ABSTRACT
A new family of non-autonomous retrotransposons with self-cleaving hammerhead ribozymes, the so called retrozymes, has recently been found encoded in diverse plant genomes. These retroelements can be actively transcribed, and their RNAs accumulate in the cells as abundant non-coding circular RNAs (circRNAs) of small size (600–1000 nt). Related circRNAs with self-cleaving ribozymes had already been described in plants, and belong to a group of infectious RNA agents with an uncertain origin: the viroids and viroid-like satellites of plant RNA viruses. These pathogenic circRNAs show many structural similarities with retrozyme circRNAs, and both have been found to occur in flowering plants as heterogeneous RNA molecules of positive and negative polarities. Taking all these data together, we hypothesize that circRNAs encoded by genomic retrozymes could have given origin to infectious circRNAs with self-cleaving ribozymes. Moreover, we propose that retrozymes in time could have evolved from the ancient family of Penelope-like retroelements, which also harbour hammerhead ribozymes. Putative retrozyme sequences with hammerhead ribozymes have been detected as well in metazoan genomes, opening the door to a common occurrence of circRNAs with self-cleaving motifs among eukaryotes.
Abbreviations
| circRNA | = | circular RNA |
| HHR | = | hammerhead ribozyme |
| LTR | = | long-terminal repeat |
| PBS | = | primer binding site |
| PPT | = | polypurine tract |
| RT | = | retrotranscriptase |
| SMART | = | small LTR retrotransposon |
| TRIM | = | terminal-repeat retrotransposon in miniature |
| TSD | = | target site duplication |
Introduction
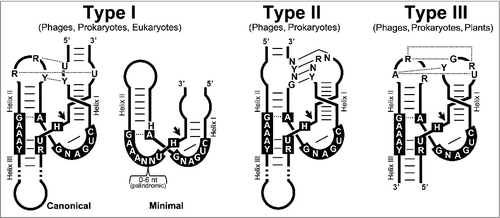
With the discovery of catalytic RNAs or ribozymes more than 30 y agoCitation1,2 we started to become aware of the hidden capabilities of the RNA molecule in biology. Moreover, the ribozymes strongly supported the hypothesis of a prebiotic RNA world where the first living entities would have been based on RNA as both the genetic material and as catalyst.Citation3-5 Only a few of those ancient ribozymes are believed to have remained in extant organisms, carrying out essential functions such as tRNA maturation by the RNAse P,Citation2 mRNA splicing by the spliceosome,Citation6 and even protein translation by the ribosome.Citation7 Among the simplest ribozymes described so far, there is an enigmatic family of small self-cleaving RNAs composed of 9 different classes: hammerhead (HHR),Citation8,9 hairpin (HPR),Citation10 human Hepatitis-δ (HDV),Citation11 Varkud-satellite (VS),Citation12 GlmS,Citation13 twister,Citation14 twister sister, hatchet and pistolCitation15 ribozymes. The HHR was the first discovered and is one of the best-known members of this family. It is composed of a catalytic core of 15 conserved nucleotides surrounded by 3 double helixes (I to III), which adopt a γ-shaped fold where helix I interacts with helix II through tertiary interactions required for efficient in vivo activity.Citation16-18 There are 3 possible circularly permuted topologies for the HHR, named type-I, -II or -III, depending on the open-ended helix (). Originally described in the genomes of infectious circular RNAs (circRNAs) of plants, such as some viroids and viral satellite RNAs, the HHR catalyzes a self-cleaving transesterification that is required during the rolling-circle replication of these molecular replicons. A few HHRs were also exceptionally described in the DNA genomes of some unrelated eukaryotes,Citation19-23 and mostly associated with repetitive sequences. In 2010, we reported the widespread occurrence of HHR motifs in genomes from bacteria to eukaryotes,Citation24 including humans.Citation25 These observations were confirmed and extended by other laboratories,Citation26-28 revealing the HHR as a ubiquitous catalytic RNA motif in all life kingdoms.Citation29 Other small self-cleaving RNAs, such as the HDVCitation30 and twister ribozymes,Citation14 have also been found widespread in DNA genomes, which corroborates that small catalytic RNAs are much more frequent than previously thought. Although the precise biologic roles of these genomic ribozymes are not well understood, a tight connection with mobile genetic elements such as retrotransposons has been reported in diverse eukaryotes.Citation31-34 Retrotransposons are major components of most eukaryotic genomes, and can be classified in autonomous and non-autonomous. Autonomous retrotransposons encode the protein factors required for their own mobilization, which includes a retrotranscriptase (RT) responsible for cDNA synthesis from an RNA transposition intermediate. Eukaryotic genomes can also contain many copies of small non-autonomous retroelements, which do not encode any protein and whose genomic mobility depends on the autonomous retrotransposons.Citation35
Figure 1. Representation of the possible hammerhead ribozyme (HHR) topologies. The conserved nucleotides of the HHR are boxed. Loop-loop interactions are also indicated. Dotted and continuous lines refer to non-canonical and Watson-Crick base pairs, respectively. The most typical lengths of the helix stems for each HHR type are drawn. N stands for any nucleotide, whereas R stands for purines (A or G), Y for pyrimidines (U or C) and H for either A, U or C.
Hammerhead ribozymes in plant genomes are part of a new family of non-autonomous retroelements: The retrozymes
We previously reported the presence in some plant genomes of HHR motifs, which in some cases occur as tandem repeats of a few hundred base pairs.Citation24 More recent and deeper bioinformatic searches have extended these observations to the genomes of more than 40 plant species.Citation34 Comparative genomic analysis revealed that the tandem HHR motifs were embedded within the sequence of what constitutes a novel family of non-autonomous retroelements, the retrozymes (retrotransposons with hammerhead ribozymes). These retroelements have sizes that range from around 1 to 1,5 kb, and show almost no sequence homology among distant plant genomes. All retrozymes, however, do display a similar structure: they are delimited by 4 bp target-site duplications (TSDs), with the HHRs embedded in direct long-terminal repeats (LTRs) of ∼300–400 bp delimiting a unique central region (∼300–600 bp), and flanked by the primer binding site or PBS (complementary to the tRNAMet sequence) and a poly-purine tract (PPT), both sequences required to prime DNA synthesis during the mobilization of LTR-retrotransposonsCitation36 (). Retrozymes are similar to other non-autonomous retroelements of plants such as TRIMsCitation37 and SMARTsCitation38 () in that they rely on the machinery encoded by autonomous retrotransposons for their mobilization, most likely of the Ty3-gypsy family in the case of retrozymes.Citation34 Plant retrozyme RNAs showed high self-cleaving activity in vitro, whereas northern blot hybridizations of RNAs from different plant tissues revealed abundant levels of circular and linear RNAs of the precise size encompassed by the HHRs,Citation34 which is an indication of self-processing activity during in vivo transcription (). Despite the lack of sequence identity between most retrozymes (with the exception of the short PPT, PBS and HHR motifs), secondary structure predictions for these circRNAs show a similar compact architecture with high stability (), suggesting a selection pressure at this level. A final feature of retrozymes is their occurrence in vivo as heterogeneous RNA sequences of both the positive and negative polarities, which suggests that retrozyme circRNAs could be undergoing RNA-to-RNA replication through a rolling-circle mechanism, similar to the ones described for viroids and viral RNA satellites.Citation34
Figure 2. Sequence features of genomic retrozymes. (A) Schematic representation (top) of a full genomic retrozyme element. Target Site Duplications (TSDs) delimiting the retrozyme are shown in gray. Long terminal repeats (LTRs) are shown in blue. The positions of the primer binding site (PBS), the polypurine tract (PPT) and the hammerhead ribozymes (HHR) are indicated. The self-cleavage sites (SC) delimiting the retrozyme RNA are indicated with arrows. The resulting self-cleaved retrozyme RNA after transcription (middle) and circularization (bottom) are indicated. (B) Schematic representation of 2 examples of plant small non-autonomous LTR-retrotransposons such as TRIMs (top) and SMARTs (bottom) retroelements.
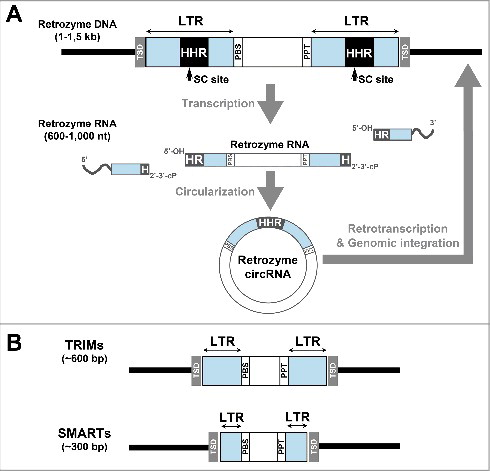
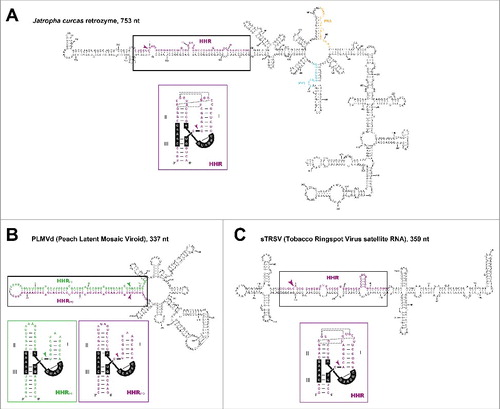
Figure 3. Minimum free energy secondary structure predictions for (A) a retrozyme circRNA of Jatropha curcas (Entry KX273075.1), (B) the avsunviroid PLMVd (Entry M83545.1) and (C) the Nepovirus satellite RNA sTRSV (Entry M14879.1). HHR sequences are shown in purple (positive polarity) and green (negative polarity), and the PBS and PPT motifs of the retrozyme are shown in orange and blue, respectively. The corresponding structures of the HHRs motifs are shown under each circRNA structure and, with the exception of PLMVd HHRs, dotted lines indicate putative tertiary interactions between HHR loops. Self-cleavage sites are indicated with arrows. Kissing-loop interaction of PLMVd is also shown. Numbering for each circRNA starts at the self-cleavage site of the positive polarity HHR.
The multiple connections between retrozyme and viroidal circRNAs
The first circRNAs reported in the literature were discovered in the 70s and called viroids.Citation39,40 Based on different features, these small (240–470 nt) infectious non-protein coding circRNAs have been classified in 2 different families: Avsunviroidae and Pospiviroidae.Citation41 Although a detailed description of the biology of these minimal plant pathogens can be found elsewhere,Citation42 here we will summarize the major attributes of these 2 families. Members of the family Avsunviroidae (4 species, ) are characterized by the presence of HHR motifs in both polarity strands and show no sequence similarity between them, whereas their RNA secondary structure can be either rod-like or highly branched () depending on the viroid size. On the other hand, members of the family Pospiviroidae (28 species) do not possess HHRs but several conserved sequence motifs, and show a rod-like secondary structure. In the 80s, a second group of infectious circRNAs with HHRs was found encapsidated in helper plant RNA viruses and called viroid-like satellite RNAs.Citation43 The 9 known species of viroid-like satellites share many similarities with viroids (), such as being composed of a small (220–460 nt) circular RNA with a high degree of base pairing (), replication through an analogous rolling-circle mechanism, and lack of protein-coding capacity (although an extraordinary case has been reported recentlyCitation44). Furthermore, as observed for the Avsunviroidae, viroid-like satellites show very little sequence similarity among them and all encode HHR self-cleaving motifs in one or both polarity strands. In this regard, retrozyme RNAs fit well the general description of viroidal RNAs with HHRs (either viroids, viral satellites, or other viroid-like RNAs. ). Genomic retrozymes are transcribed into self-processing circRNAs of slightly larger sizes (600–1000 nt), which are also predicted to have a stable branched secondary structure with a high degree (above 70%) of self-pairing (). Moreover, the circRNAs of both retrozyme and viroidal RNAs with HHRs show a long stem containing the self-cleaving HHR motif paired with a highly complementary sequence (), which most likely helps to prevent self-cleavage of the circRNA. RNAs from most retrozymes and viroid-like sequences show similar higher G-C content (∼55%, ), with guanine as the most frequent nucleotide (∼30%), and adenine and uracil around 22% each. A final point in common between retrozymes and viroidal RNAs with HHRs is the efficient in vitro circularization of the linear double self-cleaved RNAs by plant tRNA ligases, which suggests that both types of elements would require this enzyme for in vivo circRNA synthesis.Citation34,45
Table 1. Compilation of circRNAs with hammerhead ribozymes from sequence databases.
Looking for the origin of viroidal RNAs: Escaped introns, transposons or relics from the RNA world?
Since their discovery, infectious circRNAs have been considered as the lowest step of the biologic scale (so-called subviral agents) due to their minimal genome size and extreme simplicity as autonomously replicating entities. Regarding their possible origin, the first discovered viroids were proposed to be escaped spliceosomal introns as a result of some sequence similarity with snRNAs.Citation46 The discovery and analysis of more viroid-like sequences led to propose a hypothetical origin from different retrotransposable elements based again on primary sequence similarities with either Ty-1 retrotransposonsCitation47 or Group I introns.Citation48 With the landmark discovery of ribozymes as the most ancient biocatalysts, a latter hypothesis for the origin of the infectious circRNAs in the frontier of life posited that they might be “living fossils” from the primordial RNA world.Citation49,50 Among the reasons supporting this idea are that the first entities of the RNA world should also have been small RNA replicons, most probably circular, with no protein-coding capacity but having simple self-processing ribozyme activities like the HHR.Citation51 The monophyletic origin proposed for viroidal RNAs also lends support to the primordial origin hypothesis,Citation52 although, due to the extremely small and fast-evolving genomes of these pathogens, such a common evolutionary origin should be regarded with caution.Citation53 Nevertheless, the assumption of viroidal RNAs as survivors of the RNA world raises different questions difficult to explain,Citation54 particularly, the identification of a reasonable evolutionary path accounting for the presence of these putative RNA fossils only in flowering plants (originated ∼200 million years ago) but their absence in any ancestor of these plants, from algae to prokaryotes (4,100 million years ago). However, this direct connection between the RNA world and infectious circRNAs has been favored in the literatureCitation50,51,55,56 over the less fascinating hypothesis of escaped retroelements and/or introns. In this regard, the recent discovery of the widespread occurrence of small self-cleaving RNA motifs, such as the HHR or the HDV ribozymes, in viral, bacterial, archaeal, and eukaryotic genomes indicates that small ribozymes are not restricted to subviral RNA agents, such as viroidal RNAs with HHRs or the human Hepatitis-δ agent, but are very frequent components of DNA genomes. Furthermore, the discovery of a new family of retroelements with HHRs that spread through circRNAs, precisely in the genomes of flowering plants, as well as their structural similarities with infectious circRNAs, opens a more likely scenario where viral satellites and viroids with HHRs may have emerged de novo from the population of abundant retrozyme circRNAs present in plant transcriptomes. In this respect, it seems plausible that the circRNAs encoded by genomic retrozymes may be encapsidated by RNA viruses during plant infection in a similar way as reported for other host RNAs derived from Ty3-gypsy retroelements.Citation57 Viral encapsidation of a retrozyme circRNA would be the first step in the biogenesis of a viroid-like satellite RNA, which would subsequently require the acquisition of recognition signals for the viral RNA polymerase to be replicated and, eventually, a second ribozyme in the opposite polarity (). On the other hand, de novo appearance of a HHR viroid from retrozyme circRNAs seems also feasible and would entail the acquisition of recognition motifs for plant RNA polymerases (a signal that may be already present, see before andCitation34) and cell-to-cell movement, as well as the appearance of a second HHR in the negative polarity. In this regard, the architecture of retrozyme circRNAs showing a highly self-paired HHR already offers a quasi-HHR sequence in the opposite polarity (). Viroidal RNAs with HHRs would not be the first infectious agents for which a cellular origin is suspected. The discovery of a novel class of cellular RT genes present in all major taxonomic groups but absent from selfish elements indicated that retrovirus evolved from genomic LTR retrotransposons rather than in the other way around.Citation58,59 In view of all these data, an in planta origin for viroidal RNAs with HHRs would seem to us as a more realistic hypothesis than being ancient relics of precellular evolution.
Discussion
As recently reported, genomic retrozymes with HHRs show a patchy distribution among flowering plants, occurring numerously in different species, but being absent in some others.Citation34 “Canonical” retrozymes (which contain HHRs, PBS and PPT motifs) seem to be mostly restricted to dicots, although the presence of putative retrozyme sequences with the characteristic tandem HHR copies have also been detected in primitive land plants (such as the spikemoss Selaginella moellendorfii), algae (such as Chlamydomonas reinhardtii), and even protists (such as oomycetes),Citation24 as well as in many metazoans,Citation24-29,34 suggesting that genome-encoded circRNAs with HHRs could occur in eukaryotes other than angiosperms. This scenario allows to propose a much simpler evolutionary path for small infectious circRNAs with HHRs of plants, which may have come by chance from the abundant reservoirs of retrozyme circRNAs present in plant transcriptomes. An obvious counter-argument is that retrozymes themselves could have originated from viroidal RNAs. Although this possibility cannot be ruled out, a better answer to this question can be found in the ancient family of the Penelope-like elements or PLEs.Citation60 PLEs are a large family of retrotransposons found in many eukaryotes (including diatoms, algae and primitive land plants such as sellaginellas), which show phylogenetic connections with prokaryotic self-splicing introns and are believed to predate telomerases and most eukaryotic retrotransposons.Citation60,61 Interestingly, PLEs show in their LTRs the presence of conserved HHRs, which are most likely involved in the self-processing of the RNA transposition intermediates of the retroelement.Citation31 Consequently, genomic HHRs and retrozymes present in modern angiosperms could derive evolutionarily from PLEs and other related genomic HHR-sequences present in primitive selaginella plants. This possibility offers a more parsimonious path to the origin of retrozymes in higher plants, which could be regarded as simpler genomic parasites descended from PLEs, and consisting primarily of HHRs and cis signals for their recognition and genomic replication by the machinery of autonomous retrotransposons of the plant. Reduction of size and complexity are well-known trends in the evolution of parasitic and endosymbiotic genomes, and in this line, viroids (∼300 nt) would represent a step further in this genome reduction from retrozymes (∼700 nt). Of course, our proposed origin of current viroidal RNAs with HHRs from plant retrozymes does not totally rule out the primordial origin proposed by Diener.Citation49 However, while there is no reported evidence of viroidal RNAs present in cyanobacteria or any other prokaryotes, the de novo origin of viroids and viroid-like RNA satellites seems the most plausible theory, which would also imply that future threats of this type may be originated in plants at higher frequency than expected.
Concluding remarks
It has been long known that circular DNAs are common molecules in the biosphere, from prokaryotic plasmids to the genomes of most bacteriophages, bacteria, archaea and plastids. Circular RNAs, however, have been regarded as rare in biology until the recent confirmation that numerous life forms express stable circRNAs of different origins. Among them, it is noteworthy the finding of a myriad of splicing-derived circRNAs in eukaryotes with potential functions in transcription, splicing, or the biogenesis of small RNAs (for a review seeCitation62). In this regard, genome-encoded circRNAs with self-cleaving ribozymes represent a new level of complexity, whose study will offer functional clues as well as biotechnological applications in the fast-growing field of circular RNA molecules.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Funding for this work was provided by the Ministerio de Economía y Competitividad of Spain and FEDER (BFU2014–56094-P).
References
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982; 31:147-57; PMID:6297745; https://doi.org/10.1016/0092-8674(82)90414-7
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983; 35:849-57; PMID:6197186; https://doi.org/10.1016/0092-8674(83)90117-4
- Crick FH. The origin of the genetic code. J Mol Biol 1968; 38:367-79; PMID:4887876; https://doi.org/10.1016/0022-2836(68)90392-6
- Orgel LE. Evolution of the genetic apparatus. J Mol Biol 1968; 38:381-93; PMID:5718557; https://doi.org/10.1016/0022-2836(68)90393-8
- Woese CR. The fundamental nature of the genetic code: Prebiotic interactions between polynucleotides and polyamino acids or their derivatives. Proc Natl Acad Sci U S A 1968; 59:110-7; PMID:5242115; https://doi.org/10.1073/pnas.59.1.110
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. RNA catalyses nuclear pre-mRNA splicing. Nature 2013; 503:229-34; PMID:24196718; https://doi.org/10.1038/nature12734
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science 2000; 289:920-30; PMID:10937990; https://doi.org/10.1126/science.289.5481.920
- Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science 1986; 231:1577-80; PMID:17833317; https://doi.org/10.1126/science.231.4745.1577
- Hutchins CJ, Rathjen PD, Forster AC, Symons RH. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res 1986; 14:3627-40; PMID:3714492; https://doi.org/10.1093/nar/14.9.3627
- Buzayan JM, Gerlach WL, Bruening G. Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature 1986; 323:349-53; https://doi.org/10.1038/323349a0
- Kuo MY, Sharmeen L, Dinter-Gottlieb G, Taylor J. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J Virol 1988; 62:4439-44; PMID:3184270
- Saville BJ, Collins RA. A site-specific self-cleavage reaction performed by a novel RNA in neurospora mitochondria. Cell 1990; 61:685-96; PMID:2160856; https://doi.org/10.1016/0092-8674(90)90480-3
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 2004; 428:281-6; PMID:15029187; https://doi.org/10.1038/nature02362
- Roth A, Weinberg Z, Chen AG, Kim PB, Ames TD, Breaker RR. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat Chem Biol 2014; 10:56-60; PMID:24240507; https://doi.org/10.1038/nchembio.1386
- Weinberg Z, Kim PB, Chen TH, Li S, Harris KA, Lunse CE, Breaker RR. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol 2015; 11:606-10; PMID:26167874; https://doi.org/10.1038/nchembio.1846
- De la Peña M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J 2003; 22:5561-70; PMID:14532128; https://doi.org/10.1093/emboj/cdg530
- Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol 2003; 10:708-12; PMID:12881719; https://doi.org/10.1038/nsb959
- Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 2006; 126:309-20; PMID:16859740; https://doi.org/10.1016/j.cell.2006.06.036
- Przybilski R, Graf S, Lescoute A, Nellen W, Westhof E, Steger G, Hammann C. Functional hammerhead ribozymes naturally encoded in the genome of arabidopsis thaliana. Plant Cell 2005; 17:1877-85; PMID:15937227; https://doi.org/10.1105/tpc.105.032730
- Ferbeyre G, Smith JM, Cedergren R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol 1998; 18:3880-8; PMID:9632772; https://doi.org/10.1128/MCB.18.7.3880
- Rojas AA, Vazquez-Tello A, Ferbeyre G, Venanzetti F, Bachmann L, Paquin B, Sbordoni V, Cedergren R. Hammerhead-mediated processing of satellite pDo500 family transcripts from dolichopoda cave crickets. Nucleic Acids Res 2000; 28:4037-43; PMID:11024185; https://doi.org/10.1093/nar/28.20.4037
- Epstein LM, Gall JG. Self-cleaving transcripts of satellite DNA from the newt. Cell 1987; 48:535-43; PMID:2433049; https://doi.org/10.1016/0092-8674(87)90204-2
- Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature 2008; 454:899-902; PMID:18615019; https://doi.org/10.1038/nature07117
- De la Peña M, Garcia-Robles I. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA 2010; 16:1943-50; PMID:20705646; https://doi.org/10.1261/rna.2130310
- De la Peña M, Garcia-Robles I. Intronic hammerhead ribozymes are ultraconserved in the human genome. EMBO Rep 2010; 11:711-6; PMID:20651741; https://doi.org/10.1038/embor.2010.100
- Jimenez RM, Delwart E, Luptak A. Structure-based search reveals hammerhead ribozymes in the human microbiome. J Biol Chem 2011; 286:7737-43; PMID:21257745; https://doi.org/10.1074/jbc.C110.209288
- Perreault J, Weinberg Z, Roth A, Popescu O, Chartrand P, Ferbeyre G, Breaker RR. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput Biol 2011; 7:e1002031; PMID:21573207; https://doi.org/10.1371/journal.pcbi.1002031
- Seehafer C, Kalweit A, Steger G, Gräf S, Hammann C. From alpaca to zebrafish: Hammerhead ribozymes wherever you look. RNA 2011; 17:21-6; PMID:21081661; https://doi.org/10.1261/rna.2429911
- Hammann C, Luptak A, Perreault J, de la Peña M. The ubiquitous hammerhead ribozyme. RNA 2012; 18:871-85; PMID:22454536; https://doi.org/10.1261/rna.031401.111
- Webb CH, Riccitelli NJ, Ruminski DJ, Luptak A. Widespread occurrence of self-cleaving ribozymes. Science 2009; 326:953; PMID:19965505; https://doi.org/10.1126/science.1178084
- Cervera A, De la Peña M. Eukaryotic penelope-like retroelements encode hammerhead ribozyme motifs. Mol Biol Evol 2014; 31:2941-7; PMID:25135949; https://doi.org/10.1093/molbev/msu232
- Eickbush DG, Eickbush TH. R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA cotranscript. Mol Cell Biol 2010; 30:3142-50; PMID:20421411; https://doi.org/10.1128/MCB.00300-10
- Ruminski DJ, Webb CH, Riccitelli NJ, Luptak A. Processing and translation initiation of non-long terminal repeat retrotransposons by hepatitis delta virus (HDV)-like self-cleaving ribozymes. J Biol Chem 2011; 286:41286-95; PMID:21994949; https://doi.org/10.1074/jbc.M111.297283
- Cervera A, Urbina D, de la Peña M. Retrozymes are a unique family of non-autonomous retrotransposons with hammerhead ribozymes that propagate in plants through circular RNAs. Genome Biol 2016; 17:135; PMID:27339130; https://doi.org/10.1186/s13059-016-1002-4
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet 2007; 8:973-82; PMID:17984973; https://doi.org/10.1038/nrg2165
- Gorinsek B, Gubensek F, Kordis D. Evolutionary genomics of chromoviruses in eukaryotes. Mol Biol Evol 2004; 21:781-98; PMID:14739248; https://doi.org/10.1093/molbev/msh057
- Witte CP, Le QH, Bureau T, Kumar A. Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc Natl Acad Sci U S A 2001; 98:13778-83; PMID:11717436; https://doi.org/10.1073/pnas.241341898
- Gao D, Chen J, Chen M, Meyers BC, Jackson S. A highly conserved, small LTR retrotransposon that preferentially targets genes in grass genomes. PloS One 2012; 7:e32010; PMID:22359654; https://doi.org/10.1371/journal.pone.0032010
- Diener TO. Potato spindle tuber “virus.” IV. A replicating, low molecular weight RNA. Virology 1971; 45:411-28; PMID:5095900; https://doi.org/10.1016/0042-6822(71)90342-4
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976; 73:3852-6; PMID:1069269; https://doi.org/10.1073/pnas.73.11.3852
- Di Serio F, Flores R, Verhoeven JT, Li SF, Pallas V, Randles JW, Sano T, Vidalakis G, Owens RA. Current status of viroid taxonomy. Arch Virol 2014; 159:3467-78; PMID:25216773; https://doi.org/10.1007/s00705-014-2200-6
- Flores R, Minoia S, Carbonell A, Gisel A, Delgado S, Lopez-Carrasco A, Navarro B, Di Serio F. Viroids, the simplest RNA replicons: How they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res 2015; 209:136-45; PMID:25738582; https://doi.org/10.1016/j.virusres.2015.02.027
- Randles JW, Davies C, Hatta T, Gould AR, Francki RI. Studies on encapsidated viroid-like RNA I. Characterization of velvet tobacco mottle virus. Virology 1981; 108:111-22; PMID:18635027; https://doi.org/10.1016/0042-6822(81)90531-6
- AbouHaidar MG, Venkataraman S, Golshani A, Liu B, Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci U S A 2014; 111:14542-7; PMID:25253891; https://doi.org/10.1073/pnas.1402814111
- Nohales MA, Molina-Serrano D, Flores R, Daros JA. Involvement of the chloroplastic isoform of tRNA ligase in the replication of viroids belonging to the family avsunviroidae. J Virol 2012; 86:8269-76; PMID:22623792; https://doi.org/10.1128/JVI.00629-12
- Diener TO. Are viroids escaped introns? Proc Natl Acad Sci U S A 1981; 78:5014-5; PMID:16593072; https://doi.org/10.1073/pnas.78.8.5014
- Kiefer MC, Owens RA, Diener TO. Structural similarities between viroids and transposable genetic elements. Proc Natl Acad Sci U S A 1983; 80:6234-8; PMID:6312450; https://doi.org/10.1073/pnas.80.20.6234
- Dinter-Gottlieb G. Viroids and virusoids are related to group I introns. Proc Natl Acad Sci U S A 1986; 83:6250-4; PMID:3462692; https://doi.org/10.1073/pnas.83.17.6250
- Diener TO. Circular RNAs: Relics of precellular evolution? Proc Natl Acad Sci U S A 1989; 86:9370-4; PMID:2480600; https://doi.org/10.1073/pnas.86.23.9370
- Diener TO. Viroids: “living fossils” of primordial RNAs? Biol Direct 2016; 11:15; PMID:27016066; https://doi.org/10.1186/s13062-016-0116-7
- Ma W, Yu C, Zhang W. Circularity and self-cleavage as a strategy for the emergence of a chromosome in the RNA-based protocell. Biol Direct 2013; 8:21; PMID:23971788; https://doi.org/10.1186/1745-6150-8-21
- Elena SF, Dopazo J, de la Peña M, Flores R, Diener TO, Moya A. Phylogenetic analysis of viroid and viroid-like satellite RNAs from plants: A reassessment. J Mol Evol 2001; 53:155-9; PMID:11479686; https://doi.org/10.1007/s002390010203
- Jenkins GM, Woelk CH, Rambaut A, Holmes EC. Testing the extent of sequence similarity among viroids, satellite RNAs, and hepatitis delta virus. J Mol Evol 2000; 50:98-102; PMID:10654264; https://doi.org/10.1007/s002399910011
- Chela-Flores J. Are Viroids Molecular Fossils of the RNA World? J Theor Biol 1994; 166:163-6.
- Bussiere F, Lafontaine D, Cote F, Beaudry D, Perreault JP. Evidence for a model ancestral viroid. Nucleic Acids Symp Ser 1995:143-4; PMID:8643352
- Flores R, Gago-Zachert S, Serra P, Sanjuan R, Elena SF. Viroids: Survivors from the RNA world? Annu Rev microbiol 2014; 68:395-414; PMID:25002087; https://doi.org/10.1146/annurev-micro-091313-103416
- Ghoshal K, Theilmann J, Reade R, Maghodia A, Rochon D. Encapsidation of Host RNAs by cucumber necrosis virus coat protein during both agroinfiltration and infection. J Virol 2015; 89:10748-61; PMID:26269190; https://doi.org/10.1128/JVI.01466-15
- Gladyshev EA, Arkhipova IR. A widespread class of reverse transcriptase-related cellular genes. Proc Natl Acad Sci U S A 2011; 108:20311-6; PMID:21876125; https://doi.org/10.1073/pnas.1100266108
- Koonin EV, Dolja VV. A virocentric perspective on the evolution of life. Curr Opin Virol 2013; 3:546-57; PMID:23850169; https://doi.org/10.1016/j.coviro.2013.06.008
- Evgen'ev MB, Zelentsova H, Shostak N, Kozitsina M, Barskyi V, Lankenau DH, Corces VG. Penelope, a new family of transposable elements and its possible role in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci U S A 1997; 94:196-201; PMID:8990185; https://doi.org/10.1073/pnas.94.1.196
- Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci U S A 2007; 104:9352-7; PMID:17483479; https://doi.org/10.1073/pnas.0702741104
- Barrett SP, Salzman J. Circular RNAs: Analysis, expression and potential functions. Development 2016; 143:1838-47; PMID:27246710; https://doi.org/10.1242/dev.128074
- Wu Q, Wang Y, Cao M, Pantaleo V, Burgyan J, Li WX, Ding SW. Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc Natl Acad Sci U S A 2012; 109:3938-43; PMID:22345560; https://doi.org/10.1073/pnas.1117815109
- Zhang Z, Qi S, Tang N, Zhang X, Chen S, Zhu P, Ma L, Cheng J, Xu Y, Lu M, et al. Discovery of replicating circular RNAs by RNA-seq and computational algorithms. PLoS Pathog 2014; 10:e1004553; PMID:25503469; https://doi.org/10.1371/journal.ppat.1004553
- Wang WB, Fei JM, Wu Y, Bai XC, Yu F, Shi GF, Li YF, Kuai YZ. A new report of a mosaic dwarf viroid-like disease on mulberry trees in China. Pol J Microbiol 2010; 59:33-6; PMID:20568527
