has been proposed for many years. It lies in the nature of such a mechanism that it is hard to prove and specificity is always an issue, but available data suggest its existence.Citation44,45 A particularly instructive example for both the use of rare codons and the dependence of specific genes on specific tRNA modifications is illustrated by the stationary phase sigma factor rpoS (σS) in E. coli. In contrast to its related gene, the vegetative sigma factor rpoD, rpoS contains rare codons at many positions. Replacing these codons with synonymous frequent codons reduced the levels of RpoS protein by destabilizing its mRNACitation46 and it has been argued that ribosomal pausing might increase ribosomal density on the mRNA and thus protect the message from RNaseE-dependent degradation. On the other hand, one of the many mechanisms of RpoS induction in stationary growth may be increased availability of charged rare tRNAs. Recently, it was discovered that RpoS expression is reduced in miaA mutant cells.Citation47 More recently it was shown that MiaA activity was necessary for proper decoding of UUR-Leu codons in RpoS. Exchange of the UUR (Leu) codons with CUY (Leu) codons in a rpoS-lacZ reporter gene alleviated the requirement for MiaA. Expression of a RpoS-lacZ reporter protein could be increased by overexpression of a miaA-independent tRNALeu.Citation48 Translation of RpoS is thus regulated by the availability of i6A-modified tRNAs reading rare codons.
Mod5 in S. cerevisiae
The mod5–1 mutation reduced the efficiency of a tRNATyr UAA suppressor. The mutant showed decreased levels of i6A, and an altered chromatographic behavior of tRNATyr and tRNASer on benzoylated DEAE-sepharose columns.. tRNA aminoacylation and other tRNA modifications were not altered in the mutant. The lack of i6A had almost no effect on its growth rate on several complex or simple media at either 28°C and 37°C. The only finding was that mod5–1 homozygotes failed to sporulate.Citation24 lists all yeast tRNAs that are isopentenylated. The same gene is required to modify cytosolic and mitochondrial tRNAs.Citation49 Alternative usage of 2 initiation codons, at positions 1 and 12, produces a longer mitochondrial isoform containing an N-terminal mitochondrial import sequence, and a shorter cytosolic isoform.Citation50-52 The cytosolic isoform was also found in nuclei, especially in the nucleolus, and contains a nuclear localization sequence.Citation53,54 Interestingly, ms2i6A is neither found in yeast cytoplasmic nor mitochondrial tRNAs suggesting that the respective MiaB homolog is entirely absent in yeastsCitation55 ().
Tit1 in S. pombe
A Tit1 mutant strain (sin1) exhibited a loss of tRNA suppression of the UGA codon, which was translated as serine by tRNASerUCA.Citation56-59 This tRNA was not isopentenylated. Moreover, other tRNAs were found non-isopentenylated in this strain, such as tRNATyr and tRNATrp.Citation57 As with mod5–1, changes in cell growth of these mutants were hardly apparent. The sequence determinants for tRNA isopentenylation are not yet fully known. The A36A37A38 sequence in the anticodon stem loop is a necessary, but not sufficient, determinant for isopentenylation as shown in E. coli.Citation12 For example, cy-tRNATrpCCA, is isopentenylated in S. pombe, but not in S. cerevisiae. In contrast, differential modification of cy-tRNACysGCA in S. cerevisiae, but not in S. pombe, is fully explained by guanosine at position 37 in the S. pombe tRNACys. Substrates of both enzymes are cy-tRNATyrGΨA, cy-tRNASerIGA, cy-tRNASerCGA, cy-tRNASerUGA60 (). The mitochondrial isoform of Tit1 is unknown. However, Lamichhane and coworkers showed mt-tRNATrp was partially modified by Tit1 but not mt-tRNACys or mt-tRNASerCitation61 ().
Rapamycin is an inhibitor of the mTOR pathway, which controls growth and proliferation. S. pombe is insensitive to rapamycin because of partial inhibition of TORC1 activity.Citation62 However, tit1 mutants are sensitive to rapamycin and show decreased growth rate, shorten phenotype and reduced protein translation, especially of several unidentified proteins.Citation61 One could speculate that the lack of isopentenylation might change the amino acid composition of a component of TORC1 and make it more sensitive to rapamycin, since it was also demonstrated in the same publication that isopentenylation promoted cy-tRNATyrGΨA misreading of a near-cognate Cys UGC codon in a ß-galactosidase reporter.
In contrast to factor rpoS of E. coli, there is no clear example of skewed gene expression in S. pombe. Computational analysis demonstrated enrichment of mRNAs with i6A dependent codons among ribosomal proteins and certain mitochondrial enzymes. Accordingly, ribosomal proteins S14 and L12Citation61 and the elongation factor Tif512Citation55 mRNAs showed a reduced sucrose gradient sedimentation polysome profile in tit1 mutant cells. One could speculate that a global reduction of protein translation shown by tit1 mutants is due to mistranslated proteins involved in translation.
Mitochondrial dysfunction in yeasts can be unveiled by slow growth on glycerol. Tit1 mutant cells showed this phenotype. While one might have expected that hypomodification of mt-tRNATrp could explain the phenotype, it was surprisingly rescued with the cytosolic isoform of Mod5 or cy-tRNATyr overexpression. This finding is in agreement with the estimate of cy-tRNATyr as the most limiting i6A modified cytosolic tRNA in S. pombe. The authors explained their finding with mitochondrial proteins involved in energy metabolism, being translated in the cytoplasm where they depend on cy-tRNATyr.Citation55
gro-1 in Caenorhabditis elegans
The only metazoan model organism deficient in tRNA isopentenylation is the gro-1 (e2400) mutant in the roundworm C. elegans.Citation63 It was initially identified in a screen for maternally rescued mutants with extended life-span.Citation64 Maternal rescue designates the phenotypic rescue of homozygous mutants through a process when heterozygous mothers pass a (regulatory) factor on to their offspring through deposition of protein or RNA in eggs. The gro-1 mutants displayed slower growth and development at several temperatures and significantly extended life-span. Since adult life-span was not increased, this means that larval development was slower. Backcrossing to the standard N2 genetic background decreased the effect on life-span.Citation63 The protein encoded by gro-1 is the tRNA isopentenyltransferase homologous to MiaA, Tit1, and Mod5. Like Mod5, GRO-1 contains 2 alternative translational start sites. The gro-1 phenotype was only rescued by a mitochondrial targeted functional GRO-1:GFP construct, while the respective cytoplasmic GRO-1:GFP construct did not rescue the growth phenotype. The mitochondrial involvement correlated with a weak and transient respiratory phenotype,Citation65 but is consistent with mitochondrial effects of other genes mutated in slow-growing mutants (e.g., clk-1). The gro-1 mutation truncated the isopentenyltransferase by about 100 C-terminal amino acids and induced a frame-shift. Whether the maternal effect depends on the inheritance of modified tRNA or isopentenyltransferase enzyme has not been elucidated. We have not found any characterization of tRNA modification in gro-1 worms.
TRIT1 in mammals
Human TRIT1 was cloned by BLAST homology search based on the Mod5 sequence.Citation27 The mouse Trit1 gene was simply assigned because of sequence homology. Data on the occurrence of i6A and ms2i6A modifications in mammals are scattered and partially incomplete (). In contrast to yeast, most mitochondrial tRNAs that carry i6A are further modified to ms2i6A in mammalian cells.Citation8,66 Only 2 cytoplasmic tRNAs, cy-tRNASer(UCN) and tRNA[Ser]Sec, are known to carry i6A37 in human cells.Citation11,67 In mammals, ms2i6A is not reported in cytoplasmic tRNA.
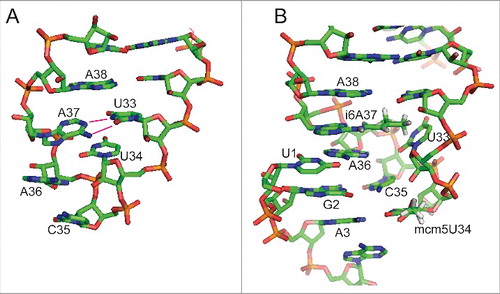
Selenoprotein expression depends on tRNA[Ser]Sec which is reading UGA codons in mRNAs carrying a selenocysteine insertion sequence (SECIS) element in their 3′ untranslated region as sense.Citation68,69 Mouse models deficient in tRNA[Ser]Sec are not able to express any selenocysteine-containing selenoproteins,Citation70,71 and a hypomorphic promoter mutant expressed at reduced levels was associated with decreased selenoprotein expression and seizures.Citation72 A single nucleotide exchange in the human tRNA[Ser]Sec impairs selenoprotein expression in a patient with a complex syndrome.Citation73 Thus, a whole class of proteins depends on the function of one tRNA which is decoding (in all but one case) only one singular UGA/Sec codon per mRNA. Modification of tRNA[Ser]Sec within the anticodon loop involves i6A37 and mcm5U(m)34.Citation74 Mutations in the anticodon loop of tRNA[Ser]Sec affect selenoprotein expression.Citation75 Lovastatin, a lipid lowering drug, is an inhibitor of HMG-CoA reductase, the enzyme catalyzing the committed step of cholesterol biosynthesis. Inhibition of this pathway not only affects cholesterol biosynthesis, but all isoprenoids, including dimethylallylpyrophosphate (DMAPP), the substrate of tRNA isopentenyl transferases. Consistent with a role for i6A in tRNA[Ser]Sec, treatment of cultured cells with lovastatin reduced selenoprotein expression in a selenoprotein-specific pattern.Citation76 tRNA[Ser]Sec carrying the A37G mutation (abolishing the i6A37 modification) supported expression of a subset of selenoproteins (including essential enzymes Txnrd1 and Gpx4), while the same tRNA[Ser]Sec seemed to act as a dominant negative on translation of a subset of stress-related selenoproteins, including Gpx1. In a more recent study, the effect of overexpression of A37G tRNA[Ser]Sec on selenoprotein expression was investigated by ribosomal profiling demonstrating the transcript-specific effects of the tRNA[Ser]Sec mutation.Citation77 Another mutant, U34A, was most likely edited in vivo to inosine (I34), and was able to sustain low levels of MsrB1 (and traces of Gpx4), while Gpx1 translation was apparently not supported by this tRNA.Citation75 The inosine is expected to read pyrimidine bases at the wobble position and adenosine, but not guanosine. Some selenoproteins were made at lower efficiency in the presence of this tRNA. It remained unclear whether the I34 in tRNA[Ser]Sec promoted Sec incorporation at UGY/Cys codons in other proteins.Citation75 We probed the role of i6A in tRNA[Ser]Sec by showing that reduced expression of Trit1 in cells reduces the expression of selenoproteins.Citation67 Structural studies suggested that the i6A37 modification in tRNA[Ser]Sec clearly promotes ordering of the anticodon stem loop. In vitro transcribed tRNA[Ser]Sec crystallized in 2 aberrant conformations - both incompatible with reading the codon, because A37 embarked on illicit intermolecular base-pairing interactions and the characteristic U-turn 3′ of the anticodon did not formCitation78 (). In similar nuclear magnetic resonance studies on oligonucleotides representing the anticodon stem loop structure of E. coli tRNAPhe, i6A alone was not able to induce a U-turn and order the anticodon,Citation79 suggesting that other modifications might contribute to anticodon structuring.
Figure 2. Effect of base 37 modification in tRNASec. (A) Unmodified murine tRNASec displays a disordered anticodon loop. Instead of forming a U-turn, U33 base pairs with unmodified A37 leading to an anticodon incapable of interaction with the codon.Citation78 (B) Model of fully modified tRNASec decoding the codon in the ribosome. The model was generated by the authors using MacPyMOL (Schrödinger) software and is based on PDB 2UUC containing a bacterial tRNAVal carrying 5-Oxoacetic acid-modified U34 and N6-Methyladenosine at position 37.Citation80
Proposed functions for TRIT1 not associated with tRNA isopentenylation include tRNA gene mediated transcriptional silencingCitation81 and a role for TRIT1 as a tumor suppressor.Citation82
Base 37 modification in human disease
Defects in tRNA modification lead to several human disorders, including mitochondrial diseases ().Citation83 In a family with clinical symptoms of mitochondrial disease, 2 patients carrying the homozygous p.Arg323Gln mutation in TRIT1 were identified.Citation84 Severe combined mitochondrial respiratory chain defects mainly affecting complex I and complex IV were found in skeletal muscle biopsy. In vitro experiments indicated reduced expression of ND1 and ND5 of complex I and COXI and COXII in complex IV. All these proteins are mitochondrial encoded and it was concluded that mitochondrial translation was affected. Since TRIT1 acts on mitochondrial tRNAs, recombinant enzyme was found to have reduced 14C-DMAPP:tRNA isopentenyl transferase activity toward oligonucleotides mimicking mt-tRNA substrates. The authors further showed that the m.7480A>G mutation that changes base A38 to G in mt-tRNASer(UCN) abolishes isopentenylation of the tRNA, because it disrupts the AAA motif in TRIT1.
Table 4. Genes involved in tRNAs modification at position 37 in eukaryotes and associated phenotypes.
Most mammalian mt-tRNAs carrying i6A are further modified to ms2i6A (). Wei et al. have proposed that oxidative stress caused by the mt-DNA mutation m.3234A>G (affecting taurinomethylation of τm5U34 in mt-tRNALeu) interferes with the function of [4Fe-4S] clusters in CDK5RAP1, the enzyme needed for methylthiolation of isopentenylated mt-tRNAs, and thus further decreases mitochondrial protein expression and activity leading to mitochondrial encephalopathy lactic acidosis and stroke-like episodes (MELAS).Citation85
Mutations in an intron of CDKAL1, encoding the enzyme mediating methylthiolation of t6A to ms2t6A in mt-tRNAs, have been associated with lowered insulin secretion and development of type 2 diabetes mellitus.Citation86,87 Accordingly, in a pancreatic β-cell specific Cdkal1-knockout mouse model, aberrant proinsulin biosynthesis and impaired glucose metabolism were described.Citation88 Since the glucose sensor in pancreatic β-cells is in essence an ATP sensor, mitochondrial dysfunction may play a role in decreased insulin secretion. The authors have proposed another potential pathomechanism, i.e., the misreading of Lys (AAA and AAG) codons in insulin by cytoplasmic tRNALys (UUU). Indeed, proteolytic processing of proinsulin depends on Lys at the cleavage site between the A chain and the C-peptide of proinsulin. 14C-Lys incorporation and C-peptide abundance were reduced in Cdkal1-deficient pancreatic β-cells explaining the hyperglycemia of the knockout mice.Citation88 Human mutations affecting formation of the t6A modification have not yet been identified, although the enzymes responsible for the modification are present in metazoans, including humans.Citation89
A biosynthetic defect in wybutosine formation has not been detected, possibly implying that this modification of tRNAPhe at base 37 is essential. Amplification of one of the genes involved in wybutosine formation (TRMT12) has been reported in breast cancer.Citation90 Given the large number of genes involved in tRNA base 37 modification (), it can be expected that more patients are discovered carrying inborn errors in base 37 modification.
The isopentenyl transferase reaction
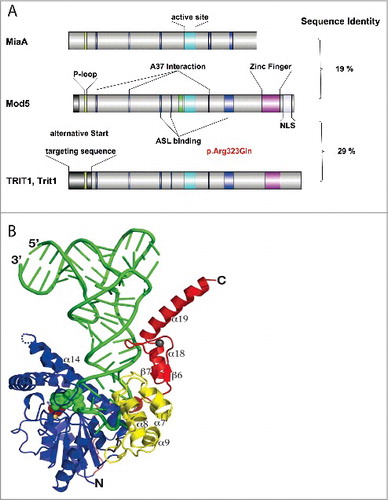
As mentioned above, all tRNA isopentenyl transferases are encoded by homologous genes. Mod5 is highly homologous to MiaA from E. coli with the notable exception that eukaryotic isopentenyl transferases contain an additional zinc-finger domain at their C-terminus, which is not present in bacteria (). To clarify the mechanism of tRNA isopentenyl transferases, bacterial MiaA and yeast Mod5 have been crystallized as apo enzymes as well as in several conformations bound to tRNA substrates and DMAPP.Citation91-93 In general, isopentenyl transferase binds the anticodon stem loop of tRNA and distorts it flipping out bases U33, G34, C35, and A37 from the anticodon loop of tRNA. Remarkably, A36 and A38 of the AAA motif engage in specific interactions within the tRNA, i.e., the AAA sequence motif is not read by the protein, but by the tRNA.Citation53 The substrate base A37 is specifically bound in a channel within the enzyme that leads to the DMAPP binding site. Here, N6 of A37 is activated by Asp46 and attacks the DMAPP molecule with pyrophosphate as excellent leaving group in a nucleophilic substitution (SN2-) reaction. The DMAPP binding site forms only upon tRNA binding (and includes direct interaction with the purine ring of A37) supporting the ordered mechanism observed before in kinetic experiments.Citation94 There exist several lines of evidence, which indicate that the C-terminal domain is necessary for substrate discrimination. First of all, the C-terminal domain interacts with other nucleotides in the anticodon stem loop of the tRNA, which are not involved in catalysis as shown in the Mod5-tRNACysGCA crystal structure. One of these nucleotides is G34.Citation93 Second, this interaction is necessary for enzyme activity as removal of this domain compromises Tit1 activity.Citation60 Third, mutation of C at position 34 of the non-Mod5 substrate, cy-tRNATrpCCA, to G converts it into a Mod5 substrate. Fourth, mutation of the G34 binding loop of Mod5, which disrupts the contact with the wobble base, selectively impairs isopentenylation of cy-tRNATyrGΨA and cy-tRNACysGCA, over tRNASerNGA, which does not carry G34.Citation60 Therefore, the wobble base is a determinant for Mod5 activity in both tRNAs. However this rule is no applicable to cy-tRNASerNGA.Citation60 A shown in , the Zn-finger domain also interacts with the D-loop of substrate tRNA.
Figure 3. Structure of isopentenyl transferases and mode of substrate binding. (A) Comparison of isopentenyl transferases from E. coli (MiaA), baker's yeast (Mod5), and mammals (TRIT1, Trit1). Stretches of sequences with amino acid identity are highlighted in color. The sequence shown in green represents the P-loop involved in DMAPP binding. The turquoise sequence is the core of the active site. The blue sequences refer to amino acids interacting with the tRNA substrate (A37 binding or anticodon stem loop (ASL)-binding). The zinc-finger present in eukaryotes is depicted in violet. NLS, nuclear localization sequence. Overall sequence identity is shown on the right. The pathogenic mutation in human TRIT1 is indicated in red. The figure was prepared with IBS software.Citation95 (B) Crystal structure of Mod5- tRNACysGCA complex. The tRNA is shown in green. The DMAPP-binding and catalytic center in green balls, the zinc-finger domain is shown in red. Reproduced with permission from Zhou C and Huang RH. Crystallographic snapshots of eukaryotic dimethylallyltransferase acting on tRNA: insight into tRNA recognition and reaction mechanism. Proc Natl Acad Sci U S A 2008; 105:16142–7.
DMAPP, an intermediate of the isoprenoid and sterol biosynthetic pathways, is the isopentenyl donor substrate used by tRNA isopentenyl transferases. Isotope labeled DMAPP can be used to directly demonstrate isopentenylation of tRNAs or oligonucleotides resembling tRNA anticodon stem loops. Competition for DMAPP between tRNA modification and sterol biosynthesis in vivo became apparent, when Erg20 (farnesyl diphosphate synthetase) overexpression decreased isopentenylated tRNAs, which were restored by overexpression of Mod5.Citation96,97 Statins are drugs widely used to decrease cholesterol biosynthesis in patients with hypercholesterolemia. As statin treatment of cells decreased selenoprotein expression,Citation76 it was speculated that statin toxicity may be mediated by a specific lack of selenoprotein N (SELENON),Citation98 a gene known to be mutated in a spectrum of congenital myopathies, selenoprotein N-related myopathy.Citation99-101 That statins, applied in therapeutic dosage, impair tRNA isopentenylation has not been demonstrated.
It is interesting that apart from the function of Mod5 as an isopentenyl transferase, other functions have been discovered recently. It was suggested that the longer eukaryotic C-terminal domain might be involved in these and thus would be exclusive to eukaryotic organisms. One of these functions is the involvement of the nuclear fraction of Mod5 in tRNA-mediated transcriptional silencing.Citation81,102 In addition, Mod5 can form prions, of which the full biologic role is not clear. Formation of MOD+ prions, however, reduced i6A in tRNA.Citation103 As both functions are not clearly linked to tRNA isopentenyl transferase activity of Mod5, they are beyond of the scope of this review.
Conclusion
Isopentenyladenosine is one of the longest known tRNA modifications and occurs only at one specific position in tRNA. More than 40 y of study in several model organisms have accumulated a wealth of data on i6A, ms2i6A, and other modifications specific for base 37 in tRNA. All these modifications are of ancient origin and are retained in prokaryots and eukaryots. Clearly, i6A and ms2i6A are functionally important for mammals, including humans. All isopentenyl transferase genes known are homologous. The database on tRNAs that carry i6A and its hypermodified derivatives is almost complete and will certainly soon be complete with ever better mass spectrometric analyses. In a next step, it would be desirable, that i6A and ms2i6A is studied in all standard organisms of molecular biology (at least E. coli, the 2 yeasts, and human and mouse) using the same technology and independent of synthetic reporter genes. Then, the effects of loss of all base 37 modifications should be studied with the same technology in the same organism. As a result of this review, we realized that not even for i6A, t6A, and related modifications there are 2 global analyses of gene expression in the same organism. Such data, in our view, would be required to fully appreciate the biologic roles of these modifications. Structural data on all relevant modified tRNAs, ideally in complex with mRNA in the context of the ribosome might finally reveal subtle differences between e.g., ms2i6A and ms2t6A modifications. The application of recently developed powerful technology like ribosomal profiling might reveal on a global, yet gene-specific, scale why these modifications have been retained through evolution.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors want to dedicate this article to the memory of Mathias Sprinzl who introduced US to translation and in particular tRNA modification.
Funding
This work was supported by Deutsche Forschungsgemeinschaft DFG under Schw914/5–1 within the priority program Chemical biology.
References
- Bjork GR, Ericson JU, Gustafsson CE, Hagervall TG, Jonsson YH, Wikstrom PM. Transfer RNA modification. Annu Rev Biochem 1987; 56:263-87; PMID:3304135; http://dx.doi.org/10.1146/annurev.bi.56.070187.001403
- El Yacoubi B, Bailly M, de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet 2012; 46:69-95; PMID:22905870; http://dx.doi.org/10.1146/annurev-genet-110711-155641
- Persson BC, Esberg B, Olafsson O, Bjork GR. Synthesis and function of isopentenyl adenosine derivatives in tRNA. Biochimie 1994; 76:1152-60; PMID:7748950; http://dx.doi.org/10.1016/0300-9084(94)90044-2
- Soll D. Enzymatic modification of transfer RNA. Science 1971; 173:293-9; PMID:4934576; http://dx.doi.org/10.1126/science.173.3994.293
- Grosjean H, Westhof E. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res 2016; 44:8020-40; PMID:27448410; http://dx.doi.org/10.1093/nar/gkw608
- Murphy FV, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNA(Lys) UUU. Nat Struct Mol Biol 2004; 11:1186-91; http://dx.doi.org/10.1038/nsmb861
- Weixlbaumer AM, F. V. IV. Crystallographic Studies of Decoding by Modified Bases: Correlation of Structure and Function. In: Grosjean H, ed. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution: Landes Bioscience, 2009:493-508
- Suzuki T, Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res 2014; 42:7346-57; PMID:24831542; http://dx.doi.org/10.1093/nar/gku390
- Deutsch C, El Yacoubi B, de Crecy-Lagard V, Iwata-Reuyl D. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J Biol Chem 2012; 287:13666-73; PMID:22378793; http://dx.doi.org/10.1074/jbc.M112.344028
- Perche-Letuvee P, Molle T, Forouhar F, Mulliez E, Atta M. Wybutosine biosynthesis: structural and mechanistic overview. RNA Biol 2014; 11:1508-18; PMID:25629788; http://dx.doi.org/10.4161/15476286.2014.992271
- Lamichhane TN, Mattijssen S, Maraia RJ. Human cells have a limited set of tRNA anticodon loop substrates of the tRNA isopentenyltransferase TRIT1 tumor suppressor. Mol Cell Biol 2013; 33:4900-8; PMID:24126054; http://dx.doi.org/10.1128/MCB.01041-13
- Motorin Y, Bec G, Tewari R, Grosjean H. Transfer RNA recognition by the Escherichia coli Delta(2)-isopentenyl-pyrophosphate:tRNA Delta 2-isopentenyl transferase: Dependence on the anticodon arm structure. RNA 1997; 3:721-33; PMID:9214656
- Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 2009; 37:D159-62; PMID:18957446; http://dx.doi.org/10.1093/nar/gkn772
- Dunin-Horkawicz S, Czerwoniec A, Gajda MJ, Feder M, Grosjean H, Bujnicki JM. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res 2006; 34:D145-9; PMID:16381833; http://dx.doi.org/10.1093/nar/gkj084
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res 2013; 41:D262-7; PMID:23118484; http://dx.doi.org/10.1093/nar/gks1007
- Zachau HG, Dutting D, Feldmann H. Nucleotide sequences of two serine-specific transfer ribonucleic acids. Angew Chem Int Ed Engl 1966; 5:422; PMID:4956644; http://dx.doi.org/10.1002/anie.196604221
- Hall RH, Robins MJ, Stasiuk L, Thedford R. Isolation of N6-(Gamma Gamma-Dimethylallyl)Adenosine from Soluble Ribonucleic Acid. JACS 1966; 88:2614; http://dx.doi.org/10.1021/ja00963a061
- Bartz J, Soll D, Burrows WJ, Skoog F. Identification of Cytokinin-Active Ribonucleosides in Pure Escherichia-Coli Transfer Rna Species. P Natl Acad Sci USA 1970; 67:1448; PMID:4922291; http://dx.doi.org/10.1073/pnas.67.3.1448
- Bartz JK, Kline LK, Soll D. N6-(Delta2-Isopentenyl)Adenosine - Biosynthesis in-Vitro in Transfer Rna by an Enzyme Purified from Escherichia-Coli. Biochem Bioph Res Co 1970; 40:1481; PMID:4326583; http://dx.doi.org/10.1016/0006-291X(70)90035-5
- Agris PF, Armstrong DJ, Schafer KP, Soll D. Maturation of a Hypermodified Nucleoside in Transfer-Rna. Nucleic Acids Res 1975; 2:691-8; PMID:49880; http://dx.doi.org/10.1093/nar/2.5.691
- Caillet J, Droogmans L. Molecular-Cloning of the Escherichia-Coli Miaa Gene Involved in the Formation of Delta-2-Isopentenyl Adenosine in Transfer-Rna. J Bacteriol 1988; 170:4147-52; PMID:3045085; http://dx.doi.org/10.1128/jb.170.9.4147-4152.1988
- Eisenberg SP, Yarus M, Soll L. The effect of an Escherichia coli regulatory mutation on transfer RNA structure. J Mol Biol 1979; 135:111-26; PMID:93644; http://dx.doi.org/10.1016/0022-2836(79)90343-7
- Landgraf BJ, McCarthy EL, Booker SJ. Radical S-Adenosylmethionine Enzymes in Human Health and Disease. Annu Rev Biochem 2016; 85:485-514; PMID:27145839; http://dx.doi.org/10.1146/annurev-biochem-060713-035504
- Laten H, Gorman J, Bock RM. Isopentenyladenosine deficient tRNA from an antisuppressor mutant of Saccharomyces cerevisiae. Nucleic Acids Res 1978; 5:4329-42; PMID:364426; http://dx.doi.org/10.1093/nar/5.11.4329
- Dihanich ME, Najarian D, Clark R, Gillman EC, Martin NC, Hopper AK. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol Cell Biol 1987; 7:177-84; PMID:3031456; http://dx.doi.org/10.1128/MCB.7.1.177
- Connolly DM, Winkler ME. Genetic and physiological relationships among the miaA gene, 2-methylthio-N6-(delta 2-isopentenyl)-adenosine tRNA modification, and spontaneous mutagenesis in Escherichia coli K-12. J Bacteriol 1989; 171:3233-46; PMID:2656644; http://dx.doi.org/10.1128/jb.171.6.3233-3246.1989
- Golovko A, Hjalm G, Sitbon F, Nicander B. Cloning of a human tRNA isopentenyl transferase. Gene 2000; 258:85-93; PMID:11111046; http://dx.doi.org/10.1016/S0378-1119(00)00421-2
- Reiter V, Matschkal DM, Wagner M, Globisch D, Kneuttinger AC, Muller M, Carell T. The CDK5 repressor CDK5RAP1 is a methylthiotransferase acting on nuclear and mitochondrial RNA. Nucleic Acids Res 2012; 40:6235-40; PMID:22422838; http://dx.doi.org/10.1093/nar/gks240
- Arragain S, Handelman SK, Forouhar F, Wei FY, Tomizawa K, Hunt JF, Douki T, Fontecave M, Mulliez E, Atta M. Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA. J Biol Chem 2010; 285:28425-33; PMID:20584901; http://dx.doi.org/10.1074/jbc.M110.106831
- Armstrong DJ, Burrows WJ, Skoog F, Roy KL, Soll D. Cytokinins - Distribution in Transfer Rna Species of Escherichia Coli. Proc Natl Acad Sci USA 1969; 63:834; PMID:4899879; http://dx.doi.org/10.1073/pnas.63.3.834
- Schön A, Böck A, Ott G, Sprinzl M, Söll D. The selenocysteine-inserting opal suppressor serine tRNA from E. coli is highly unusual in structure and modification. Nucleic Acids Res 1989; 17:7159-65; PMID:2529478; http://dx.doi.org/10.1093/nar/17.18.7159
- Gefter ML, Russell RL. Role of Modifications in Tyrosine Transfer Rna - a Modified Base Affecting Ribosome Binding. J Mol Biol 1969; 39:145; PMID:4938812; http://dx.doi.org/10.1016/0022-2836(69)90339-8
- Buck M, Griffiths E. Iron Mediated Methylthiolation of Transfer-Rna as a Regulator of Operon Expression in Escherichia-Coli. Nucleic Acids Res 1982; 10:2609-24; PMID:7043398; http://dx.doi.org/10.1093/nar/10.8.2609
- Mclennan BD, Buck M, Humphreys J, Griffiths E. Iron-Related Modification of Bacterial Transfer-Rna. Nucleic Acids Res 1981; 9:2629-40; PMID:6792594; http://dx.doi.org/10.1093/nar/9.11.2629
- Buck M, Griffiths E. Regulation of Aromatic Amino-Acid-Transport by Transfer-Rna - Role of 2-Methylthio-N6-(Delta-2-Isopentenyl)-Adenosine. Nucleic Acids Res 1981; 9:401-14; PMID:7010315; http://dx.doi.org/10.1093/nar/9.2.401
- Griffiths E, Humphreys J. Alterations in Transfer-Rnas Containing 2-Methylthio-N6-(Delta-2-Isopentenyl)-Adenosine during Growth of Enteropathogenic Escherichia-Coli in Presence of Iron-Binding Proteins. Euro J Biochem 1978; 82:503-13; PMID:342239; http://dx.doi.org/10.1111/j.1432-1033.1978.tb12044.x
- Buck M, Ames BN. A modified nucleotide in Transfer-Rna as a possible regulator of aerobiosis - synthesis of Cis-2-methylthioribosylzeatin in the transfer-Rna of salmonella. Cell 1984; 36:523-31; PMID:6362893; http://dx.doi.org/10.1016/0092-8674(84)90245-9
- Diaz I, Pedersen S, Kurland CG. Effects of miaA on translation and growth rates. Mol Gen Genet 1987; 208:373-6; PMID:3312947; http://dx.doi.org/10.1007/BF00328126
- Ericson JU, Bjork GR. Pleiotropic effects induced by modification deficiency next to the anticodon of transfer-Rna from salmonella-typhimurium-Lt2. J Bacteriol 1986; 166:1013-21; PMID:2423501; http://dx.doi.org/10.1128/jb.166.3.1013-1021.1986
- Bouadloun F, Srichaiyo T, Isaksson LA, Bjork GR. Influence of modification next to the anticodon in Transfer-Rna on codon context-sensitivity of translational suppression and accuracy. J Bacteriol 1986; 166:1022-7; PMID:3086285; http://dx.doi.org/10.1128/jb.166.3.1022-1027.1986
- Diaz I, Ehrenberg M. Ms2i6a deficiency enhances proofreading in translation. J Mol Biol 1991; 222:1161-71; PMID:1762149; http://dx.doi.org/10.1016/0022-2836(91)90599-2
- Urbonavicius J, Qian O, Durand JMB, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. Embo J 2001; 20:4863-73; PMID:11532950; http://dx.doi.org/10.1093/emboj/20.17.4863
- Durand JMB, Bjork GR, Kuwae A, Yoshikawa M, Sasakawa C. The modified nucleoside 2-methylthio-N-6-isopentenyladenosine in tRNA of Shigella flexneri is required for expression of virulence genes. J Bacteriol 1997; 179:5777-82; PMID:9294434; http://dx.doi.org/10.1128/jb.179.18.5777-5782.1997
- Duechler M, Leszczynska G, Sochacka E, Nawrot B. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell Mol Life Sci 2016; 73:3075-95; PMID:27094388; http://dx.doi.org/10.1007/s00018-016-2217-y
- Endres L, Dedon PC, Begley TJ. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol 2015; 12:603-14; PMID:25892531; http://dx.doi.org/10.1080/15476286.2015.1031947
- Kolmsee T, Hengge R. Rare codons play a positive role in the expression of the stationary phase sigma factor RpoS (sigma(S)) in Escherichia coli. RNA Biol 2011; 8:913-21; PMID:21788735; http://dx.doi.org/10.4161/rna.8.5.16265
- Thompson KM, Gottesman S. The MiaA tRNA modification enzyme is necessary for robust RpoS expression in Escherichia coli. J Bacteriol 2014; 196:754-61; PMID:24296670; http://dx.doi.org/10.1128/JB.01013-13
- Aubee JI, Olu M, Thompson KM. The i6A37 tRNA modification is essential for proper decoding of UUX-Leucine codons during rpoS and iraP translation. RNA 2016; 22:729-42; PMID:26979278; http://dx.doi.org/10.1261/rna.053165.115
- Martin NC, Hopper AK. Isopentenylation of both cytoplasmic and mitochondrial tRNA is affected by a single nuclear mutation. J Biol Chem 1982; 257:10562-5; PMID:7050116
- Najarian D, Dihanich ME, Martin NC, Hopper AK. DNA sequence and transcript mapping of MOD5: features of the 5′ region which suggest two translational starts. Mol Cell Biol 1987; 7:185-91; PMID:3031457; http://dx.doi.org/10.1128/MCB.7.1.185
- Slusher LB, Gillman EC, Martin NC, Hopper AK. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proc Natl Acad Sci U S A 1991; 88:9789-93; PMID:1946403; http://dx.doi.org/10.1073/pnas.88.21.9789
- Gillman EC, Slusher LB, Martin NC, Hopper AK. MOD5 translation initiation sites determine N6-isopentenyladenosine modification of mitochondrial and cytoplasmic tRNA. Mol Cell Biol 1991; 11:2382-90; PMID:1850093; http://dx.doi.org/10.1128/MCB.11.5.2382
- Boguta M, Hunter LA, Shen WC, Gillman EC, Martin NC, Hopper AK. Subcellular locations of MOD5 proteins: mapping of sequences sufficient for targeting to mitochondria and demonstration that mitochondrial and nuclear isoforms commingle in the cytosol. Mol Cell Biol 1994; 14:2298-306; PMID:8139535; http://dx.doi.org/10.1128/MCB.14.4.2298
- Tolerico LH, Benko AL, Aris JP, Stanford DR, Martin NC, Hopper AK. Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics 1999; 151:57-75; PMID:9872948
- Lamichhane TN, Arimbasseri AG, Rijal K, Iben JR, Wei FY, Tomizawa K, Maraia RJ. Lack of tRNA-i6A modification causes mitochondrial-like metabolic deficiency in S. pombe by limiting activity of cytosolic tRNATyr, not mito-tRNA. RNA 2016; 22:583-96; PMID:26857223; http://dx.doi.org/10.1261/rna.054064.115
- Egel R, Kohli J, Thuriaux P, Wolf K. Genetics of the fission yeast Schizosaccharomyces pombe. Ann Rev Genet 1980; 14:77-108; PMID:7011176; http://dx.doi.org/10.1146/annurev.ge.14.120180.000453
- Janner F, Vogeli G, Fluri R. The antisuppressor strain sin1 of Schizosaccharomyces pombe lacks the modification isopentenyladenosine in transfer RNA. J Mol Biol 1980; 139:207-19; PMID:7411631; http://dx.doi.org/10.1016/0022-2836(80)90305-8
- Kohli J, Kwong T, Altruda F, Soll D, Wahl G. Characterization of a UGA-suppressing serine tRNA from Schizosaccharomyces pombe with the help of a new in vitro assay system for eukaryotic suppressor tRNAs. J Biol Chem 1979; 254:1546-51; PMID:762155
- Rafalski A, Kohli J, Agris P, Soll D. The nucleotide sequence of a UGA suppressor serine tRNA from Schizosaccharomyces pombe. Nucleic Acids Res 1979; 6:2683-95; PMID:461200; http://dx.doi.org/10.1093/nar/6.8.2683
- Lamichhane TN, Blewett NH, Maraia RJ. Plasticity and diversity of tRNA anticodon determinants of substrate recognition by eukaryotic A37 isopentenyltransferases. RNA 2011; 17:1846-57; PMID:21873461; http://dx.doi.org/10.1261/rna.2628611
- Lamichhane TN, Blewett NH, Crawford AK, Cherkasova VA, Iben JR, Begley TJ, Farabaugh PJ, Maraia RJ. Lack of tRNA modification isopentenyl-A37 alters mRNA decoding and causes metabolic deficiencies in fission yeast. Mol Cell Biol 2013; 33:2918-29; PMID:23716598; http://dx.doi.org/10.1128/MCB.00278-13
- Takahara T, Maeda T. TORC1 of fission yeast is rapamycin-sensitive. Genes Cells 2012; 17:698-708; PMID:22762302; http://dx.doi.org/10.1111/j.1365-2443.2012.01618.x
- Lemieux J, Lakowski B, Webb A, Meng Y, Ubach A, Bussiere F, Barnes T, Hekimi S. Regulation of physiological rates in Caenorhabditis elegans by a tRNA-modifying enzyme in the mitochondria. Genetics 2001; 159:147-57; PMID:11560893
- Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science 1996; 272:1010-3; PMID:8638122; http://dx.doi.org/10.1126/science.272.5264.1010
- Braeckman BP, Houthoofd K, De Vreese A, Vanfleteren JR. Apparent uncoupling of energy production and consumption in long-lived Clk mutants of Caenorhabditis elegans. Curr Biol 1999; 9:493-6; PMID:10330373; http://dx.doi.org/10.1016/S0960-9822(99)80216-4
- Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet 2011; 45:299-329; PMID:21910628; http://dx.doi.org/10.1146/annurev-genet-110410-132531
- Fradejas N, Carlson BA, Rijntjes E, Becker NP, Tobe R, Schweizer U. Mammalian Trit1 is a tRNA([Ser]Sec)-isopentenyl transferase required for full selenoprotein expression. Biochem J 2013; 450:427-32; PMID:23289710; http://dx.doi.org/10.1042/BJ20121713
- Lee BJ, Worland PJ, Davis JN, Stadtman TC, Hatfield DL. Identification of a selenocysteyl-tRNA(Ser) in mammalian cells that recognizes the nonsense codon, UGA. J Biol Chem 1989; 264:9724-7; PMID:2498338
- Schweizer U, Fradejas-Villar N. Why 21? The significance of selenoproteins for human health revealed by inborn errors of metabolism. FASEB J 2016; 30(11):3669-81; PMID:27473727; http://dx.doi.org/10.1096/fj.201600424
- Carlson BA, Novoselov SV, Kumaraswamy E, Lee BJ, Anver MR, Gladyshev VN, Hatfield DL. Specific excision of the selenocysteine tRNA[Ser]Sec (Trsp) gene in mouse liver demonstrates an essential role of selenoproteins in liver function. J Biol Chem 2004; 279:8011-7; PMID:14660662; http://dx.doi.org/10.1074/jbc.M310470200
- Schweizer U, Streckfuss F, Pelt P, Carlson BA, Hatfield DL, Köhrle J, Schomburg L. Hepatically derived selenoprotein P is a key factor for kidney but not for brain selenium supply. Biochem J 2005; 386:221-6; PMID:15638810; http://dx.doi.org/10.1042/BJ20041973
- Carlson BA, Schweizer U, Perella C, Shrimali RK, Feigenbaum L, Shen L, Speransky S, Floss T, Jeong SJ, Watts J, et al. The selenocysteine tRNA STAF-binding region is essential for adequate selenocysteine tRNA status, selenoprotein expression and early age survival of mice. Biochem J 2009; 418:61-71; PMID:18973473; http://dx.doi.org/10.1042/BJ20081304
- Schoenmakers E, Carlson B, Agostini M, Moran C, Rajanayagam O, Bochukova E, Tobe R, Peat R, Gevers E, Muntoni F, et al. Mutation in human selenocysteine transfer RNA selectively disrupts selenoprotein synthesis. J Clin Invest 2016; 126:992-6; PMID:26854926; http://dx.doi.org/10.1172/JCI84747
- Diamond AM, Choi IS, Crain PF, Hashizume T, Pomerantz SC, Cruz R, Steer CJ, Hill KE, Burk RF, McCloskey JA, et al. Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine transfer RNA[Ser]Sec. J Biol Chem 1993; 268:14215-23; PMID:8314785
- Carlson BA, Moustafa ME, Sengupta A, Schweizer U, Shrimali R, Rao M, Zhong N, Wang S, Feigenbaum L, Lee BJ, et al. Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J Biol Chem 2007; 282:32591-602; PMID:17848557; http://dx.doi.org/10.1074/jbc.M707036200
- Warner GJ, Berry MJ, Moustafa ME, Carlson BA, Hatfield DL, Faust JR. Inhibition of selenoprotein synthesis by selenocysteine tRNA[Ser]Sec lacking isopentenyladenosine. J Biol Chem 2000; 275:28110-9; PMID:10821829
- Howard MT, Carlson BA, Anderson CB, Hatfield DL. Translational redefinition of UGA codons is regulated by selenium availability. J Biol Chem 2013; 288:19401-13; PMID:23696641; http://dx.doi.org/10.1074/jbc.M113.481051
- Ganichkin OM, Anedchenko EA, Wahl MC. Crystal structure analysis reveals functional flexibility in the selenocysteine-specific tRNA from mouse. PLoS One 2011; 6:e20032; PMID:21629646; http://dx.doi.org/10.1371/journal.pone.0020032
- Cabello-Villegas J, Winkler ME, Nikonowicz EP. Solution conformations of unmodified and A(37)N(6)-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNA(Phe). J Mol Biol 2002; 319:1015-34; PMID:12079344; http://dx.doi.org/10.1016/S0022-2836(02)00382-0
- Weixlbaumer A, Murphy FV 4th, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat Struct Mol Biol 2007; 14:498-502; PMID:17496902; http://dx.doi.org/10.1038/nsmb1242
- Smaldino PJ, Read DF, Pratt-Hyatt M, Hopper AK, Engelke DR. The cytoplasmic and nuclear populations of the eukaryote tRNA-isopentenyl transferase have distinct functions with implications in human cancer. Gene 2015; 556:13-8; PMID:25261850; http://dx.doi.org/10.1016/j.gene.2014.09.049
- Spinola M, Galvan A, Pignatiello C, Conti B, Pastorino U, Nicander B, Paroni R, Dragani TA. Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer. Oncogene 2005; 24:5502-9; PMID:15870694; http://dx.doi.org/10.1038/sj.onc.1208687
- Torres AG, Batlle E, de Pouplana LR. Role of tRNA modifications in human diseases. Trends Mol Med 2014; 20:306-14; PMID:24581449; http://dx.doi.org/10.1016/j.molmed.2014.01.008
- Yarham JW, Lamichhane TN, Pyle A, Mattijssen S, Baruffini E, Bruni F, Donnini C, Vassilev A, He L, Blakely EL, et al. Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA. PLoS Genet 2014; 10:e1004424; PMID:24901367; http://dx.doi.org/10.1371/journal.pgen.1004424
- Wei FY, Zhou B, Suzuki T, Miyata K, Ujihara Y, Horiguchi H, Takahashi N, Xie P, Michiue H, Fujimura A, et al. Cdk5rap1-mediated 2-methylthio modification of mitochondrial tRNAs governs protein translation and contributes to myopathy in mice and humans. Cell Metab 2015; 21:428-42; PMID:25738458; http://dx.doi.org/10.1016/j.cmet.2015.01.019
- Steinthorsdottir V, Reynisdottir I, Thorleifsson G, Ghosh S, Benediktsson R, Sigurdsson G, et al. The recently identified type 2 diabetes gene CDKAL1 is widely expressed and its expression in pancreatic beta cells is affected by glucose concentration. Diabetologia 2007; 50(Suppl 1): S129; http://dx.doi.org/10.1007/s00125-007-0809-7
- Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet 2007; 39:770-5; PMID:17460697; http://dx.doi.org/10.1038/ng2043
- Wei FY, Suzuki T, Watanabe S, Kimura S, Kaitsuka T, Fujimura A, Matsui H, Atta M, Michiue H, Fontecave M, et al. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J Clin Invest 2011; 121:3598-608; PMID:21841312; http://dx.doi.org/10.1172/JCI58056
- Thiaville PC, Iwata-Reuyl D, de Crecy-Lagard V. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t(6)A), a universal modification of tRNA. RNA Biol 2014; 11:1529-39; PMID:25629598; http://dx.doi.org/10.4161/15476286.2014.992277
- Rodriguez V, Chen Y, Elkahloun A, Dutra A, Pak E, Chandrasekharappa S. Chromosome 8 BAC array comparative genomic hybridization and expression analysis identify amplification and overexpression of TRMT12 in breast cancer. Genes Chromosomes Cancer 2007; 46:694-707; PMID:17440925; http://dx.doi.org/10.1002/gcc.20454
- Chimnaronk S, Forouhar F, Sakai J, Yao M, Tron CM, Atta M, Fontecave M, Hunt JF, Tanaka I. Snapshots of dynamics in synthesizing N(6)-isopentenyladenosine at the tRNA anticodon. Biochemistry 2009; 48:5057-65; PMID:19435325; http://dx.doi.org/10.1021/bi900337d
- Seif E, Hallberg BM. RNA-protein mutually induced fit: structure of Escherichia coli isopentenyl-tRNA transferase in complex with tRNA(Phe). J Biol Chem 2009; 284:6600-4; PMID:19158097; http://dx.doi.org/10.1074/jbc.C800235200
- Zhou C, Huang RH. Crystallographic snapshots of eukaryotic dimethylallyltransferase acting on tRNA: insight into tRNA recognition and reaction mechanism. Proc Natl Acad Sci U S A 2008; 105:16142-7; PMID:18852462; http://dx.doi.org/10.1073/pnas.0805680105
- Moore JA, Poulter CD. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: A binding mechanism for recombinant enzyme. Biochemistry 1997; 36:604-14; PMID:9012675; http://dx.doi.org/10.1021/bi962225l
- Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, Lahrmann U, Zhao Q, Zheng Y, Zhao Y, et al. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015; 31:3359-61; PMID:26069263; http://dx.doi.org/10.1093/bioinformatics/btv362
- Benko AL, Vaduva G, Martin NC, Hopper AK. Competition between a sterol biosynthetic enzyme and tRNA modification in addition to changes in the protein synthesis machinery causes altered nonsense suppression. Proc Natl Acad Sci U S A 2000; 97:61-6; PMID:10618371; http://dx.doi.org/10.1073/pnas.97.1.61
- Kaminska J, Grabinska K, Kwapisz M, Sikora J, Smagowicz WJ, Palamarczyk G, Zoładek T, Boguta M. The isoprenoid biosynthetic pathway in Saccharomyces cerevisiae is affected in a maf1-1 mutant with altered tRNA synthesis. FEMS Yeast Res 2002; 2:31-7; PMID:12702319; http://dx.doi.org/10.1111/j.1567-1364.2002.tb00066.x
- Moosmann B, Behl C. Selenoprotein synthesis and side-effects of statins. Lancet 2004; 363:892-4; PMID:15031036; http://dx.doi.org/10.1016/S0140-6736(04)15739-5
- Ferreiro A, Quijano-Roy S, Pichereau C, Moghadaszadeh B, Goemans N, Bonnemann C, Jungbluth H, Straub V, Villanova M, Leroy JP, et al. Mutations of the selenoprotein N gene, which is implicated in rigid spine muscular dystrophy, cause the classical phenotype of multiminicore disease: reassessing the nosology of early-onset myopathies. Am J Hum Genet 2002; 71:739-49; PMID:12192640; http://dx.doi.org/10.1086/342719
- Ferreiro A, Ceuterick-de Groote C, Marks JJ, Goemans N, Schreiber G, Hanefeld F, Fardeau M, Martin JJ, Goebel HH, Richard P, et al. Desmin-related myopathy with Mallory body-like inclusions is caused by mutations of the selenoprotein N gene. Ann Neurol 2004; 55:676-86; PMID:15122708; http://dx.doi.org/10.1002/ana.20077
- Moghadaszadeh B, Petit N, Jaillard C, Brockington M, Roy SQ, Merlini L, Romero N, Estournet B, Desguerre I, Chaigne D, et al. Mutations in SEPN1 cause congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome. Nat Genet 2001; 29:17-8; PMID:11528383; http://dx.doi.org/10.1038/ng713
- Pratt-Hyatt M, Pai DA, Haeusler RA, Wozniak GG, Good PD, Miller EL, McLeod IX, Yates JR 3rd, Hopper AK, Engelke DR. Mod5 protein binds to tRNA gene complexes and affects local transcriptional silencing. Proc Natl Acad Sci U S A 2013; 110:E3081-9; PMID:23898186; http://dx.doi.org/10.1073/pnas.1219946110
- Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 2012; 336:355-9; PMID:22517861; http://dx.doi.org/10.1126/science.1219491
- Yamaizumi Z, Kuchino Y, Harada F, Nishimura S, McCloskey JA. Primary structure of Escherichia coli tRNA UUR Leu. Presence of an unknown adenosine derivative in the first position of the anticodon which recognizes the UU codon series. J Biol Chem 1980; 255:2220-5; PMID:6986390
- Barrell BG, Sanger F. The sequence of phenylalanine tRNA from E. coli. FEBS Lett 1969; 3:275-8; PMID:11947028; http://dx.doi.org/10.1016/0014-5793(69)80157-2
- Ishikura H, Yamada Y, Nishimura S. The nucleotide sequence of a serine tRNA from Escherichia coli. FEBS Lett 1971; 16:68-70; PMID:11945903; http://dx.doi.org/10.1016/0014-5793(71)80688-9
- Hirsh D. Tryptophan transfer RNA as the UGA suppressor. J Mol Biol 1971; 58:439-58; PMID:4933412; http://dx.doi.org/10.1016/0022-2836(71)90362-7
- Goodman HM, Abelson JN, Landy A, Zadrazil S, Smith JD. The nucleotide sequences of tyrosine transfer RNAs of Escherichia coli. Eur J Biochem 1970; 13:461-83; PMID:4315419; http://dx.doi.org/10.1111/j.1432-1033.1970.tb00950.x
- Etcheverry T, Colby D, Guthrie C. A precursor to a minor species of yeast tRNASer contains an intervening sequence. Cell 1979; 18:11-26; PMID:389430; http://dx.doi.org/10.1016/0092-8674(79)90349-0
- Madison JT, Kung HK. Large oligonucleotides isolated from yeast tyrosine transfer ribonucleic acid after partial digestion with ribonuclease T1. J Biol Chem 1967; 242:1324-30; PMID:6023574
- Keith G, Roy A, Ebel JP, Dirheimer G. The nucleotide sequences of two tryptophane-tRNAs from brewer's yeast. FEBS Lett 1971; 17:306-8; PMID:11946053; http://dx.doi.org/10.1016/0014-5793(71)80171-0
- Wilson RK, Roe BA. Presence of the hypermodified nucleotide N6-(delta 2-isopentenyl)-2-methylthioadenosine prevents codon misreading by Escherichia coli phenylalanyl-transfer RNA. Proc Natl Acad Sci U S A 1989; 86:409-13; PMID:2643111; http://dx.doi.org/10.1073/pnas.86.2.409
- Bouadloun F, Srichaiyo T, Isaksson LA, Bjork GR. Influence of modification next to the anticodon in tRNA on codon context sensitivity of translational suppression and accuracy. J Bacteriol 1986; 166:1022-7; PMID:3086285; http://dx.doi.org/10.1128/jb.166.3.1022-1027.1986
- Thiaville PC, Legendre R, Rojas-Benitez D, Baudin-Baillieu A, Hatin I, Chalancon G, Glavic A, Namy O, de Crécy-Lagard V. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microbial Cell 2016; 3:29-45; PMID:26798630; http://dx.doi.org/10.15698/mic2016.01.473
- Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 2005; 44:8078-89; PMID:15924427; http://dx.doi.org/10.1021/bi050343f
- Subramanian M, Srinivasan T, Sudarsanam D. Examining the Gm18 and m(1)G Modification Positions in tRNA Sequences. Genomics Inform 2014; 12:71-5; PMID:25031570; http://dx.doi.org/10.5808/GI.2014.12.2.71
- Wang M, Peng Y, Zheng J, Zheng B, Jin X, Liu H, Wang Y, Tang X, Huang T, Jiang P, et al. A deafness-associated tRNAAsp mutation alters the m1G37 modification, aminoacylation and stability of tRNAAsp and mitochondrial function. Nucleic Acids Res 2016; 44:10974-85; PMID:27536005; http://dx.doi.org/10.1093/nar/gkw726
- Tuorto F, Lyko F. Genome recoding by tRNA modifications. Open Biol 2016; 6:pii: 160287; PMID:27974624; http://dx.doi.org/10.1098/rsob.160287
- Waas WF, Druzina Z, Hanan M, Schimmel P. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J Biol Chem 2007; 282:26026-34; PMID:17623669; http://dx.doi.org/10.1074/jbc.M703391200
