ABSTRACT
PIWI-interacting RNAs (piRNAs) and associated proteins comprise a conserved pathway for silencing transposons in metazoan germlines. piRNA pathway components are also expressed in multipotent somatic stem cells in various organisms. piRNA functions have been extensively explored in bilaterian model systems, however, comprehensive studies in non-bilaterian phyla remain limited. Here we investigate the piRNA pathway during the development of Nematostella vectensis, a well-established model system belonging to Cnidaria, the sister group to Bilateria. To date, no population of somatic stem cells has been identified in this organism, despite its long life-span and regenerative capacities that require a constant cell-renewal. We show that Nematostella piRNA pathway components are broadly expressed in early developmental stages, while piRNAs themselves show differential expression, suggesting specific developmental roles of distinct piRNA families. In adults, piRNA associated proteins are enriched in the germline but also expressed in somatic cells, indicating putative stem cell properties. Furthermore, we provide experimental evidence that Nematostella piRNAs cleave transposable elements as well as protein-coding genes. Our results demonstrate that somatic expression of piRNA associated proteins as well as the roles of piRNAs in transposon repression and gene regulation are likely ancestral features that evolved before the split between Cnidaria and Bilateria.
Introduction
The discovery of small RNAs has greatly altered our understanding of gene regulation.Citation1 Yet, PIWI-interacting RNAs (piRNAs) remain a relatively enigmatic group of small RNAs. These 24–31 nucleotides-long small RNAs guide PIWI proteins, a subclade of the Argonaute (AGO) family, to repress transposons and safeguard the genome from their deleterious defects. piRNAs are transcribed as long, single-stranded precursors from piRNA clusters, genomic loci enriched with truncated transposon sequences, or active transposons.Citation2,3 Precursor transcripts are processed into primary piRNAs with a U at position 1 in distinct cytoplasmic perinuclear foci collectively called nuage (“cloud” in French), due to their appearance in TEM images.Citation4,5 There, secondary piRNAs complementary in 10 nucleotides with primary piRNA and a corresponding A at position 10 are generated via the ping pong cycle, a mechanism combining transposon destruction and amplification of piRNAs.Citation4 The DEAD box helicase Vasa is a conserved player in this cycle facilitating the transfer of precursor piRNAs between PIWI proteins in the nuage.Citation6,7 Like PIWI proteins, Vasa is required for germ cell specification and maintenance. Thus, zebrafish, mouse and Drosophila mutant for piwi or vasa genes are sterile.Citation8-10 Consequently, PIWI and Vasa proteins are reliable markers for the germline in these model systems. In addition, piwi and vasa expression is commonly found in multipotent somatic stem cells in bilaterian animals with a high regeneration capacity such as planarians and polychaetes but also in early branching, morphologically simple animals including sponges and cnidarians.Citation9,10 Therefore, these piRNA pathway components have been suggested to be part of an ancestral gene repertoire conserved from sponges to vertebrates responsible for maintenance of “stemness” in germline stem cells as well as somatic stem cells, referred to as germline mulitpotency program (GMP).Citation11,12
The phylum Cnidaria, which includes corals, sea anemones, jellyfish and hydroids, is the sister group to BilateriaCitation13 and thus provides a unique opportunity to study the evolution of shared mechanisms and pathways. Recently, piRNAs and PIWI proteins were identified in the cnidarian Hydra.Citation14-16 Hywi and Hyli (the two paralogous Hydra PIWI proteins) are predominantly expressed in the germline as well as in the interstitial cell lineage (the so-called i-cells) including multipotent progenitor stem cells and germline stem cells, but also in epithelial stem cells. Hydra piRNAs comprise a large fraction among their entire repertoire of small RNAs and map to transposons. Interestingly, transposon targeting seems to be largely restricted to the i-cell population. However, i-cells appear to be specific to hydrozoans and are not found in other members of the diverse phylum Cnidaria, such as the Anthozoa (sea anemones and corals).
The sea anemone Nematostella vectensis has emerged as a leading anthozoan model organism. Its ability to readily complete its life cycle under laboratory conditions makes it perfectly suitable to study the piRNA pathway and its implications during cnidarian development. Here, we identify and quantify key components of the piRNA pathway known from Bilateria and characterize their expression patterns at different developmental stages as well as in different tissues of adult female and male sea anemones. The somatic expression of piwi and vasa genes suggests multipotency and stem cell character of adult somatic cells. In addition, we show that the highly abundant piRNAs in Nematostella are differentially expressed across development and provide insights into their function in transposon repression and regulation of protein-coding genes.
Results
piRNAs are an extremely abundant class of small RNAs in Nematostella
We previously reported a data set of Nematostella small RNAs obtained from eight developmental stages including female and male adults.Citation17 For each stage two libraries were generated: One untreated and one sodium periodate-treated library to remove small RNA classes lacking 2′-O-methylation. We considered reads as piRNAs if they mapped to no other annotated small RNAs or coding regions in the Nematostella genome (see Materials and Methods for details) and confirmed that these are indeed piRNAs by showing that they exhibit a so-called “ping-pong signature” (Supplemental Fig. S1).
In a pioneering study of small RNAs in basally-branching metazoans, Grimson and colleagues identified piRNAs as the most abundant small RNAs in Nematostella.Citation18 To assess what fraction of small RNAs is represented by piRNAs in different species, we compared small RNA reads from Nematostella, third trimester human fetal ovaries,Citation19 Drosophila melanogaster adult ovariesCitation20 and Hydra magnipapillata whole adult polypsCitation16 (Supplemental Fig. S2). We chose Drosophila and human ovaries as relevant comparative tissues for our analysis since piRNAs are generally enriched in gonads in Bilateria.Citation2,21
Clearly, the piRNA fraction is highest in Nematostella at every developmental stage compared with other species and tissues. The proportion of piRNAs among small RNAs in Hydra is slightly smaller but comparable to that of Nematostella, indicating that the high proportion of piRNAs among small RNAs is probably common to Cnidaria.
Nematostella encodes 2 bona fide PIWI proteins and a degenerated homolog
Driven by the finding of a very large piRNA population among small RNAs, we decided to study the Nematostella homologs of the major players in the bilaterian piRNA pathway. PIWI proteins consist of three functional domains that are highly conserved across Metazoa: The PAZ and the MID domain are responsible for anchoring the incorporated piRNA and the PIWI domain performs the endonucleolytic cleavage of the target transcript. Using BLAST searches, we identified candidate gene models,Citation22 initially corresponding to 3 piwi genes in Nematostella. Piwi1 and Piwi2 encode bona fide PIWI proteins both containing the PAZ and PIWI domains. The MID domain, which is usually situated between the PAZ and PIWI domain, was not identified via motif search programs, but we detected identities of 43–56% between the MID domains of the Hydra PIWIs Hywi and Hyli and the putative MID domains of Nematostella PIWIs. The third piwi gene, however, encodes a truncated PIWI protein harboring a stop codon that interrupts the PAZ domain, and a PIWI domain, which is split between two reading frames. Consequently, the protein product of this gene is not a functional PIWI protein. To reconstruct the phylogenetic relationship of the bona fide Nematostella PIWIs (Piwi1 and Piwi2), we performed maximum likelihood and Bayesian analyses that both resulted in the same tree topology of metazoan PIWI proteins (). PIWIs of the ctenophore Pleurobrachia bachei (PbaPiwi1 and PbaPiwi2) form a sister group to all other animal PIWI proteins. Nematostella PIWIs fall into two clearly distinct clades and cluster with other cnidarian homologs. The cnidarian Piwi2 clade is more closely related to the vertebrate PiwiL2, the lophotrochozoan Piwi2 and the ecdysozoan Ago3 group. The other clade includes cnidarian Piwi1 and sponge PiwiA and Piwi-like1 homologs.
Figure 1. Phylogenetic relationship of Nematostella PIWI proteins. Phylogenetic tree of full sequences of metazoan PIWI proteins. Maximum likelihood was calculated with the LG model (+I, +G, +F). Bootstrap support values above 50% appear next to the respective node. Bayesian analysis was performed by the WAG model. Posterior probabilities (PP) are indicated by green (PP = 1), blue (0.95 ≤ PP < 1.0) or red (0.7 ≤ PP < 0.95) asterisks. Species abbreviations: Porifera (sponges): Aqu, Amphimedon queenslandica; Efl, Ephydatia fluviatilis; Ctenophora (comb jellies): Pba, Pleurobrachia bachei (sea gooseberry); Cnidaria: Ami, Acropora millepora (staghorn coral); Apa, Aiptasia pallida (brown anemone); Aau, Aurelia aurita (moon jelly); Che; Clytia hemisphaerica; Hvu; Hydra vulgaris (fresh water polyp); Nbi, Nanomia bijuga; Nve, Nematostella vectensis (starlet sea anemone); Pca, Podocoryne carnea. Lophotrochozoa: Lgi, Lottia gigantea (owl limpet); Vli, Villosa lienosa (little spectaclecase). Ecdysozoa: Bmo, Bombyx mori (silkworm); Dme, Drosophila melanogaster (fruit fly); Ptr, Portunus trituberculatus (horse crab). Vertebrata: Hsa, Homo sapiens (human); Dre, Danio rerio (zebrafish); Mmu, Mus musculus (mouse).
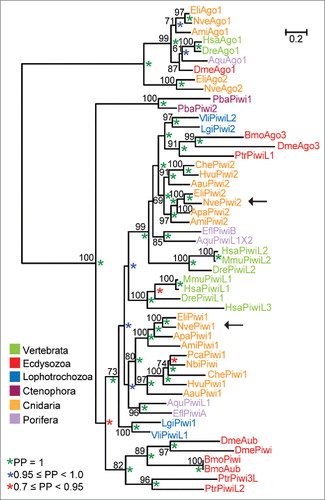
We also determined the phylogenetic position of the truncated Nematostella PIWI candidate and thereby found that it is a lineage specific duplication of Piwi1 (Supplemental Fig. S3). It is conceivable that this is either an example of a transition from a protein-coding gene to a pseudogene, which acquired nonsense and frameshift mutations in functional domains after duplication, or, alternatively, underwent neo-functionalization. To emphasize its origin, we thus termed this variant ExPiwi1.
Transcripts of piRNA pathway components are dynamically expressed during development and enriched in adult mesenteries
To obtain an initial overview of the spatiotemporal expression of key bilaterian piRNA pathway components in different developmental stages and various body regions of Nematostella, we determined their transcript levels on the nCounter platform, which enables the direct measurement and quantification of RNA molecules (). We investigated the transcript abundance of Piwi1, Piwi2, ExPiwi1, Vasa1, Vasa2, Armitage, Maelstrom and Spindle-E. The helicases Armitage and Spindle-E are involved in biogenesis and loading of piRNAs into PIWI proteins as well as transposon silencing.Citation23,24 Maelstrom is fundamental for transcriptional transposon silencing and conserved in Hydra, Drosophila and mouse.Citation4,25,26 For comparison, we also included genes that encode microRNA machinery proteins (AGO1, AGO2, Dcr1, Dcr2 and GW182) in our analysis. According to their temporal expression patterns across developmental stages, the investigated genes cluster into three groups (). The first group contains GW182, Dicer2, Piwi1 and Vasa2 and shows strongest expression early in development from the gastrula stage onwards, presumably indicating developmental roles of these genes. The high expression of Piwi1 and Vasa2 in the adult male suggests possible involvement in spermatogenesis. The second group exclusively consists of piRNA pathway components (Piwi2, Vasa1, Maelstrom, Armitage and Spindle-E), whose expression peaks in the unfertilized egg and blastula pointing to maternal inheritance of these factors. Armitage is weakly expressed in subsequent developmental stages but is enriched again in the male adult. The genes AGO1, AGO2, Dicer1 and ExPiwi1 form an outgroup and are strongly expressed in later developmental stages and in adults.
Figure 2. Spatiotemporal expression of transcripts of piRNA pathway proteins. Heat map displaying normalized read counts of homologs of bilaterian genes involved in the piRNA pathway created by direct quantification of transcripts from different developmental stages (A) and various body parts of adult animals (B) by the nCounter platform. Investigated stages include unfertilized egg (UE), blastula (BL), gastrula (GA), 2 d old (early) planula (2d), 4 d old (late) planula (4d), 6 d old metamorphosing planula (6d), 10 d old primary polyp (10d), 2 and 4 months old polyp (2m, 4m) and male and female adult (male and female). Body parts contained mesenteries, pharynx, tentacles and physa (B). Argonaute1 (AGO1), Argonaute2 (AGO2), Dicer1 (Dcr1), Dicer2 (Dcr2) and GW182 comprise the outgroup. Scale bar depicts fold change of expression.
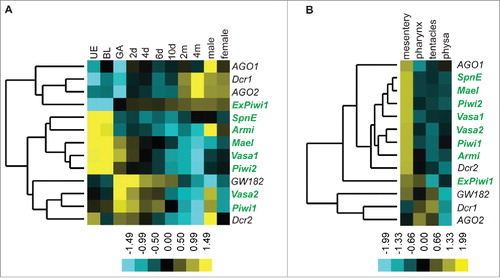
For the expression analysis in adult animals, we used mesenteries (endodermal folds harboring the gonads), pharynges, tentacles and physae (the aboral portion of the body column pinching off during asexual reproduction and capable of rebuilding an entire animal) of females. All investigated piRNA pathway components cluster together and are strongly enriched in the mesenteries () hinting to an enrichment in the germ cells. In the pharynx, tentacles and physa, only low to medium expression levels were detected. ExPiwi1 differs from the rest exhibiting similar expression intensities in the pharynx and the mesenteries (). Piwi1, Piwi2, Vasa1 and Vasa2 show overall stronger expression than other piRNA pathway components and peak in mesenteries (Supplemental Fig. S4A). In addition, they show substantial expression in somatic tissues such as the pharynx, the tentacles and the physa, demonstrating that somatic cells express these factors in the adult stage (Supplemental Fig. S4B).
Nematostella piRNA pathway genes are expressed in broad and overlapping regions during early development
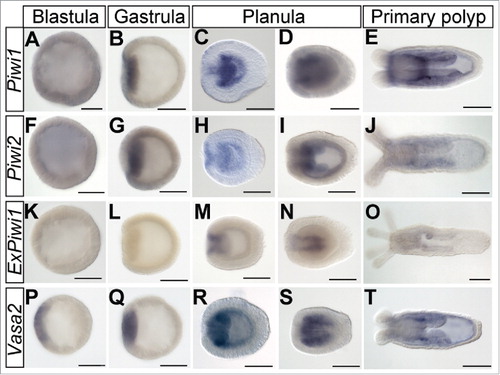
To gain a better spatial resolution of the expression of the canonical piRNA pathway components, we investigated the expression of piwi and vasa homologs by in situ hybridization at different developmental stages (). Piwi1 is weakly expressed throughout the blastula (). In the gastrula stage Piwi1 expression is strongest in the pre-endodermal plate (). It remains strongly expressed throughout the endoderm in the early planula () and accumulates in longitudinal stripes in the pre-pharyngeal endoderm in late planulae () corresponding to the developing mesenteries. This pattern also persists in the primary polyp () in addition to an overall endodermal staining. Within the mesenteries, small areas of stronger expression can be detected. Piwi2 is expressed very similarly to Piwi1 throughout development (). ExPiwi1 is not detectable before the early planula stage, where it is expressed in the pharyngeal ectoderm (). As development progresses, ExPiwi1 staining gets weaker, so that in the polyp it is barely detectable in the pharynx and mesenteries (). An earlier studyCitation27 and our nCounter data ( and Supplemental Fig. S4) showed that Nematostella expresses two vasa genes: Vasa1 and Vasa2. We were not able to obtain expression patterns clearly discernible from background signal for Vasa1 at early developmental stages, however, according to a previous study, Vasa1 is expressed in the oral endoderm and ectoderm of the gastrula and in the forming mesenteries at the planula stage.Citation27 In polyps, the expression is confined to specific areas in the mesenteries.Citation27 Vasa2 is expressed in a patch in the blastula () and later in the pre-endodermal plate of the gastrulating embryo () and the endoderm of the early planula (). At this stage also weak ectodermal expression is detectable. In later planulae and primary polyps, the staining is prominent in the mesenteries and in the entire endoderm. Like for Piwi1, we detect stronger stained patches of cells in the prepharyngeal endoderm. Taken together, piwi and vasa genes are expressed in vast and similar domains throughout early developmental stages of Nematostella.
Figure 3. piwi genes and Vasa2 exhibit broad expression domains in early developmental stages. Spatial expression patterns of Piwi1 (A-E), Piwi2 (F-J), ExPiwi1 (K-O) and Vasa2 (P-T) in early developmental stages ranging from blastula to primary polyp obtained by in situ hybridization. Lateral views, oral pole is oriented to the left. Scale bars represent 100 µm.
piwi and vasa genes are enriched in putative germline stem cells and their division products
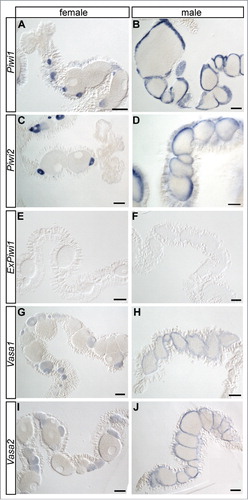
Next, we localized Piwi1, Piwi2, ExPiwi1, Vasa1 and Vasa2 in dissected tissues of adult animals. In females, Piwi1, Piwi2, Vasa1 and Vasa2 are strongly expressed in early oocytes (, , , ). The signal fades with advancing size and maturation stage of the oocytes and cannot be detected in mature oocytes by in situ hybridization. However, our quantification analysis detected transcripts of these piRNA pathway genes in unfertilized eggs ( and Supplemental Fig. S4A). Thus, the lack of signal in mature oocytes likely represents an artifact due to signal dilution or, alternatively, due to impeding yolk content. Besides stained oocytes, small Piwi1- and Piwi2-positive cells are found in the somatic gonad, the epithelium surrounding and nourishing the gonad. These might represent oogonia, female germline stem cells (Supplemental Fig. S5). In male gonads, the expression of Piwi1, Piwi2, Vasa1 and Vasa2 is confined to the outer margins of the gonadal compartments (, , , ). Like in many other animals, the male gonad in Nematostella is stratified reflecting the maturation stage of the germ cells. Spermatogonia occupy the gonadal margin, surrounding different maturation stages of sperm cells with mature sperm cells in the center. The localization of the staining and the general organization of male gonads in various animals implicate that the stained cells in Nematostella are male germline stem cells, spermatogonia, and their early division products. In contrast to Piwi1 and Piwi2, ExPiwi1 is not expressed in female or male gonads. Taken together, apart from ExPiwi1, all piwi and vasa genes are expressed in putative germline stem cells and their division products in the reproductive tissue of Nematostella, consistent with the well characterized role of piwi and vasa homologs in germline establishment and maintenance in numerous bilaterian species.Citation10 Other investigated tissues such as the pharynx and the physa did not show any in situ staining for these genes discernible from background signal (data not shown). In contrast to that, as described above, a direct quantification of transcripts of piRNA pathway components detected substantial expression in somatic tissues like the pharynx, the tentacles and the physa in addition to mesenteries. This discrepancy highlights the detection limit of in situ hybridization (as described in [Citation28]) and shows that although the enrichment of respective genes in the gonads is readily detectable by in situ hybridization, their expression in the soma is too low for this technique.
Figure 4. piwi and vasa genes are expressed in germ cells in adult animals. Spatial expression patterns of Piwi1, Piwi2, ExPiwi1, Vasa1 and Vasa2 in female (A, C, E, G, I) and male gonads (B, D, F, H, J) determined by in situ hybridization. Shown are mesenteries with the gonadal compartments. All genes except for ExPiwi1 are expressed in the early germ cells (early oocytes and cells at the margin of spermaries). Scale bars represent 100 µm.
Nematostella Vasa2 protein is expressed in granules around nuclei
To learn about the intracellular localization of the Vasa protein, we designed two independent custom antibodies directed against 2 different epitopes of Nematostella Vasa2 (see Materials and Methods). We performed Western blot on protein extracts of various tissues obtained from female adults. Both antibodies detected the same molecular weight bands in the physa, mesenteries and the pharynx as well as in whole adult animals (Supplemental Fig. S6A, E). The calculated molecular weight of Vasa2 is 83 kDa and Western blotting revealed bands of approximately 60 and 70 kDa for both antibodies. The higher band falls into the commonly observed divergence between expected and detected protein sizes in Western blots, while the lower band might represent a splicing variant. Clearly, Vasa2 is strongest expressed in the mesenteries compared with the pharynx and physa (Supplemental Fig. S6A, E). In tentacles however, Vasa2 expression is barely detectable. In immunohistochemistry, both antibodies stained the same subcellular structures (Supplemental Fig. S6B-D, F-H), also confirming their specificity.
Vasa2 protein staining is detected in granules surrounding the nuclei throughout the whole development of Nematostella, aside of early oocytes, which show protein foci distributed throughout the ooplasm (, Supplemental Fig. S7). With growing size of the oocytes the signal fades, recapitulating the artifact of impeding yolk content or signal dilution encountered by in situ hybridization. piRNA pathway proteins and associated piRNAs are commonly maternally inherited in other animalsCitation3 and consistently, mass spectrometry analysis identified PIWI and Vasa proteins in mature Nematostella eggs.Citation29,30
Figure 5. Vasa2 protein accumulates around nuclei. (A), (E), (I) and (M) show schematic representations of a blastula (A), gastrula (E), planula (I) and a mesentery (M). Vasa2 antibody staining of a blastula (B-D), gastrula (F-H; oral view), planula (J-L; lateral view), cross sections of female (N-P) and male mesenteries with gonads (Q-S) are shown. The staining in blastulae (B) and gastrulae (F) is ubiquitously distributed throughout the embryos, whereas the planula (J) displays a stronger staining in the endoderm in the position of the future mesenteries. In female gonads only the early oocytes are Vasa2 positive (N). Male gonads show a strong staining in the marginal zone of the spermaries (Q). Counterstaining with DAPI reveals a perinuclear staining of Vasa2 (D, H, L, and S). The arrowhead in (D) indicates an M-phase chromosome with no detectable Vasa2 staining. White: Vasa2; red: DAPI. Abbreviations: ec, ectoderm; en, endoderm; g, gonad; m, mesentery; pm, parietal muscle; rm, ring muscle; sf, septal filament. Asterisks in (E), (F), (I) and (J) mark the oral pole. All pictures are single optical sections except for (N) displaying a z-projection. Scale bars in (B), (F), (J), (N) represent 50 µm, in (C), (G), (K), (R) 10 µm and in (Q) and (O) 100 µm.
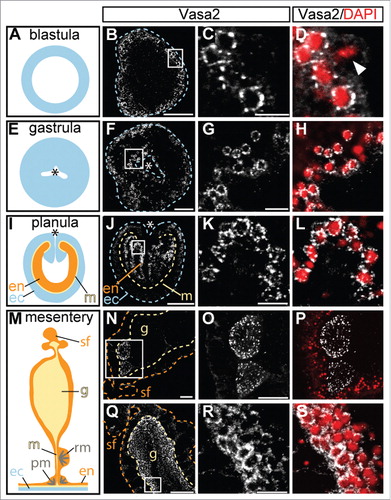
It seems that nuage is transiently dissolved during mitosis, as we detected elongated DAPI-positive M-phase chromosomes without surrounding Vasa2 staining (; arrowhead).
Nematostella piRNAs are developmentally regulated and differentially target transposable elements (TEs)
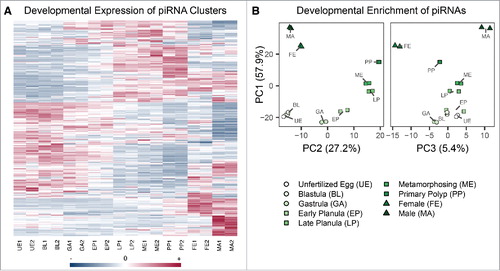
In light of the differences in piwi and vasa expression across Nematostella development, we were curious whether developmental regulation of piRNAs occurs as well. To answer this question, we first determined clusters of piRNA sequences in the Nematostella genome representing putative transcriptional loci of primary piRNAs. Based on this, we estimate that Nematostella harbors at least 457 piRNA clusters. Interestingly, these clusters are differentially expressed across developmental stages. This phenomenon is replicated in the untreated libraries (). The vast majority (402) of the piRNA clusters are unistranded, being transcribed from one strand of the piRNA locus. Fifty-five are dual-stranded piRNA clusters, with both DNA strands of the piRNA locus being transcribed into piRNAs and which also exhibit differential expression patterns across development (Supplemental Fig. S8). We confirmed that ping-pong signature was detectable across all developmental stages as shown in Supplemental Fig. S9, indicating these are functional piRNAs. As shown in , piRNA cluster transcription levels can be grouped into windows of development: in early developmental stages ranging from unfertilized egg to early planula, particular piRNA clusters are active compared with older stages including late and metamorphosing planulae and primary polyps. piRNA cluster expression in adult females and males greatly differs from younger stages.
Figure 6. Developmental regulation of Nematostella piRNA expression. (A) Heat map of piRNA cluster expression across developmental stages. Each row denotes one piRNA cluster, and the blue-red scale represents the standardized total reads that were assigned across developmental stages. (B) Plots of the first 3 principal components of individual piRNA enrichment across developmental stages, which account for 90.5% of the variation. Points cluster by developmental stage in all components, indicating the inter-stage variability is replicated. Principal components 1 and 2 separate early-adult and mid stages, respectively, whereas principal component 3 accounts for the differences between adult male and female piRNAs.
Not only piRNA clusters, but also distinct individual piRNAs within the identified piRNA clusters exhibit differential enrichment with respect to the coverage profiles in other developmental stages (e.g., Supplemental Fig. S10). As piRNA clusters are thought to be transcribed as a single unit comprising several piRNAs, this differential enrichment could be explained by differential activity of a distinct individual piRNA in the ping-pong cycle due to a temporal over-abundance of the target substrate. To uncover patterns of developmental enrichment of these piRNAs, individual loci were extracted from clusters by applying a custom algorithm to the clusters' coverage patterns. Using differential expression and principal components analysis, we detected that differential enrichment of individual piRNAs was replicated in the untreated Nematostella libraries and that consecutive developmental stages cluster together (). We applied DESeq2Citation31 to read counts of piRNA loci in all pairs of sequenced developmental stages. As shown in Supplemental Table S5, between <1 – 10% of the annotated piRNAs are differentially enriched from one developmental stage to the next. This suggests functional relevance of piRNAs independent of the cluster expression level at the observed developmental stage.
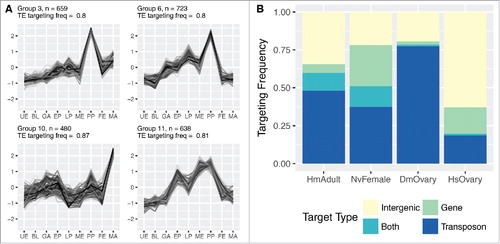
To deduce functions of differentially enriched piRNAs across developmental stages, we analyzed the enrichment profiles of each piRNA locus. We grouped the piRNA loci based on the similarity of their relative expression levels at each developmental stage, independent of the expression levels of neighboring piRNAs or the originating primary piRNA cluster. The enrichment profiles of these groups are illustrated in Supplemental Fig. S11. Among those groups, we then surveyed the proportion of piRNAs within each group that were predicted to target transposable elements. We observed significant enrichment in TE-targeting piRNAs in 4 groups shown in . Group 5 contains piRNAs enriched in the primary polyp stage and female adults. Group 8 shows distinct enrichment in the planula and primary polyp stages. piRNAs of group 9 show enrichment from the early planula stage on and are highly enriched in primary polyps, but are underrepresented in adults. Group 11 piRNAs are enriched in metamorphosing planulae and peak in the primary polyp stage. In the adult female, the enrichment of transposon targeting piRNAs could be connected to the process of oogenesis and serve the maintenance of the germline genome. The group 4 profile, which peaks in the sampled adult males, also represents enriched TE-targeting piRNAs, although not at a significant level (Supplemental Fig. S11). Taken together, piRNA clusters as well as individual piRNAs targeting TEs exhibit differential enrichment across development of Nematostella.
Figure 7. Targeting potential of Nematostella piRNAs. (A) Groups of Nematostella piRNAs significantly enriched for targeting TEs have distinct enrichment profiles during development. The number of associated piRNA loci contributing to the respective groups is depicted with an “n.” Group 3, group 6 and group 11 are characterized by enrichment of TE-targeting piRNAs in the primary polyp stage, with only group 10 reaching an average of more than 2-fold greater enrichment in the adult male stage. (B) Almost half of Nematostella piRNAs (45%) potentially target transposable elements. piRNA targets were classified into 4 classes: “intergenic regions,” “genes,” “transposons” and “both,” combining genes and transposons.
Nematostella piRNAs target TEs and protein-coding genes
We then sought to determine the TE-targeting potential of Nematostella piRNAs in comparison to human, Drosophila and Hydra piRNAs. To this end, we aligned the piRNA fraction of the reads of each library to their respective genome with relaxed parameters and filtered for alignments which showed a 18-base perfect match and had no more than two mismatches downstream, as previously suggested.Citation32 We assessed the frequency of reads binding different elements in each genome to those found in the adult female library of Nematostella and grouped the reads into 4 categories: (I) piRNAs targeting intergenic regions, (II) piRNAs targeting protein-coding genes, (III) piRNAs targeting transposons and (IV) piRNAs targeting transcripts of both protein-coding genes and transposons. We observed the ping-pong signature in piRNAs belonging to categories II-IV (Supplemental Figs. S1 and S9). The majority of Nematostella piRNAs (51%) targets TEs (; categories III and IV, i.e. “transposon” and “both” transposons and protein-coding genes are counted). This finding is in line with the conserved role of piRNAs in transposon surveillance in numerous animals of different phyla.Citation2-4 Notably, in Nematostella the fraction of piRNAs targeting protein-coding genes (41%) is significantly larger compared with investigated samples of humans and Drosophila (18.4% and 3.2%, respectively, shown in ), possibly implying a role of piRNAs in the regulation of gene expression in Nematostella.
To support the functionality of Nematostella piRNAs we compared the sequenced piRNAs with our previously reported degradome, a set of experimentally identified cleaved mRNA fragments.Citation17 While computational mapping of piRNAs to genomic loci hints to potential interactions, this approach enabled us to verify the interaction of piRNAs with their actual targets, more specifically their piRNA-guided slicing. We searched the degradome for RNAs that terminated exactly 10 bases antisense and upstream to the 5´- end of the target. As shown in Supplemental Table S6, 10,261 distinct degradation products are validated piRNA targets within annotated loci including 9,129 products of transposable elements and 742 products corresponding to 62 non-transposon protein-coding genes. This indicates that Nematostella piRNAs act predominantly in transposon silencing but also have a role in the regulation of protein-coding genes. A similar observation was made in Hydra: immunoprecipitation experiments against Hywi and Hyli detected physical association of these PIWI proteins and piRNAs mainly with transposon sequences but also, to a smaller extent, with protein-coding genes.Citation25,33 Approximately 5% and 12% of total and Hywi-bound piRNAs mapped to transcripts of protein-coding genes respectively.Citation25 To investigate the overlap of protein-coding genes as piRNA targets between Hydra and Nematostella, we performed a reciprocal BLAST analysis and found that no transcripts targeted by piRNAs are shared in these two species.
Discussion
The piRNA pathway has essential roles in transposon repression and fertility in all metazoan species investigated so far. However, it has mainly been exploited in bilaterian animals and the knowledge about its evolution and roles in non-bilaterians remains scarce. This study constitutes the first deep investigation of piRNAs and associated proteins in the sea anemone Nematostella vectensis, a member of Cnidaria, the sister group of Bilateria.
By comparing the fraction of piRNAs among small RNA complements in various animals, we show that Nematostella piRNAs are the most abundant class of small RNAs among investigated species. In bilaterian model organisms piRNAs are enriched in the germline. Nevertheless, the fraction of Nematostella piRNAs substantially exceeds the proportion of piRNAs in human and Drosophila ovaries (Supplemental Fig. S2 and Supplemental Table S4). This high abundance in all developmental stages could be connected to the wide expression of piRNA pathway components in early developmental stages and in adult sea anemones not restricted to the gonads (, Supplemental Figs. S4 and S6A, E). Hence, Nematostella piRNAs and their partner proteins possibly silence TEs in the soma in addition to the germline. Interestingly, the fact that piRNAs and piRNA-related proteins are also highly and widely expressed in Hydra,Citation25,33 and that piRNAs constitute the highest fraction of small RNAs in the sponge Amphimedon queenslandicaCitation34 as well, suggest that these aspects represent shared traits of early-branching metazoans.
Components of the GMP such as piwi and vasa genes are expressed in somatic stem cells in addition to the germline in a wide range of animals including early branching phyla such as sponges, ctenophores and hydrozoan cnidarians.Citation3,9,10 In Hydra, the two PIWI proteins are expressed in multipotent i-cells and germline stem cells, but also in mitotic epithelial cells with stem cell character in the ectoderm and endoderm.Citation15,16 vasa expression is as well detected in i-cells and epithelial cells in addition to the gonads.Citation35 To date, no stem cells have been identified in Nematostella, but the expression of these markers connected to “stemness” detected in body regions outside the gonads by direct quantification of transcripts and by western blot may hint to the presence of somatic stem cells.
Somatic expression of these genes seems to correlate with the ability to regenerate body parts in some animals of various phyla including the salamander Ambystoma mexicanum,Citation36 annelidsCitation37-40 or planarians.Citation41 Cnidarians are well known for their remarkable regeneration capacity. Recent studies in Hydractinia echinata show that this capacity is impaired upon piwi knockdown.Citation42 Although further studies to support this hypothesis have to be conducted, a connection of the somatic expression of piwi and vasa genes and the regenerative capacity in Nematostella seems to be a valid possibility.
In Hydra, the somatic expression of PIWI proteins and thus the likely silencing of transposons in the soma was linked to the ability of asexual reproduction, since mutational load could be passed on through the soma.Citation15 Consistently, the expression of piRNA pathway components in somatic regions in Nematostella and the assumption that the high fraction of piRNAs is caused by an expression not restricted to the gonads could support a relationship between expression patterns and mode of reproduction as well. The prevalent mode of asexual reproduction in Nematostella is physal pinching. In line with this, we detect substantial expression of piRNA pathway proteins in the physa, possibly counteracting the propagation of transposons (, Supplemental Fig. S4B).
Localization of piwi and vasa genes by in situ hybridization across Nematostella development revealed wide and overlapping expression patterns. These genes are also expressed in broad regions at early stages of development in multiple species across animal phylogeny including flatworms, mollusks, annelids and arthropods and involvement of these factors in embryogenesis has been proposed.Citation38,43-50 The highly similar expression domains of Nematostella piwi and vasa homologs () are in line with their clustering in the nCounter analysis () and imply a conservation of Vasa as a factor in the piRNA pathway between Cnidaria and Bilateria.
Vasa is a conserved component of nuage structures, electron dense aggregates of ribonucleoproteins important for mRNA regulation and the site of piRNA production found in germ cells and multipotent somatic cells.Citation5,28,51 Besides Vasa, nuage contains several other conserved components such as Nanos, Mael, PIWI and Tudor proteins. Most of these factors were shown to be essential in piRNA amplification and transposon silencing.Citation2,3,51 The perinuclear granules positive for Vasa2 found throughout the development of Nematostella () are highly reminiscent of nuage. Interestingly, we sometimes detect M-phase chromosomes whose surroundings were not stained by the anti-Vasa2 antibody (). The absence of Vasa2 staining around M-phase chromosomes (, arrowhead) suggests the association of Nematostella Vasa2 with the nuclear envelope, as in bilaterian model systems.Citation51 In Cnidaria, nuage has been identified in Clytia hemisphaerica in growing oocytes and in i-cells and epithelial stem cells of Hydra.Citation25,28,33 Our data confirm the notion that the association of the piRNA pathway to nuage is ancestral.
In an attempt to tackle the question about developmental regulation of piRNAs in Nematostella we assigned piRNAs to piRNA clusters and detected their differential expression across development. This implies a role of piRNAs in Nematostella development and is reminiscent of the situation in mouse, where dynamic transcription of piRNA clusters during development was observed as well.Citation52 This observation raises intriguing possibilities regarding differential control by the host over the activity of its repetitive elements, which potentially can have important genetic and evolutionary effects.Citation53,54 However, more research is required to assess this notion. The dynamic expression of individual TE-targeting piRNAs () could reflect the changing threat of transposons and herein the variable necessity of silencing them during different stages of development. Specifically, the enrichment of piRNAs targeting transposons at the primary polyp stage () could be connected to precursor germ cell formation and the requirement to safeguard the germline from the deleterious effects of transposons. Consistently, we detect cell clusters of stronger staining for piwi1 and vasa2 by in situ hybridization in the mesenteries of primary polyps (). The corresponding regions in the mesenteries were shown to be positive for the homolog of Nanos, another bilaterian germline marker, and due to characteristic morphological features were suggested to be primordial germ cells.Citation27 These cells could represent the onset of a detectable germline vulnerable to transposons in Nematostella. However, aside from one studyCitation27 and the presented data here, we currently lack insights into germline establishment in Nematostella. Thus, the proposed link between enrichment of TE-targeting piRNAs and strong expression of piRNA pathway components at the future site of gonads in the primary polyp stage remains speculative. As far as we are aware, this is the first study investigating the developmental enrichment of individual TE-targeting piRNAs.
To determine the function of piRNAs in Nematostella, we identified targets of clustered piRNAs. These include predominantly TEs, but also a substantial fraction of protein-coding genes, which clearly outreaches the equivalent in other investigated species (). Although studies of the function of piRNAs have focused primarily on their transposon silencing activity, there is accumulating evidence implicating piRNAs in the regulation of protein-coding genes. This is supported by several works in a variety of species highlighting that the phenomenon is more common than previously appreciated.Citation2,3,55,56 A class of piRNAs originates from the 3´ UTRs of protein-coding genes (thus termed genic piRNAs) and is suspected to target their host gene in cis or other transcripts in trans. Gene-derived piRNAs were studied in Drosophila, Xenopus, mice, pig and primates including human.Citation56-59 However, these studies identify protein-coding genes as piRNA targets computationally with little or without any experimental validation. In contrast, with the analysis of our degradome library we provide the first direct biochemical observation of the actual slicing of protein-coding gene transcripts and TEs mediated by piRNAs in a non-bilaterian animal. The biologic significance of the piRNA mediated regulation of protein-coding genes in Nematostella needs to be addressed in future studies.
It is known that piRNAs are rapidly evolving and do not share any sequence homology even between related species across the animal kingdom. The comparison of protein-coding genes as piRNA targets between Nematostella and Hydra did not yield common targets and thus implies that also target mRNAs have a high turn-over rate in Cnidaria and did not co-evolve with their piRNAs to confer their own silencing.
Taken together our data represent a comprehensive characterization of the piRNA pathway during the development of the sea anemone Nematostella vectensis and provide insights into the functions of its piRNAs. Our findings suggest that the adult expression of piRNA pathway components in somatic regions in addition to the gonads as well as the transposon silencing and the regulation of protein-coding genes by piRNAs are ancestral traits.
Materials and methods
Sea anemone husbandry
Adult sea anemones were cultured at 18°C in water in 16 ‰ sea salt (Nematostella medium, NM). To obtain early developmental stages, spawning was induced as described elsewhere.Citation60
Identification and cloning of Nematostella piRNA pathway components
Nematostella PIWI protein sequences were identified by running tblastn searchesCitation61 of PIWI proteins of Drosophila melanogaster, Homo sapiens or Mus musculus against the non-redundant (nr) nucleotide collection of N. vectensis of the National Center for Biotechnology Information (NCBI). Best gene model hits were subjected to reciprocal blastp searches against the non-redundant NCBI protein database. Gene models were verified by PCR on a cDNA template generated from RNA from mixed developmental stages from N. vectensis by SuperScript III reverse transcriptase (Thermo Fisher Scientific) according to the manufacturer's protocol. Full length sequences of Nematostella PIWI proteins were amplified by using the primers 5′- GGTAAGTTTTGGTCATGACTGGAAGGGC-3′ and 5′-GATGGGGAGGACCTCTACTAAGTAGTGCC-3′ for NvePiwi1 (GenBank MF683122), 5′- GGCACTGAAAGGGAGTAGAACTGTAGC-3′ and 5′- GATGTTGGACCAATCTTTGTTGGATGGC-3′ for NvePiwi2 (Genbank MF683123) and 5′-GGTGAAGGAGGCTACGACTACGATTGG-3′ and 5′-CCATGAATCTGGTGAGCTCAGTGGC-3′ for NveExPiwi1 (GenBank MF683124). The Nematostella sequences of Armitage (GenBank MF683126), Maelstrom (GenBank MF683127) and Spindle-E (GenBank MF683125) were retrieved by using the Drosophila melanogaster and Homo sapiens homologs as queries. Partial Vasa sequences were taken from Extavour et al. 2005 (GenBank AY730696 and AY730697).
Phylogenetic analysis of PIWI proteins
The sequences of the Edwardsiella lineata Argonaute (AGOs) and PIWI proteins were retrieved by a tblastn search of the Nematostella PIWI and Ago proteins against the E. lineata transcriptomeCitation62 and translating the best hits. Other homologous protein sequences were obtained from the NCBI (protein or transcriptome shot gun assembly data set, accession numbers can be found in Supplemental Table S1). Full PIWI sequences were aligned using MUSCLE and regions with low-quality alignments were trimmed by TrimAl 1.2 rev 59 using the option “automated1” for the trimming.Citation63 ProtTest 2.4Citation64 analysis retrieved LG (+I+G+F) as the best-fitting model for the phylogenetic reconstruction. The maximum likelihood tree was then generated with PhyML 3.0Citation66 BIONJ input tree, optimized tree topology, 4 substitution rate categories, best of NNI and SPR, 100 non-parametric bootstrap replicates. Bayesian analysis was performed by MrBayes version 3.2.2Citation66 WAG, 5,000,000 generations, every 100th generation reported). The tree log likelihood function was asymptotic after 100,000 generations, and so many generations were discarded as the burn in fraction of the search. Phylogenetic analysis including the ExPiwi1 (Supplemental Fig. S1) was performed as described above, yet the sequence of the ExPiwi1 was modified by removal of the 2 stop codons in the PAZ domain and setting it in frame.
Nanostring nCounter assay
Total RNA from different developmental stages and body parts of the adult sea anemone was extracted with Tri-Reagent (Sigma-Aldrich) according to manufacturer's protocol, treated with Turbo DNAse (Thermo Fisher Scientific) and then re-extracted with Tri-Reagent. RNA quality was assessed on standard sensitivity Bioanalyzer Nanochip (Agilent) and only samples with RNA Integrity Number (RIN) ≥ 8.0 were used. Those samples were analyzed on the nCounter platform (NanoString performed by Agentek Ltd.) in triplicates as described previously.Citation67 In brief, for each transcript to be tested, 2 probes were generated and hybridized to the respective mRNA. The mRNAs were immobilized on a cartridge and the barcodes on one of the probes were counted by an automated fluorescent microscope. For normalization, 5 reference genes with stable expression across development were selected as follows: we calculated the Shannon entropy for each gene (as described inCitation68 based on normalized transcript abundance estimates for 6 time-points of Nematostella development.Citation69 We then ranked the genes by entropy (indicating minimal temporal change in abundance), and from the top 20 chose 5 genes (NVE3324, NVE5273, NVE11206, NVE22425, NVE14488 fromCitation22) with complete gene models and mean abundance levels spanning the expected experimental range.
In situ hybridization
In situ hybridization of young stages such as blastulae and gastrulae was essentially performed as described.Citation70 Zygotes were separated from the gelatinous egg mass by incubation in 4% L-cysteine in NM with adjusted pH of 7.4 for 30–45 minutes on a rotary shaker until the egg mass was dissolved. Afterwards the zygotes were washed 5 times in NM. Blastulae and gastrulae were fixed in 3.7% formaldehyde and 2.5% glutaraldehyde in NM for 90 seconds on ice. The solution was then exchanged with 3.7% formaldehyde in NM and animals were fixed for an additional hour under gentle rotation at 4°C. Subsequently, embryos were washed five times in PTw (1x PBS with 0.1% Tween-20). Planula larvae were fixed the same way but in 3.7% formaldehyde and 0.25% glutaraldehyde in NM was used in the 90-second fixation step. Primary polyps were incubated in 0.5% dimethyl sulfoxide (DMSO) in NM for 24 hours before fixation, anesthetized by adding 1 M MgCl2 drop by drop into the Petri dish with NM to prevent muscle contraction, and subsequently fixed in 3.7% formaldehyde/0.25% glutaraldehyde/0.5% DMSO in NM for 1 minute before fixation in 3.7% formaldehyde in NM for 1 hour at 4°C. For staining of adult body tissues the animals were also anesthetized by adding MgCl2 into the Petri dish as well as into the gastric cavity by introducing it through the mouth opening. The polyps were fixed in 3.7% formaldehyde and 0.25% glutaraldehyde for 90 seconds on ice and then with 3.7% formaldehyde in NM for 40 minutes at 4°C. Subsequently the heads and the physae were cut off to facilitate better fixative penetration, and the body parts were fixed for another 40 minutes in 3.7% formaldehyde in NM at 4°C. The tissues were washed five times in PTw and dissected into small pieces. These were washed several times in methanol until no animal pigments were washed out anymore. In situ hybridization with DIG labeled RNA probes for early stages was performed as describedCitation70 with the following modifications: the embryos were treated with 80 µg/mL proteinase K in PTw for 20 minutes. The triethanolamine washes were done with 1% triethanolamine in PTw. The concentration of the anti-DIG/AP antibody was raised to 1:2000 in blocking buffer (1% blocking reagent (Roche) in maleic acid buffer, pH 7.5). For primary polyps an incubation step with RNAseT1 (1U/µl; Thermo Fisher Scientific) in 2x SSC was introduced before the 0.05x SSCT washes. For adult tissue pieces and primary polyps the protocol was performed with using PTx (1x PBS with 0.3% Triton X-100) instead of PTw. After the rehydration, a bleaching step with 0.5% H2O2/5% formamide/0.5x SSC in H2O for 5 minutes was introduced. The triethanolamine concentration was changed to 1% in PTx. The prehybridization step was performed overnight in hybridization buffer containing additionally 10% dextrane sulfate (Sigma-Aldrich) and 1% blocking reagent (Roche). The probe concentration was raised to 0.75 ng/µl in hybridization buffer supplemented with dextrane sulfate and blocking reagent. Also here an RNAseT1 treatment (1U/µl) was performed for 40 minutes before the 0.2 × SSC washes. The washes before and after antibody incubation were performed in PBT × (1 × PBS with 0.3% Triton X-100 and 0.1% bovine serum albumin). The anti-DIG/AP antibody concentration was raised to 1:2000 in PTx. Adult pieces were washed in PTx after the ethanol incubation, embedded in 10% gelatine in PBS and sectioned on a vibratome (see below). Embryos and sections were mounted in 85% glycerol and photographed with a Nikon Eclipse 80i microscope connected to a Nikon Digital Sight DS-U2 camera.
Vibratome sectioning
Fixed body parts were embedded in 10% gelatine in PBS. For tissues with in situ hybridization staining, samples were refixed in 3.7% formaldehyde in PBS, for antibody staining in 4% paraformaldehyde (PFA) in PBS overnight. After several washes, 50 µm sections were generated on a Leica VT 1200S vibratome. These were either mounted directly on microscope slides and visualized (in situ hybridization samples) or subjected to the antibody staining procedure.
Western blotting
Custom mouse monoclonal antibodies were produced by Abmart against short synthetic peptides (AGEDGDRPKP and SAGGGDDDWE) corresponding to Nematostella Vasa2. Nematostella adult polyps were flash-frozen with liquid nitrogen and homogenized in protein extraction buffer (150 mM KCl, 0.5% NP-40, 10% Glycerol and 50 mM Tris, pH 7.4) containing 1:100 diluted Halt™ Protease Inhibitor Cocktail (Thermo Fisher Scientific). Protein concentration was determined with the BCA kit (Thermo Fisher Scientific) and the lysate was then mixed with SDS sample buffer (New England Biolabs, USA) and boiled for 8 min at 100°C. Samples were run on precast polyacrylamide gradient gel (4–15%; Bio-Rad, USA) and blotted to a nitrocellulose membrane (Bio-Rad). The membrane was washed with TBST buffer (20 mM Tris pH 7.6, 150 mM NaCl, 0.1% Tween 20) and then blocked with blocking buffer (5% skim milk in TBST) for 1 hour at room temperature. 5 µl of purified monoclonal antibody against NvVasa2 were added to 5 ml of TBST containing 5% BSA, on the membrane in a sealed sterile plastic bag and incubated at 4°C overnight. After 16 hours, the membrane was washed 3x with TBST and then incubated for 1 hour with Goat anti-mouse IgG conjugated to horseradish peroxidase (Jackson ImmunoResearch) diluted to a concentration of 0.1 µg/ml in 5% skim milk in TBST. The membrane was washed 3x with TBST and detection was performed with the Clarity™ ECL kit (Biorad) according to the manufacturer's instructions with a CCD camera of the Odyssey Fc imaging system (Licor). Size determination was performed by simultaneously running the PageRuler Prestained NIR Protein Ladder (Thermo Fisher) and scanning the same blot on the same system at 700 nm.
Antibody staining
Blastulae, gastrulae and planulae were fixed in 4% paraformaldehyde (PFA) in MEM buffer or 1x PBS for one hour at 4°C under gentle rotation. Subsequently, the embryos were washed 5 times in MEM buffer or PTw. For staining on mesenteries, adult animals were anesthetized and fixed in 4% PFA in PBT (1 × PBS and 0.2% Tween-20) for 40 minutes at 4°C. Heads and physae were cut off and fixed for another 40 minutes at 4°C and washed 5 times in PBT. The body parts were then cut into small pieces, embedded in gelatine and sectioned on a vibratome. The obtained sections and the early stages were incubated for 2 hours in blocking solution (20% sheep serum and 1% bovine serum albumin in PBS) at room temperature (RT) before overnight incubation in primary monoclonal mouse antibodies against NvVasa2 (Abmart, China) diluted 1:500 in blocking buffer at 4°C. The next day, sections and embryos were washed 8x in PBT, blocked again for 2 hours at RT and the secondary antibody (for mesenteries goat anti-mouse Alexa Fluor® 488, and for early stages goat anti-mouse Alexa Fluor® 568 (Thermo Fisher Scientific)) in a dilution of 1:1000 and 5 µg/mL DAPI were added and incubated overnight at 4°C. On the next day, the samples were washed 8x in PBT. Early stages were mounted in SlowFade® (Thermo Fisher Scientific), vibratome sections were mounted in ProLong® Gold (Thermo Fisher Scientific). The staining was visualized by a Leica SP5 X Confocal Laser Scanning Microscope.
Data curation of small RNA reads
H. sapiens genome build hg38 and D. melanogaster build dm6 were downloaded from the UCSC genome browser.Citation71 N. vectensis genome build Nemvec1 and H. magnipapillata build Hydra_RP_1.0 were used.Citation72,73 Small RNA data sets were downloaded from the NCBI Sequence Read Archive and derive from references,Citation16,17,19,20 summarized in Supplemental Table S2. 3′ adaptors (also listed in Supplemental Table S2) were trimmed using trimmomaticCitation74 using the options trimomatic_adapt (Supplemental Table S3). rRNA sequences were removed using SortMeRNA 2.0 by aligning against the RFAM 5.8S, Archaea, Bacterial and Eukaryotic LSU rRNA SILVA databases provided with the distribution.Citation75 Reads potentially deriving from non-piRNA sequence were filtered first by mapping the reads to the genome using the settings star_noMM (Supplemental Table S3). Where available, miRNA, tRNA, lincRNA and snoRNA annotations were downloaded from the UCSC genome browser. miRNA annotations for N. vectensis were taken from,Citation17 and other annotations were downloaded from the JGI Genome Portal.Citation76 H. magnipapillata annotations were obtained from NCBI. Reads that mapped to snoRNA, miRNA, tRNA, lincRNAs, tRNAs, and CDS loci, where available, were filtered out (Supplemental Fig. S2).
Assessment of the ping-pong signal
All clustered piRNAs among all developmental stages and adults were mapped to the Nematostella genome using STAR using settings star_3 MM (Supplemental Table S3). Each read which overlapped divergently at most 20 bases was counted and removed from further analysis. Each pair of reads was plotted once in a histogram (Figs. S1A and S9). Positional weight matrices starting from the 5′ ends of reads were calculated among four categories: “ping-pong” (overlapping 10 bases) mapping to the sense strand or antisense strand, and non-“ping-pong” reads mapping to the sense strand or antisense strand of either transposable elements or genes. The resulting matrices were plotted using DiffLogo.Citation77
Prediction of piRNA clusters and loci from sequencing data
piRNA cluster detection was performed on Nematostella piRNAs using the proTRAC pipeline version 2.1Citation78 with the options protrac_nv (Supplemental Table S3) for each developmental stage. Clusters were then merged using the bedtools suite,Citation79 resulting in 457 clusters. Unassigned reads were aligned to the Nematostella genome with the STAR aligner, using the settings star_noMM (Supplemental Table S3), and those mapping within the boundaries of detected piRNA clusters were designated as “clustered piRNA.” piRNA loci within the detected piRNA clusters were determined via a custom peak finding method. Briefly, the number of “cluster piRNA” reads from all developmental stages mapping to each genomic position within the cluster were smoothed using a Gaussian kernel with a window size of 5. The first derivative was estimated as the difference between the smoothed signal current and following position. Peaks were determined by selecting pairs of maximal ascending and descending positions which were between 23 and 35 base pairs long. This yielded 33,734 loci. Targets of these loci were detected using a custom script, filter_targets.py, which requires an 18-base perfect match region in which all but the first nucleotide need to match, and the non-perfect match portion of the alignment to have at most 2 mismatches. These criteria follow suggestions from.Citation80
Detection of differential enrichment of piRNAs between developmental stages
Expression profiles of piRNA clusters among libraries of 8 developmental stages including female and male adults (hereafter referred to as 8 developmental stages) were then determined. To measure the reproducibility of the piRNA cluster expression, untreated libraries corresponding to each developmental stage was used as replicates. As piRNA reads are prone to map to several locations, we chose to limit our analysis to reads mapping to a single location within the piRNA cluster space. Read count data were normalized using the DESeq2 methodCitation31 and standardized such that each locus' enrichment mean was 0 and standard deviation was 1 across stages. Heat maps were generated using the gplots package in R, and only included the clusters for which there were unique piRNAs mapped in all stages. To confirm that the difference between individual piRNA loci were statistically significant, we tested the differential enrichment between the developmental stages using DESeq2. Over- and under- enrichment of piRNAs were defined with p < .05 adjusted using the Bonferroni correction, with the additional requirement of a minimum fold-change of 2 and an automated Cook's distance cutoff to reduce false positives, as implemented in DESeq2. Additionally, principal component analysis of the counts was visualized after variance stabilization was applied.Citation31
Detection of Nematostella piRNA targets
Cluster piRNAs among all developmental stages and adults were mapped to the Nematostella genome using STAR with settings star_3 MM (Supplemental Table S3). Mappings were subsequently filtered using the filter_targets.py script. Mappings were associated with Nematostella protein-coding genesCitation22 and TEs if they mapped to the antisense strand of the locus. To assess degradome support of the putative targets, reads from the degradome libraries were mapped to the Nematostella genome using STAR with settings star_DEG. Targets were considered degradome-supported if at least one read in either of the two libraries mapped in the sense direction with the 5′ end mapping 10 base pairs downstream and antisense to the mapped putatively targeting piRNA read. Protein coding targets were filtered against a library of transposable element proteins and using RepeatProteinMask (81) with the arguments listed in repeat_protein_mask (Supplemental Table S3).
We compared these targets to a list of targets found in a previously assembled Hydra magnipapillata transcriptome (33 and provided via personal communication with Celina Juliano). To detect corresponding transcripts we subjected them to reciprocal BLAST analysis. First we found longest ORFs in the sequences from the Hydra and Nematostella transcriptomes using TransDecoder.Citation82 The full transcriptomes were compared reciprocally using BLASTP with a maximum e-value of 1e-5. Matches which were the top hit in both Hydra-Nematostella and Nematostella-Hydra comparisons with respect to e-value or bit score were considered to be orthologs. We then checked if any of the orthologs which were found to be targets of Hyli or Hywi-bound piRNAs in Hydra were also confirmed targets in our set, and found no overlap.
Enrichment of individual piRNAs in temporal profiles
Enrichment profiles from developmental stages of Nematostella from each of the detected piRNA loci were clustered using Mfuzz.Citation83 Loci were required to have at least one read associated per stage, a between-stage variance of at least two and an associated target outside of any piRNA cluster, leaving 7,351 loci. Read count data were similarly normalized and standardized as described above. To estimate the number of cluster centers, we measured the minimum inter-cluster Euclidian center distance when the number of clusters 50 ≥ c ≥ 9, with up to 1000 iterations, repeating the analysis ten times. The greatest drop in minimum distance was observed when c = 12. TE targets were partitioned among the clusters according to cluster membership as defined by its highest membership value. α-core members were plotted where membership values were ≥ .95. To determine which of the clusters were enriched in piRNA loci-targeting TEs, χ2 tests were performed using a Bonferroni correction, yielding 4 significant clusters.
Cross-species TE targeting comparison
The surviving set was considered as piRNAs and analyzed for their potential to target TEs in their respective genomes. Repeat annotations for D. melanogaster, H. sapiens and N. vectensis were retrieved from the repeatmasker.org website.Citation81 H. magnipapillata repeats were annotated using the standard RepBase library as well as a custom library obtained from Oleg Simakov, originally described in.Citation73 All repeat annotations attributed to the LINE, LTR, RC and DNA classes were considered as TEs. For the custom H. magnipapillata library, all repeats were considered as TEs except for those attributed to the families “PSEUDO,” “Non-LTR,” “nonLTR,” “non-LTR,” “dUTPase,” “ARTEFACT,” “Unspecified,” “UNKNOWN” and “SINE.” Putative piRNA targets were detected using a seed-and-score strategy: first alignments were seeded with the STAR aligner version 2.5.1bCitation84 using the options star_3 MM (Supplemental Table S3), allowing for at most three mismatches; second, using a custom script filter_targets.py, described above. The fraction of sequenced reads from the piRNAs for which at least one identified target mapped to either strand of a TE was used in estimating the rate of TE targeting.
Disclosure of potential conflicts of interests
No potential conflicts of interest were disclosed.
Supplementa_Materials.zip
Download Zip (16.1 MB)Acknowledgments
The protocol for in situ hybridization of adult sea anemones was established by Patrick Steinmetz. We thank Stefan Jahnel for sharing his expertise in the procedures of dissecting and sectioning adult animal parts. We thank Sabrina Kaul-Strehlow for her support in figure assembly.
Small RNAs of different stages of Nematostella vectensis were sequenced by Hervé Seitz.
We thank the Core Facility for Cell Imaging and Ultrastructure Research of the University of Vienna for the access to the confocal microscope.
Funding
This work was supported by grants of the Austrian Science Fund FWF (P22618, P24858) to U.T., European Research Council Starting Grant (CNIDARIAMICRORNA, 637456) to Y.M. and a PhD completion fellowship of the University of Vienna to D.P.
References
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10(2):94–108. doi:10.1038/nrg2504.
- Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: its biogenesis and functions. Annu Rev Biochem. 2015;84:405–33. doi:10.1146/annurev-biochem-060614-034258.
- Lim RSM, Kai T. A piece of the pi(e): the diverse roles of animal piRNAs and their PIWI partners. Semin Cell Dev Biol. 2015;47–48:17–31. doi:10.1016/j.semcdb.2015.10.025.
- Czech B, Hannon GJ. One loop to rule them all : the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci. 2016;41(4):324–337. doi:10.1016/j.tibs.2015.12.008.
- Ketting RF. The many faces of RNAi. Dev. Cell. 2011;20(2):148–161. doi:10.1016/j.devcel.2011.01.012.
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24(9):887–892. doi:10.1101/gad.1902110.
- Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, Couté Y, Conn S, Kadlec J, Sachidanandam R, et al. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell. 2014;157(7):1698–1711. doi:10.1016/j.cell.2014.05.018.
- Hartung O, Forbes MM, Marlow FL. Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol Reprod and Dev. 2014;81(10):946–961. doi:10.1002/mrd.22414.
- Gustafson EA, Wessel GM. Vasa genes: emerging roles in the germ line and in multipotent cells. Bioessays. 2010;32(7):626–637. doi:10.1002/bies.201000001.
- Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Ann Rev Genet. 2011;45:447–469. doi:10.1146/annurev-genet-110410-132541.
- Juliano CE, Swartz SZ, Wessel GM. A conserved germline multipotency program. Development. 2010;137(24):4113–4126. doi:10.1242/dev.047969.
- Fierro-Constaín L, Schenkelaars Q, Gazave E, Haguenauer A, Rocher C, Ereskovsky A, Borchiellini C, Renard E. The conservation of the germline multipotency program, from sponges to vertebrates: a stepping stone to understanding the somatic and germline origins. Genome Biol Evol. 2017;9(3):474–488. doi:10.1093/gbe/evw289.
- Cannon JT, Vellutini BC, Smith J, Ronquist F, Jondelius U, Hejnol A. Xenacoelomorpha is the sister group to Nephrozoa. Nature. 2016;530(7588):89–93. doi:10.1038/nature16520.
- Krishna S, Nair A, Cheedipudi S, Poduval D, Dhawan J, Palakodeti D, Ghanekar Y. Deep sequencing reveals unique small RNA repertoire that is regulated during head regeneration in Hydra magnipapillata. Nucleic Acids Res. 2013;41(1):599–616. doi:10.1093/nar/gks1020.
- Lim RSM, Anand A, Nishimiya-Fujisawa C, Kobayashi S, Kai T. Analysis of Hydra PIWI proteins and piRNAs uncover early evolutionary origins of the piRNA pathway. Dev Biol. 2014;386(1):237–251. doi:10.1016/j.ydbio.2013.12.007.
- Juliano CE, Reich A, Liu N, Gotzfried J, Zhong M, Uman S, Reenan RA, Wessel GM, Steele RE, Lin H. PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. Proc Natl Acad Sci U S A. 2014;111(1):337–342. doi:10.1073/pnas.1320965111.
- Moran Y, Fredman D, Praher D, Li XZ, Wee LM, Rentzsch F, Zamore PD, Technau U, Seitz H. Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res. 2014;24(4):651–663. doi:10.1101/gr.162503.113.
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Supplemnent - Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455(7217):1193–1197. doi:10.1038/nature07415.
- Roovers EF, Rosenkranz D, Mahdipour M, Han CT, He N, Chuva de Sousa Lopes SM, van der Westerlaken LAJ, Zischler H, Butter F, Roelen BAJ, et al. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 2015;10(12):2069–2082. doi:10.1016/j.celrep.2015.02.062.
- Song J, Liu J, Schnakenberg SL, Ha H, Xing J, Chen KC. Variation in piRNA and transposable element content in strains of Drosophila melanogaster. Genome Biol Evol. 2014;6(10):2786–2798. doi:10.1093/gbe/evu217.
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi:10.1146/annurev.cellbio.24.110707.175327.
- Fredman D, Schwaiger M, Rentzsch F, Technau U. Nematostella vectensis transcriptome and gene models v2.0. 2013.
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Vladimir A, Gvozdev VA. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol. 2004;24(15):6742–6750. doi:10.1128/MCB.24.15.6742-6750.2004.
- Vagin V V, Sigova A, Li C, Gvozdev V, Zamore PD. A distinct small RNA Pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320–324. doi:10.1126/science.1129333.
- Lim RSM, Anand A, Nishimiya-Fujisawa C, Kobayashi S, Kai T. Analysis of Hydra PIWI proteins and piRNAs uncover early evolutionary origins of the piRNA pathway. Dev Biol. 2014;386(1):237–251. doi:10.1016/j.ydbio.2013.12.007.
- Sato K, Siomi MC. Functional and structural insights into the piRNA factor Maelstrom. FEBS Lett. 2015;589(14):1688–1693. doi:10.1016/j.febslet.2015.03.023.
- Extavour CG, Pang K, Matus DQ, Martindale MQ. Vasa and nanos expression patterns in a sea anemone and the evolution of bilaterian germ cell specification mechanisms. Evol Dev. 2005;7(3):201–215. doi:10.1111/j.1525-142X.2005.05023.x.
- Leclère L, Jager M, Barreau C, Chang P, Le Guyader H, Manuel M, Houliston E. Maternally localized germ plasm mRNAs and germ cell/stem cell formation in the cnidarian Clytia. Dev Biol. 2012;364(2):236–248. doi:10.1016/j.ydbio.2012.01.018.
- Levitan S, Sher N, Brekhman V, Ziv T, Lubzens E, Lotan T. The making of an embryo in a basal metazoan: Proteomic analysis in the sea anemone Nematostella vectensis. Proteomics. 2015;15(23–24):4096–4104. doi:10.1002/pmic.201500255.
- Lotan T, Chalifa-Caspi V, Ziv T, Brekhman V, Gordon MM, Admon A, Lubzens E. Evolutionary conservation of the mature oocyte proteome. EuPA Open Proteomics. 2014;3:27–36. doi:10.1016/j.euprot.2014.01.003.
- Love MI, Huber W, Anders S, Lönnstedt I, Speed T, Robinson M, Smyth G, McCarthy D, Chen Y, Smyth G, et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi:10.1186/s13059-014-0550-8.
- Zhang P, Kang JY, Gou LT, Wang J, Xue Y, Skogerboe G, Dai P, Huang DW, Chen R, Fu XD, et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015;25(2):193–207. doi:10.1038/cr.2015.4.
- Juliano CE, Reich A, Liu N, Götzfried J, Zhong M, Uman S, Reenan RA, Wessel GM, Steele RE, Lin H. PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. Proc Natl Acad Sci U S A. 2014;111(11):337–342. doi:10.1073/pnas.1320965111.
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455(7217):1193–1197. doi:10.1038/nature07415.
- Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T. Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev Genes Evol. 2001;211(6):299–308. https://doi.org/10.1007/s004270100156.
- Zhu W, Pao GM, Satoh A, Cummings G, Monaghan JR, Harkins TT, Bryant SV, Randal Voss S, Gardiner DM, Hunter T. Activation of germline-specific genes is required for limb regeneration in the Mexican axolotl. Dev Biol. 2012;370(1):42–51. doi:10.1016/j.ydbio.2012.07.021.
- Kozin VV, Kostyuchenko RP. Vasa, PL10, and Piwi gene expression during caudal regeneration of the polychaete annelid Alitta virens. Dev Genes Evol. 2015;225:129–138. doi:10.1007/s00427-015-0496-1.
- Giani VC, Yamaguchi E, Boyle MJ, Seaver EC. Somatic and germline expression of piwi during development and regeneration in the marine polychaete annelid Capitella teleta. Evodevo. 2011;2(1):10. doi:10.1186/2041-9139-2-10.
- Rebscher N, Zelada-González F, Banisch TU, Raible F, Arendt D. Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev Biol. 2007;306(2):599–611. 10.1016/j.ydbio.2007.03.521.
- Rebscher N. Establishing the germline in spiralian embyos. Int J Dev Biol. 2014;58(6–8):403–411. doi:10.1387/ijdb.140125nr.
- Gehrke AR, Srivastava M. Neoblasts and the evolution of whole-body regeneration. Curr Opin Genet Dev. 2016;40:131–137. doi:10.1016/j.gde.2016.07.009.
- Bradshaw B, Thompson K, Frank U. Distinct mechanisms underlie oral vs aboral regeneration in the cnidarian hydractinia echinata. Elife. 2015;2015(4):1–19. doi:10.7554/eLife.05506.
- Shibata N, Umesono Y, Orii H, Sakurai T, Watanabe K, Agata K. Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev Biol. 1999;206(1):73–87. doi:10.1006/dbio.1998.9130.
- Nakao H, Hatakeyama M, Lee JM, Shimoda M, Kanda T. Expression pattern of Bombyx vasa-like (BmVLG) protein and its implications in germ cell development. Dev Genes Evol. 2006;216(2):94–99. doi:10.1007/s00427-005-0033-8.
- Pfister D, De Mulder K, Hartenstein V, Kuales G, Borgonie G, Marx F, Morris J, Ladurner P. Flatworm stem cells and the germ line: Developmental and evolutionary implications of macvasa expression in Macrostomum lignano. Dev Biol. 2008;319(1):146–59. doi:10.1016/j.ydbio.2008.02.045.
- Swartz SZ, Chan XY, Lambert JD. Localization of Vasa mRNA during early cleavage of the snail Ilyanassa. Dev Genes Evol. 2008;218(1–2):107–113. doi:10.1007/s00427-008-0203-6.
- Özhan-Kizil G, Havemann J, Gerberding M. Germ cells in the crustacean Parhyale hawaiensis depend on Vasa protein for their maintenance but not for their formation. Dev Biol. 2009;327(1):230–239. doi:10.1016/j.ydbio.2008.10.028.
- Kranz AM, Tollenaere A, Norris BJ, Degnan BM, Degnan SM. Identifying the germline in an equally cleaving mollusc: Vasa and Nanos expression during embryonic and larval development of the vetigastropod Haliotis asinina. J Exp Zool Mol Dev Evol. 2010;314(4):267–279. doi:10.1002/jez.b.21336.
- Rebscher N, Lidke A, Ackermann C. Hidden in the crowd: primordial germ cells and somatic stem cells in the mesodermal posterior growth zone of the polychaete Platynereis dumerillii are two distinct cell populations. Evodevo. 2012;(3):9. doi:10.1186/2041-9139-3-9.
- Cho SJ, Vallès Y, Weisblat DA. Differential expression of conserved germ line markers and delayed segregation of male and female primordial germ cells in a hermaphrodite, the leech helobdella. Mol Biol Evol. 2014;31(2):341–354. doi:10.1093/molbev/mst201.
- Voronina E, Seydoux G, Sassone-Corsi P, Nagamori I. RNA granules in germ cells. Cold Spring Harb Perspect Biol. 2011;3(12). doi:10.1101/cshperspect.a002774.
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to De Novo DNA Methylation in Mice. Mol Cell. 2008;31(6):785–799. doi:10.1016/j.molcel.2008.09.003.
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nature Rev Genet. 2009;10(10):691–703. doi:10.1038/nrg2640.
- Britten RJ. Transposable element insertions have strongly affected human evolution. Proc Natl Acad Sci U S A. 2010;107(46):19945–8. doi:10.1073/pnas.1014330107.
- Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27(20):2702–2711. doi:10.1038/emboj.2008.204.
- Gebert D, Ketting RF, Zischler H, Rosenkranz D. piRNAs from pig testis provide evidence for a conserved role of the Piwi pathway in post-transcriptional gene regulation in mammals. PLoS One. 2015;10(5):1–22. doi:10.1371/journal.pone.0124860.
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol. 2009;19(24):2066–2076. doi:10.1016/j.cub.2009.11.064.
- Hirano T, Iwasaki YW, Lin ZY, Imamura M, Seki NM, Sasaki E, Saito K, Okano H, Siomi MC, Siomi H. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA. 2014;20(8):1223–1237. doi:10.1261/rna.045310.114.
- Ha H, Song J, Wang S, Kapusta AA, Feschotte CC, Chen KC, Xing J. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics. 2014;15(1):545. doi:10.1186/1471-2164-15-545.
- Genikhovich G, Technau U. Induction of spawning in the starlet sea anemone Nematostella vectensis, in vitro fertilization of gametes, and dejellying of zygotes. Cold Spring Harb Pprotoc. 2009;(9):pdb.prot5281. doi:10.1101/pdb.prot5281.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol Biol. 1990;215(3):403–410. doi:10.1016/S0022-2836(05)80360-2.
- Stefanik DJ, Lubinski TJ, Granger BR, Byrd AL, Reitzel AM, DeFilippo L, Lorenc A, Finnerty JR. Production of a reference transcriptome and transcriptomic database (EdwardsiellaBase) for the lined sea anemone, Edwardsiella lineata, a parasitic cnidarian. BMC Genomics. 2014;15(1):71. doi:10.1186/1471-2164-15-71.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi:10.1093/bioinformatics/btp348.
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–2105. doi:10.1093/bioinformatics/bti263.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biol. 2010;59(3):307–321. doi:10.1093/sysbio/syq010.
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi:10.1093/bioinformatics/btg180.
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. doi:10.1038/nbt1385.
- Schug J, Schuller W-P, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6(4):R33. doi:10.1186/gb-2005-6-4-r33.
- Helm RR, Siebert S, Tulin S, Smith J, Dunn CW. Characterization of differential transcript abundance through time during Nematostella vectensis development. BMC genomics. 2013;14(1):266. doi:10.1186/1471-2164-14-266.
- Genikhovich G, Technau U. In situ hybridization of starlet sea anemone (Nematostella vectensis) embryos, larvae, and polyps. Cold Spring Harb Protoc. 2009;2009(9):pdb.prot5282. doi:10.1101/pdb.prot5282.
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler AD. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi:10.1101/gr.229102.
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov V V, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007(6);317(5834):86–94. doi:10.1126/science.1139158.
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, et al. The dynamic genome of Hydra. Nature. 2010;464(7288):592–596. doi:10.1038/nature08830.
- Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–6. doi:10.1093/nar/gks1219.
- Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, Smirnova T, Grigoriev IV, Dubchak I. The genome portal of the department of energy joint genome institute: 2014 updates. Nucleic Acids Res. 2014;42(Database issue):D26–31. doi:10.1093/nar/gkt1069.
- Nettling M, Treutler H, Grau J, Keilwagen J, Posch S, Grosse I. DiffLogo: a comparative visualization of sequence motifs. BMC bioinformatics. 2015;16(1):387. doi:10.1186/s12859-015-0767-x.
- Rosenkranz D, Rudloff S, Bastuck K, Ketting RF, Zischler H. Tupaia small RNAs provide insights into function and evolution of RNAi-based transposon defense in mammals. RNA. 2015;21(5):911–922. doi:10.1261/rna.048603.114.
- Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi:10.1093/bioinformatics/btq033.
- Zhang P, Kang J-Y, Gou L-T, Wang J, Xue Y, Skogerboe G, Dai P, Huang D-W, Chen R, Fu X-D, et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015;25(2):193–207. doi:10.1038/cr.2015.4.
- Smit A, Hubley R, Green P. RepeatMasker Open-4.0. 2013–2015. http://www.repeatmasker.org. 2013.
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi:10.1038/nprot.2013.084.
- Kumar L, E Futschik M. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2(1):5–7. doi:10.6026/97320630002005.
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi:10.1093/bioinformatics/bts635.
