ABSTRACT
High-fidelity translation and a strictly accurate proteome were originally assumed as essential to life and cellular viability. Yet recent studies in bacteria and eukaryotic model organisms suggest that proteome-wide mistranslation can provide selective advantages and is tolerated in the cell at higher levels than previously thought (one error in 6.9 × 10−4 in yeast) with a limited impact on phenotype. Previously, we selected a tRNAPro containing a single mutation that induces mistranslation with alanine at proline codons in yeast. Yeast tolerate the mistranslation by inducing a heat-shock response and through the action of the proteasome. Here we found a homologous human tRNAPro (G3:U70) mutant that is not aminoacylated with proline, but is an efficient alanine acceptor. In live human cells, we visualized mistranslation using a green fluorescent protein reporter that fluoresces in response to mistranslation at proline codons. In agreement with measurements in yeast, quantitation based on the GFP reporter suggested a mistranslation rate of up to 2–5% in HEK 293 cells. Our findings suggest a stress-dependent phenomenon where mistranslation levels increased during nutrient starvation. Human cells did not mount a detectable heat-shock response and tolerated this level of mistranslation without apparent impact on cell viability. Because humans encode ∼600 tRNA genes and the natural population has greater tRNA sequence diversity than previously appreciated, our data also demonstrate a cell-based screen with the potential to elucidate mutations in tRNAs that may contribute to or alleviate disease.
Introduction
A highly accurate proteome is not required for life. In the years preceding and immediately following the elucidation of the genetic code,Citation1,Citation2 it was argued that mis-interpretation of the genetic code and the resulting mistranslation would lead to an error catastropheCitation3 in the proteome that could not support life. Crick used the same logic to argue that the code could not evolve further or tolerate significant errors because modern proteomes are composed of “so many highly evolved protein molecules that any change to these would be highly disadvantageous unless accompanied by many simultaneous mutations to correct the ‘mistakes’ produced by altering the code.Citation 4 ” There is now ample evidence that the code does indeed continue to evolve through codon reassignments, codon recoding, and natural genetic code expansion in species representing the complete diversity of life.Citation5
In contrast to the concept of a highly accurate proteome, diverse organisms tolerate mistranslation. A series of landmark studies, in Escherichia coli,Citation6 yeast,Citation7 and mammalian cellsCitation8 reshaped our view of the level of accuracy required to produce a functional proteome. Errors in protein synthesis are estimated to occur at a rate of 1 mis-incorporated amino acid in 104 to 105 codons,Citation9,Citation10 which suggests that between 0.01 and 0.001% of codons are misread in the human proteome. E. coli can tolerate remarkably high levels of mistranslation. Indeed, Ruan et al.Citation6 showed that E. coli grow similarly to wildtype even when 10% of genetically encoded asparagine residues were mistranslated as aspartic acid. The heat-shock response, and the expression of cellular proteases were essential to maintain wild-type like growth in these cells despite proteome-wide mistranslation.Citation6
In Saccharomyces cerevisiae, we selected a mutant tRNA that suppressed a stress-sensitive phenotype by inducing proline to alanine mistranslation at a rate of ∼6%.Citation11 Yeast cells tolerated this tRNA-dependent mistranslation by inducing heat-shock, again without a significant growth defect. In the mistranslating yeast strain, we also observed synthetic slow growth with a deletion of a gene (rpn4) encoding a transcription factor that regulates proteasome gene expression, implicating the proteasome and protein degradation as critical mechanisms the cell uses to tolerate mistranslation.Citation11 In mammalian cells, tyrosine limitation leads to proteome-wide mistranslation of tyrosine codons with phenylalanine at a rate of ∼1%, again without a significant impact on cellular viability.Citation12
Not only can cells tolerate mistranslation, but mistranslation may also be advantageous in conditions of stress. Mistranslation in bacteriaCitation13 and mammalian cellsCitation14,Citation15 protects cells from reactive oxygen species. In an E. coli strain with a ribosomal protein mutant (RpsD) that enhances mistranslation, a stop codon suppression assay indicated ∼5-fold increase in translation errors above the basal level. This mistranslation increased expression of a general stress response activator (RpoS) that in turn up-regulated antioxidant gene expression, making the cell more resistant to peroxide treatment.Citation13 In HEK 293T cells, mis-aminoacylation of tRNAGlu with methionine accounts for a ∼0.5% basal level of mistranslation.Citation16 Immune stimulation or chemical induction of oxidative stress increased this rate up to 10-fold.Citation17 The current model suggests that oxidative stress leads to extracellular signal–regulated kinase (ERK) dependent phosphorylation of the methionine-tRNA synthetase, which makes the enzyme more promiscuous for non-cognate tRNAs.Citation18 The additional methionine incorporated in the proteome at non-methionine positions is thought to act as a ‘sink’ for reactive oxygen species (ROS), and methionine mistranslation increased resistance to ROS in cell culture.Citation17 These studies demonstrate that cells are capable not only of tolerating proteome-wide mistranslation, but can derive a selective advantage from mistranslation under certain stress conditions.
We recently devised a selection in yeast that requires mistranslation at proline codons for the organism to survive under stress. The selection relied on a stress-sensitive allele of the PIK kinase chaperon, Tti2.Citation11,Citation19,Citation20 The selection produced four independent strains with a single base mutation (C70U) in the acceptor stem of tRNAPro, yielding a tRNAPro with the essential G3:U70 identity element for alanyl-tRNA synthetase (AlaRS). Unlike many tRNA synthetases, AlaRS does not recognize the anticodon of its cognate tRNA (). Rather it establishes critical interactions with both major and minor groove sides of the G3:U70 base pair in the acceptor stem of tRNAAla.Citation21 These interactions orient the 5′ amino acid accepting CCA-end of the tRNA in the AlaRS active site (). Although AlaRS does not tolerate the reverse U:G pair or other mutations at this position,Citation22 the human AlaRS shows significant activity with tRNAs that encode a G4:U69 pair,Citation23 of which there are several in the human reference genome. Taken together, these studies suggest that because of its minimal identity requirements, the generation of tRNA mutations that are mischarged with Ala may be more prevalent in nature, including in humans, than previously assumed.
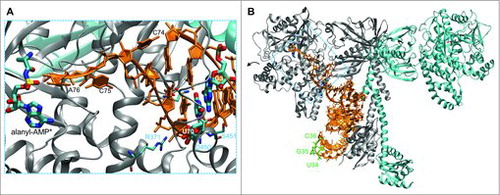
Figure 1. Structure of AlaRS and tRNAAla complex. The zoomed in view (A) focuses on the interaction of the tRNAAla acceptor stem with the AlaRS active site (PDB code 3WQYCitation21). The 3′ terminal CCA bases of the tRNA are labeled. An alanyl-adenylate analog (*) is also shown. The major AlaRS identity element G3:U70 is highlighted. AlaRS residues (R371, N359, D450, S451) form a hydrogen bond network (gray dashes) that contacts and ‘reads’ the GU pair from both the major and minor grove sides of the tRNA. The complete dimeric AlaRS is shown in complex with tRNAAla (B). The three bases of the anticodon (U34, G35, C36; green) are not recognized by the AlaRS.
Here we produced and characterized a mutant human tRNAPro with the G3:U70 base pair (). With biochemical experiments and live cell imaging, we quantified and visualized tRNA-dependent mistranslation in human cells, finding that the mutant tRNAPro is an efficient alanine acceptor that promotes mistranslation in human cells in culture.
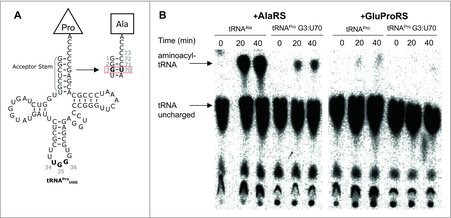
Figure 2. In vitro amino-acylation of tRNAPro (G3:U70). (A) Cloverleaf diagrams of human tRNAPro UGG WT and G3:U70. Mutations in the acceptor stem are outlined. (B) Purified 32P-radiolabeled tRNAs were aminoacylated with Ala by human AlaRS or Pro by human GluProRS as indicated in Methods. The autoradiographs show reaction progress (accumulation of aminoacylated-tRNA) for each reaction over a 40-minute time course.
Materials and methods
Plasmids and strains. Plasmid manipulations were performed with E. coli DH5α cells (Invitrogen, Carlsbad, CA, USA). For in vitro aminoacylation assays, tRNA genes were ordered as single stranded DNA oligomers from Sigma-Aldrich (Oakville, ON, Canada) with overhangs to BamHI/XbaI sites. Single-stranded oligomers were phosphorylated using T4 polynucleotide kinase (New England Biolabs (NEB), Ipswich, MA, USA) and annealed by gradual cooling from 95°C to 55°C. Annealed oligomers were cloned into pUC19 BamHI/XbaI sites using BamHI, XbaI, and T4 DNA ligase (NEB). pUC19-derived plasmids were purified by mini-prep (GeneAid, New Taipei City, Taiwan) and used as template for in vitro transcription. For expression in human cell culture, EGFP D129A or D129P genes were cloned into pcDNA3.1 HindIII/EcoRI sites. Transfer RNA genes were fused to a human U6 promoter sequence and 6x thymidine termination sequence by overlap extension PCR (pfu polymerase, Agilent Technologies), then ligated into pcDNA3.1 at the NruI site. Plasmid DNA was isolated from 100 ml E. coli cultures by midi-prep (GeneAid) followed by phenol chloroform extraction and ethanol precipitation, then re-suspended in sterile Milli-Q water. All plasmids were diluted to equimolar concentrations before transfection.
In vitro tRNA transcription and radiolabeling. In vitro transcription and radiolabeling of tRNAs for in vitro aminoacylation experiments was performed as previously describedCitation24 (see Supporting Information for tRNA sequences and details).
In vitro tRNA aminoacylation. For the aminoacylation assays we purchased recombinant human AlaRS (Abcam, ab73442, Cambridge, UK) and human glutamyl-prolyl-tRNA synthetase (EPRS, Origene, TP317559, Rockville, MD). 50 μl reactions with 5 μM tRNA, 300 nM 32P-labelled tRNA, 10 mM amino acid (Ala or Pro as indicated), 5 mM ATP and 1 μM tRNA synthetase were incubated at 37°C for 0, 20 and 40 minutes. Reactions were digested with nuclease P1, spotted on a polyethyleneimine-cellulose thin layer chromatography (TLC) plate (EMD Millipore, Billerica, MA, USA) and chromatographed in 5% acetic acid and 100 mM ammonium acetate. TLC plates were exposed to a phosphor screen, which was imaged using a Storm 860 Phosphorimager (GE Healthcare Life Science, Little Chalfont, UK).
HEK 293 cell transfection. Transfections were performed in 24-well or 6-well plates using 1.25 μg or 2.5 μg of DNA, respectively. Cells were cultured in high glucose Dulbecco's modified Eagle medium (DMEM, 4.5 g/L glucose) containing penicillin, streptomycin (P/S), and 10% fetal bovine serum (FBS) (Gibco by Life Technologies, Carlsbad, MA). At 70–80% cell confluency, media was replaced with standard DMEM (no FBS, P/S) and cells transfected with pcDNA3.1-derived plasmids using lipofectamine 2000 (Invitrogen). Cells were returned to +P/S, +10% FBS media to recover for at least 1 day before analysis. Cell fluorescence was quantitated daily as described below. For experiments in low serum, low glucose media, after ‘day 1’ quantitation, media was replaced with 1% FBS, 1 g/L glucose DMEM (Corning Cellgro, Corning, NY, USA). Geneticin selections were maintained in DMEM (4.5 g/L glucose, 10% serum, P/S) containing 500 µg/ml geneticin (G418; Gibco by Life Technologies).
Fluorescence quantitation of enhanced GFP (EGFP) reporter. To quantify EGFP fluorescence, images were captured using an EVOS FL auto fluorescent microscope (Thermo Fisher Scientific, Waltham, MA, USA) at 20X magnification and GFP foci outlined using the ellipse measurement tool. The EVOS microscope measures GFP fluorescence with 470/525 nm excitation/emission wavelengths. In each field of view, the 15 brightest foci (i.e., cells) were outlined and data collected as mean pixel intensity within each ellipse. At least 135 foci were measured from each experiment daily across three biological replicates and three fields-of-view per replicate. On the final day, an additional two fields-of-view were collected per transfection, totaling 225 foci. The average of five background measurements was subtracted from all values in each field of view to account for inconsistencies in light scattering between fields. Percentage mistranslation was estimated using the means of each data set and p-values were calculated pairwise using single-factor ANOVA with Microsoft Excel.
Western blotting. Cells were harvested by pipetting and centrifuged in 1.5 ml microcentrifuge tubes at 1500 × g for 3 min at 4°C. Supernatant was removed and cells washed with ice cold Phosphate Buffered Saline (1 × PBS pH 7.4; Corning Cellgro) and centrifuged again. PBS was aspirated off and cells were suspended in 50 μl of ice-cold Lysis buffer: 50 mM Tris-HCl (pH 7.4), 1% Triton X-100, 150 mM NaCl, 0.1% sodium dodecyl sulfate (SDS), and 1 mM Phenylmethylsulfonyl fluoride. The re-suspended cells were incubated on ice for 5 min then centrifuged at 4°C, 30,000 × g for 10 minutes. Lysates were separated on standard SDS-polyacrylamide gel electrophoresis (PAGE) (15% acrylamide) and transferred to Polyvinylidene fluoride membranes using a Trans-Blot Turbo Transfer System (BioRad, Hercules, CA, USA). Membranes were incubated for 1 hr in blocking solution (3% bovine serum albumin (BSA), 0.1% Tween 20, 1% PBS) before adding primary antibodies at a 1:5000 final dilution (α-HSP70, Invitrogen, MA3-006; α-HSP90, Protein Tech, Rosemont, IL, USA, 13171-1-AP; α-GFP, abcam, ab32146; α-GAPDH, Sigma-Aldrich, MAB374). Membranes were incubated overnight at 4°C, washed for 3 × 10 min in washing solution (1% BSA, 0.1% Tween 20, 1% PBS), then incubated with anti-mouse (Thermo Fisher Scientific, MA1-21315) or anti-rabbit (Sigma, GENA9340) horse radish peroxidase-linked secondary antibodies for 2 hr at 1:2000 final dilution. Membranes were then washed in 1 × PBS with 0.1% Tween 20 for 3 × 10 minutes, followed one wash for 10 minutes in 1 × PBS. Protein markers were visualized using Clarity Western enhanced chemiluminescence (ECL) Substrates (Bio-Rad) following the manufacturer's instructions and imaged with a ChemiDoc XRS+ System (Bio-Rad).
Cell viability assay. HEK 293 cell cultures were seeded at equivalent cell densities and grown overnight in high glucose DMEM, then transfected in triplicate and assayed either one day post-transfection or switched to low serum, low glucose DMEM for 4 days. Cellular viability was determined using a CellTiter-Glo Luminescent Cell Viability Assay following the manufacturer's instructions (Promega, Madison, WI). Each transfection was assayed in triplicate, totaling three biological and nine technical replicates per experiment.
Results
Biochemical characterization of a mistranslating human tRNAPro (G3:U70). We first investigated human tRNAPro G3:U70 for mis-acylation activity with Ala (). We produced and purified human tRNAPro, tRNAAla, and tRNAPro G3:U70 by in vitro transcription using recombinant T7 RNA polymerase. Recombinant human AlaRS (Abcam) and human glutamyl-prolyl-tRNA synthetase (GluProRS, Origene) were purchased. The ProRS activity in human cells is encoded as a single polypeptide fused to GluRS. We performed aminoacylation assays according to standard protocols (see Methods) as previously.Citation24 The level of aminoacyl-tRNA formed at specific time points was measured using 32P-radiolabeled tRNA variants (see SI Methods). Following nuclease P1 digestion, unreacted tRNA and aminoacylated-tRNA product were separated by thin layer chromatography and visualized by autoradiography (). The aminoacylation assays confirmed significant activity of human GluProRS in Pro-tRNAPro formation, and efficient aminoacylation by human AlaRS of tRNAAla with Ala (). Like our previous findings with the homologous yeast AlaRS and ProRS,Citation11 ProRS activity is weaker than that observed with AlaRS. Pro accepting activity was not detected with the tRNAPro G3:U70 mutant and human GluProRS, indicating that the single base pair mutation is a sufficient anti-determinant for GluProRS. In the presence of AlaRS and Ala, however, a gain-of-function was observed as the mutant tRNA was charged with Ala, though to a lesser extent than tRNAAla (). These results demonstrate tRNAPro (G3:U70) is an efficient Ala acceptor in vitro and the mutation is sufficient to disable ProRS activity.
GFP reporter illuminates mistranslation in living cells. We recently established a novel EGFP mistranslation reporter in yeast.Citation11 The GFP reporter contains a mutation, D129P, which is thought to cause a kink in the backbone of GFP's β-barrel structure.Citation11 When the Pro codon at position 129 in the EGFP reporter is translated accurately as Pro, the resulting EGFP, although stably produced, lacks fluorescence. The mutant D129A, however, fluoresces similarly to wild type EGFP. In our studies in yeast, this reporter was a sensitive measure of mistranslation activity in live cells. We reported a level of ∼6% mistranslation of the critical Pro129 codon to Ala in a yeast strain that co-expressed the mistranslating tRNAPro (G3:U70).Citation11
To define how human cells in culture respond to mistranslation, we conducted a series of transient transfection experiments in HEK 293 cells. Each cell line harbored a pCDNA3.1 plasmid containing separate EGFP and tRNA expression cassettes. Our EGFP reporter (D129A or D129P) was expressed under the control of a pCMV constitutive promoter. We chose to test a CCA codon for mistranslation by tRNAPro UGG G3:U70, consistent with our study in yeast.Citation11 The tRNA expression cassette was designed to include an upstream U6 constitutive promoter and followed by a downstream poly-T terminator sequence (see Methods).
Having established our Pro-codon sensitive EGFP mistranslation reporter for use in human cells, we transfected HEK 293 cells in triplicate with plasmids co-expressing EGFP D129P or D129A and the wild type (C3:G70) or mistranslating (G3:U70) human tRNAPro (, S3). Based on 3 biological replicates, transfection efficiencies were initially ∼50-60% as determined from the number of cells fluorescing in our EGFP D129A positive control, but declined to ∼20-40% by the end of our multi-day experiment due to plasmid dilution by cell division. Mistranslation was quantified by fluorescence microscopy (, see Methods).
Figure 3. tRNA-dependent mistranslation increases under glucose and serum starvation. HEK 293 cell cultures were transfected in triplicate with plasmid harboring tRNAPro and EGFP D129P (WT); tRNAPro G3:U70 and EGFP D129P (G3:U70); or tRNAPro and EGFP D129A (+). Cells were grown for 1 day post-transfection in high glucose media (high), then media was replaced with low serum, low glucose (low) and fluorescence was measured daily by fluorescence microscopy (see Methods). Box and whisker plots (A) of EGFP foci intensity. Horizontal demarcations depict quartiles with median centered. Means are depicted with an X. Dots represent the general distribution of at least 135 foci measured in each plot across 3 biological and at least 9 technical replicates. Mistranslation levels were estimated based on means of the three populations on each day and the p-value of a difference between the WT and G3:U70 populations is reported base on single-factor ANOVA. Representative images (B) of mistranslating cells. HEK 293 cell images were captured at 20X magnification using light (phase) or fluorescence (GFP; ex/em = 470/525 nm) microscopy. Scale bars depict 200 µm.
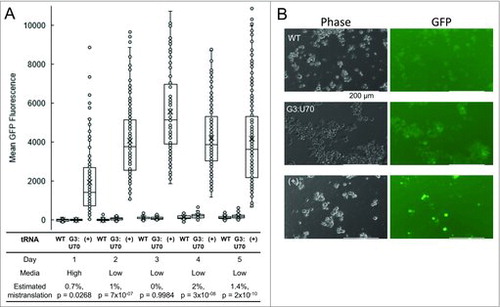
In standard, high glucose media, we did not detect a significant difference in EGFP D129P fluorescence between cells expressing tRNAPro versus tRNAPro (G3:U70). Given the rapid division of cells in rich media, we were unable to measure mistranslation rates in this condition beyond day 1 due to dilution of the transfected plasmid. Serum starvation promotes differentiation and prevents rapid cell division. We also reasoned, based on the literature (e.g., [Citation13–Citation15]), that mistranslation may be enhanced in conditions of stress. Therefore, the transfected cell lines were transferred to a low serum and low glucose medium and monitored daily. For cells expressing wild type and mutant tRNAPro, we analyzed GFP fluorescence up to 5 days following transfection. Under glucose and serum starvation, a trend of accumulating mistranslation was observed in cells expressing the mutant tRNAPro over the course of the experiment. The estimate of mistranslation was calculated based on the relative fluorescence of EGFP D129P and D129A expressing controls (, S4). Following recovery (∼1 day after switching media), we observed a reproducible, and statistically significant, 1.4–2% (p-value < 10−8) increased rate of mistranslation on days 4 and 5 in the cells expressing tRNAPro (G3:U70). In 10% of the cell population, we observed mistranslation levels of 5% or greater. Equivalent GFP expression in all cell lines was confirmed by immunoblotting and compared to a glyceraldehyde 3-phosphate dehydrogenase loading control (). The mistranslation-dependent effect began to taper on day 5 due to gradual plasmid loss. Cells were lysed at this stage for further analysis by western blotting.
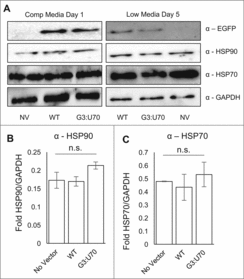
Figure 4. Mistranslation caused no detectable induction of heat shock response. Western blotting of HEK 293 cell lysates (A) with antibodies detecting EGFP, heat shock markers (HSP70, HSP90), and a GAPDH loading control. Lysates were harvested after one day incubation in high glucose media (A, left panel) or changed to low serum, low glucose media (A, right panel) for four days before harvesting. Lysates were separated by SDS-PAGE; loading quantities were balanced according to triplicate Bradford assays. Densitometry based on triplicate western blots of HSP90 (B) and HSP70 (C) levels normalized to the GAPDH control. Error bars indicated 1 standard deviation of the mean. (B, C) Lysates harvested on day 5 low serum, low glucose media from three independent transfections. None of the values reported were statistically significantly different (ANOVA).
Mistranslation following antibiotic selection. In an attempt to control for transfection efficiency in these experiments, we also observed elevated mistranslation in cells following selection on geneticin. The selected cells maintained the pCDNA plasmid bearing the GFP reporter and either the wild type or mistranslating tRNAPro. Following selection, there was no obvious change in morphology when the tRNAPro (G3:U70) mutant was expressed compared to tRNAPro, though morphological heterogeneity could be seen in both cell lines (Fig S1). Geneticin inhibits bacterial growth by interfering with ribosome function, and similar aminoglycosides impair codon-anti-codon pairing in bacterialCitation25 and mammalianCitation26 translation systems. We considered the possibility that cells treated with aminoglycosides may have overall elevated mistranslation rates and this could potentially impact our GFP reporter if the Pro codon was decoded by endogenous near-cognate tRNAs. Although we did not observe elevated mistranslation in cells expressing the wild type tRNAPro, analysis by fluorescence microscopy suggested that GFP D129P fluorescence was restored at a low level (∼1%, p-value = 1.2 × 10−8) by tRNAPro (G3:U70)-dependent mistranslation with Ala (Fig S2). This result is concordant with our observations in nutrient deprived cells ().
Mistranslation by tRNAPro (G3:U70) does not induce a heat-shock response. In our previous studies of mistranslation induced by tRNAPro (G3:U70) in yeast, the heat-shock response was observed in mistranslating cells.Citation11 HEK 293 cells were harvested following 4 days of growth on low serum and low glucose medium where mistranslation was observed. Using GAPDH as a loading control, we immunoblotted to detect markers for the heat shock response (HSP70, HSP90). Despite the fact that cells were mistranslating Pro codons with Ala at an average rate of ∼2%, we were unable to detect significant differences in the levels of HSP70 or HSP90 in comparison to cells expressing the wild type tRNAPro ().
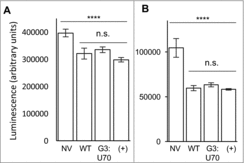
Viability of mistranslating cells. Using a standard assay for cell viability, based on the production of ATP in metabolically active cells (see Methods), we determined the impact of tRNAPro and tRNAPro G3:U70 expression on the viability of HEK 293 cells in culture. Cells were transfected with the indicated EGFP and tRNAPro expressing pCDNA 3.1 plasmids () and grown in rich media or in low nutrient conditions. Expression of the EGFP protein reduced cell viability marginally in comparison to the un-transfected control cells (p < 0.0001, ). In agreement with our measurements of the mistranslation as indicated by the GFP reporter, cell viability in rich media was unaffected by expression of tRNAPro in comparison to the mistranslating tRNAPro G3:U70 mutant (). In low serum, low glucose medium, the viability of all cultures decreased, yet cells expressing tRNAPro G3:U70 were equally as viable as cells expressing the wildtype tRNAPro ().
Figure 5. Viability of mistranslating cells. HEK 293 cell cultures were transfected in triplicate with lipofectamine only (No Vector, NV), or plasmid harboring tRNAPro and EGFP D129P (WT); tRNAPro G3:U70 and EGFP D129P (G3:U70); or tRNAPro and EGFP D129A (+). Transfected cells were assayed one day post-transfection (A) or after four days incubation in low serum, low glucose media (B). Viability assays were performed following manufacturer's instructions (see Methods) using three technical replicates per transfection. Plots show mean luminescence, which correlates with cellular ATP levels. Error bars indicated 1 standard deviation of the mean. Stars depict the indicated statistically significant differences according to single-factor ANOVA (**** = p < 0.0001; *** = p < 0.001; n.s. = p > 0.05).
Discussion
Translation of the 61 sense codons in the genetic code table requires a theoretical minimum of 32 tRNAs to translate all 20 amino acids.Citation27 There are examples of organelles (human mitochondria, 22 tRNAs), parasites (Borrelia burgdorferi, 32 tRNAs; Mycoplasma pulmonis, 28 tRNAs), and obligate symbiotic microbes (Sulcia muelleri, 31 tRNAs) with a small number of tRNA genes, some of which fall below the ‘theoretical’ limit based on wobble decoding.Citation28 In the case of organelles, a complete set of tRNAs for protein synthesis is achieved via tRNA import from nuclear encoded tDNAsCitation29 or perhaps super-wobbling (i.e., one tRNA reading 4 codons) as demonstrated with tRNAGly in tobacco plants.Citation30 The fact that no free-living organisms has less than 32 tRNAs already indicates that the coding problem is more complex or more elaborate than the simple mechanics of matching codon to anticodon and wobble decoding. In free living microbes, the smallest sets of tRNA genes are found in the model minimal genome of Mycoplasma genitalium (36 tRNA genes), while the hyperthemophile Methanopyrus kandleri encodes just 34 tRNAs genes.
The number and diversity of tRNA genes expands in correlation with the complexity of the organism.Citation31 For example, yeast encodes 275 tRNAs whereas humans have 610 tRNA genes.Citation28 None of the human tRNA genes are predicted as pseudogenes.Citation28 Most human tRNA genes are expressed in some cells or tissues,Citation28 and tRNA genes show tissueCitation32 and cell-line specific expression patterns.Citation33 Even individual tissues express a large number of these tRNA genes. For example, in human primary liver tissue, 223 tRNA genes are actively transcribed.Citation34 The reason for the over-abundance of tRNA genes in human and higher eukaryotes has remained a mystery.
A single tRNA mutant induces mistranslation in human cells. Among the human population, tRNA mutations are common. Based on sequence data from the 1000-genomes project, more than half of individuals (54%) contain at least 1 mutant tRNA and 20% of individuals harbor two or more mutant tRNAs.Citation35 Fascinatingly, tRNA gene copy number also varies even among a small group of individuals.Citation36
Mistranslating tRNAs are present in the reference human genome. Recent work demonstrated tRNA variants with a G4:U69 pair enable aminoacylation with Ala. G4:U69 mutants are common in eukaryotic genomes and 1 of the 30 copies of tRNACys in the human genome contains such a GU pair. The mutation confers Ala accepting identity on this tRNACys variant in vitro and in HEK 293 cells in culture.Citation23
Given the complexity of the human tRNAome and its diversity in the human population, we set out to determine if a single tRNA mutant would induce mistranslation in human cells in culture, and to characterize the consequences of this mistranslation. In our previous studies in yeast, expression of a mutant tRNA caused ∼6% mis-incorporation at Pro codons in cells grown in nutrient rich conditions.Citation11 Here we expressed a homologous mistranslating tRNAPro mutant in human cells in culture, and we were unable to detect mistranslation in rich media. Only upon subjecting the cells to stress, including nutrient deprivation or antibiotic selection, did we observe mistranslation induced by the tRNAPro G3:U70 mutant. The results suggest that the human cell is potentially more resistant to mistranslation caused by a single tRNA gene, however, under conditions of stress or nutrient deprivation, mis-made protein could be readily detected. We speculate that under stress conditions human cells are either less able to degrade mis-made protein, deacylate mis-charged tRNAsCitation37,Citation38 at a lower frequency, or more readily use the mis-aminoacylated tRNA in translation.
tRNA-dependent mistranslation linked to disease. There is growing evidence connecting mistranslation to disease.Citation19,Citation39-Citation41 Many efforts focus on AARSs, either affected by mutations or oxidative damage,Citation13 that promote mis-aminoacylation of their cognate tRNAsCitation12,Citation42 or of non-cognate tRNAs,Citation43 both of which can then lead to mistranslation. The relevance of analyzing the role of tRNA mutants in disease was highlighted by a recent study that identified a mutation in a single tRNA gene specifically expressed in the central nervous system. The tRNA mutation caused a synthetic neurodegenerative phenotype in mice also lacking the gene GTPB2, which is involved in ribosome recycling.Citation44 In agreement with our findings, the impact of the tRNA mutation alone, in the absence of stress or a synthetic defect, was not sufficient to cause significant mistranslation. This recently established link between tRNA mutations and disease suggests that further studies will uncover a greater role for human tRNA variants in health and disease.
Detecting tRNA-dependent mistranslation in live cells. A key difference between our work and previous work with editing defective tRNA synthetases is that the AARS mutants mis-aminoacylate (to some level) all tRNA isoacceptors for the cognate amino acid. In our experiments, we expressed a single tRNAPro mutant. Although this tRNA is specifically mis-aminoacylated with Ala, it then competes with properly aminoacylated tRNAPro isoacceptors expressed from 46 genetic loci in the diploid human genome. This contrasts with our previous studies in yeast, where mistranslating tRNAPro competes with only 10 tRNAPro genes. This difference in tRNA copy number may explain why the tRNAPro G3:U70 readily mistranslates in yeast cells in rich media, whereas in human cells, stress was required to enhance tRNAPro G3:U70 induced mistranslation. It may be that the large number of human tRNA genes serves as a ‘buffer’ against mistranslation induced by single gene tRNA mutants.
In this work, we demonstrated the use of a novel mistranslation reporter that is specifically sensitive to mistranslation at Pro codons in human cells. Given the sensitivity of GFP to mutations at key positions, a number of other groups have developed GFP-based reporters for different types of mistranslation. GFP (M72Q) has been used extensively in mammalian cells to detect natural levels of methionine mis-incorporation, which increases 10-fold in response to oxidative stress.Citation16,Citation43 GFP (E222Q)Citation45 and GFP (T65V)Citation46 are examples of reporters that detect mistranslation of glutamine and valine codons, respectively. Our GFP (D129P) reporter extends the range of GFP-based reporters for use in live human cells to include a new type of mistranslation.
A complementary approach involves using quantitative mass spectrometry to identify mistranslation in a GFP reporter.Citation47 In this approach, the fluorescence output of GFP was not used to detect mistranslation, rather a particular residue in GFP was chosen such that tryptic peptides that include the site are ideal for detection by MS. The technique, referred to as MS-READ, is unbiased as it can detect any type of mistranslation event, including detection of exceedingly low levels of amino acid mis-incorporation. Although this quantitative method cannot be applied in live cell imaging, it can be used in a kinetic mode to chart mistranslation in cells at specified time points. Taken together, these mistranslation reporters provide the opportunity to comprehensively identify mistranslation induced by natural and disease-linked tRNA variants.
Cellular adaptation to mistranslation. In yeast, tRNAPro G3:U70 induces a heat shock response.Citation11 Robust proteasomal activity was also required to maintain wild-type like growth in the context of proteome-wide mistranslation.Citation11 In the current study, we were unable to detect a significant impact of tRNA-dependent mistranslation on the heat shock response or on cell viability in HEK 293 cells. In contrast to the view of a highly accurate proteome, there are many examples in bacteria and eukaryotic cells (see Introduction), where levels of mistranslation of 1–10% have a negligible impact on cell growth and viability. For example, yeast cells expressing an editing defective phenylalanyl-tRNA synthetase (PheRS) in complete media showed no phenotypic defect, despite mis-incorporation of tyrosine at phenylalanine codonsCitation7. When these mistranslating yeast cells were grown under conditions of amino acid stress, i.e., in media containing a 1/400 ratio of Phe/Tyr, ∼5% of phenylalanine codons were mistranslated, which resulted in viable cells with a 50% reduction in growth rate. The strain failed to mount a heat shock response despite proteome wide mistranslation. The growth defect resulted from the fact that the PheRS editing defective yeast strain was ineffective in activating amino acid and protein stress response pathways to this particular type of mistranslation.Citation7 These striking observations reveal the complex and varied cellular responses that manifest as a result of distinct types of mistranslation, and highlight the need for a greater understanding of mistranslation in the context of cellular physiology and disease.
Conclusion
All cells have an inherent level of mistranslation.Citation8,Citation14 Cells, from bacteria to humans, have evolved mechanisms to cope with not only this inherent level of mistranslation, but, also elevated levels resulting from a variety of sources including oxidative and nutrient stress as well as AARS and tRNA mutations. The study noted aboveCitation7 and similar studies (reviewed inCitation48) are beginning to shed light on the pathways activated in response to mistranslation, and of particular interest is the interaction of mistranslation with cell stress. These advances are motivating our on-going work to further characterize the mammalian cellular response to Pro codon mistranslation in stress conditions and cellular models of disease. In addition, our approach represents a feasible high-throughput cell-based screen for tRNA function. We anticipate that the GFP reporter we developed here will have applications in mapping the function of human tRNA variants in the natural population and in elucidating tRNA mutations implicated as drivers or perhaps suppressors of disease.
krnb_a_1379645_sm9794.pdf
Download PDF (3.4 MB)Acknowledgments
We are grateful to Laszlo Gyenis, Michael Ibba, and David Litchfield for critical discussions, suggestions, and technical assistance. This work was supported from the Natural Sciences and Engineering Research Council of Canada [RGPIN-2015-04394 to C.J.B., RGPIN 04282-2014 to P.O.]; Canada Foundation for Innovation [229917 to P.O.]; the Ontario Research Fund [229917 to P.O.]; Canada Research Chairs [950-229917 to P.O.]; the Canadian Cancer Society Research Institute innovation grant [704324 to P.O.]; and generous donations from Graham Wright and James Robertson. M.D.B holds an Ontario Graduate Scholarship.
Additional information
Funding
References
- Söll D, Ohtsuka E, Jones DS, Lohrmann R, Hayatsu H, Nishimura S, Khorana HG. Studies on polynucleotides, XLIX. Stimulation of the binding of aminoacyl-sRNA's to ribosomes by ribotrinucleotides and a survey of codon assignments for 20 amino acids. Proc Natl Acad Sci U S A. 1965;54:1378–85. doi:10.1073/pnas.54.5.1378.
- Brimacombe R, Trupin J, Nirenberg M, Leder P, Bernfield M, Jaouni T. RNA codewords and protein synthesis, 8. Nucleotide sequences of synonym codons for arginine, valine, cysteine, and alanine. Proc Natl Acad Sci U S A. 1965;54:954–60. doi:10.1073/pnas.54.3.954
- Orgel LE. The maintenance of the accuracy of protein synthesis and its relevance to ageing. Proc Natl Acad Sci U S A. 1963;49:517–21. doi:10.1073/pnas.49.4.517
- Crick FH. The origin of the genetic code. J Mol Biol. 1968;38:367–79. doi:10.1016/0022-2836(68)90392-6
- Ling J, O'Donoghue P, Söll D. Genetic code flexibility in microorganisms: novel mechanisms and impact on physiology. Nat Rev Microbiol. 2015;13:707–21. doi:10.1038/nrmicro3568
- Ruan B, Palioura S, Sabina J, Marvin-Guy L, Kochhar S, Larossa RA, Söll D. Quality control despite mistranslation caused by an ambiguous genetic code. Proc Natl Acad Sci U S A. 2008;105:16502–7. doi:10.1073/pnas.0809179105
- Mohler K, Mann R, Bullwinkle TJ, Hopkins K, Hwang L, Reynolds NM, Gassaway B, Aerni HR, Rinehart J, Polymenis M, et al. Editing of misaminoacylated tRNA controls the sensitivity of amino acid stress responses in Saccharomyces cerevisiae. Nucleic Acids Res. 2017;45:3985–96. doi:10.1093/nar/gkx077
- Schwartz MH, Pan T. Function and origin of mistranslation in distinct cellular contexts. Crit Rev Biochem Mol Biol. 2017;52:205–19. doi:10.1080/10409238.2016.1274284
- Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 2009;10:715–24. doi:10.1038/nrg2662
- Kramer EB, Vallabhaneni H, Mayer LM, Farabaugh PJ. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA. 2010;16:1797–808. doi:10.1261/rna.2201210
- Hoffman KS, Berg MD, Shilton BH, Brandl CJ, O'Donoghue P. Genetic selection for mistranslation rescues a defective co-chaperone in yeast. Nucleic Acids Res. 2017;45:3407–21. doi:10.1093/nar/gkw1021
- Raina M, Moghal A, Kano A, Jerums M, Schnier PD, Luo S, Deshpande R, Bondarenko PV, Lin H, Ibba M. Reduced amino acid specificity of mammalian tyrosyl-tRNA synthetase is associated with elevated mistranslation of Tyr codons. J Biol Chem. 2014;289:17780–90. doi:10.1074/jbc.M114.564609
- Fan Y, Wu J, Ung MH, De Lay N, Cheng C, Ling J. Protein mistranslation protects bacteria against oxidative stress. Nucleic Acids Res. 2015;43:1740–8. doi:10.1093/nar/gku1404
- Wang X, Pan T. Stress Response and Adaptation Mediated by Amino Acid Misincorporation during Protein Synthesis. Adv Nutr. 2016;7:773S–9S. doi:10.3945/an.115.010991
- Ling J, Söll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci U S A. 2010;107:4028–33. doi:10.1073/pnas.1000315107
- Gomes AC, Kordala AJ, Strack R, Wang X, Geslain R, Delaney K, Clark WC, Keenan R, Pan T. A dual fluorescent reporter for the investigation of methionine mistranslation in live cells. RNA. 2016;22:467–76. doi:10.1261/rna.054163.115
- Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–6. doi:10.1038/nature08576
- Lee JY, Kim DG, Kim BG, Yang WS, Hong J, Kang T, Oh YS, Kim KR, Han BW, Hwang BJ, et al. Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation to protect cells against oxidative stress. J Cell Sci. 2014;127:4234–45. doi:10.1242/jcs.152470
- Hoffman KS, O'Donoghue P, Brandl CJ. Mistranslation: from adaptations to applications. Biochim Biophys Acta. 2017: doi: 10.1016/j.bbagen.2017.01.031. doi:10.1016/j.bbagen.2017.01.031
- Berg MD, Hoffman KS, Genereaux J, Mian S, Trussler RS, Haniford DB, O'Donoghue P, Brandl CJ. Evolving Mistranslating tRNAs Through a Phenotypically Ambivalent Intermediate in Saccharomyces cerevisiae. Genetics. 2017;206:1865–79. doi:10.1534/genetics.117.203232
- Naganuma M, Sekine S, Chong YE, Guo M, Yang XL, Gamper H, Hou YM, Schimmel P, Yokoyama S. The selective tRNA aminoacylation mechanism based on a single G•U pair. Nature. 2014;510:507–11. doi:10.1038/nature13440
- Park SJ, Hou YM, Schimmel P. A single base pair affects binding and catalytic parameters in the molecular recognition of a transfer RNA. Biochemistry. 1989;28:2740–6. doi:10.1021/bi00432a056
- Sun L, Gomes AC, He W, Zhou H, Wang X, Pan DW, Schimmel P, Pan T, Yang XL. Evolutionary Gain of Alanine Mischarging to Noncognate tRNAs with a G4:U69 Base Pair. J Am Chem Soc. 2016;138:12948–55. doi:10.1021/jacs.6b07121
- O'Donoghue P, Sheppard K, Nureki O, Söll D. Rational design of an evolutionary precursor of glutaminyl-tRNA synthetase. Proc Natl Acad Sci U S A. 2011;108:20485–90. doi:10.1073/pnas.1117294108
- Ling J, Cho C, Guo LT, Aerni HR, Rinehart J, Söll D. Protein aggregation caused by aminoglycoside action is prevented by a hydrogen peroxide scavenger. Mol Cell. 2012;48:713–22. doi:10.1016/j.molcel.2012.10.001
- Manuvakhova M, Keeling K, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6:1044–55. doi:10.1017/S1355838200000716
- Crick FH. Codon–anticodon pairing: the wobble hypothesis. J Mol Biol. 1966;19:548–55. doi:10.1016/S0022-2836(66)80022-0
- Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44:D184–9. doi:10.1093/nar/gkv1309
- Schneider A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu Rev Biochem. 2011;80:1033–53. doi:10.1146/annurev-biochem-060109-092838
- Rogalski M, Karcher D, Bock R. Superwobbling facilitates translation with reduced tRNA sets. Nat Struct Mol Biol. 2008;15:192–8. doi:10.1038/nsmb.1370
- Goodenbour JM, Pan T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006;34:6137–46. doi:10.1093/nar/gkl725
- Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi:10.1371/journal.pgen.0020221
- Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–80. doi:10.1093/nar/gkp787
- Kutter C, Brown GD, Goncalves A, Wilson MD, Watt S, Brazma A, White RJ, Odom DT. Pol III binding in six mammals shows conservation among amino acid isotypes despite divergence among tRNA genes. Nat Genet. 2011;43:948–55. doi:10.1038/ng.906
- Parisien M, Wang X, Pan T. Diversity of human tRNA genes from the 1000-genomes project. RNA Biol. 2013;10:1853–67. doi:10.4161/rna.27361
- Iben JR, Maraia RJ. tRNA gene copy number variation in humans. Gene. 2014;536:376–84. doi:10.1016/j.gene.2013.11.049
- Ruan LL, Zhou XL, Tan M, Wang ED. Human cytoplasmic ProX edits mischarged tRNAPro with amino acid but not tRNA specificity. Biochem J. 2013;450:243–52. doi:10.1042/BJ20121493
- Vargas-Rodriguez O, Musier-Forsyth K. Exclusive use of trans-editing domains prevents proline mistranslation. J Biol Chem. 2013;288:14391–9. doi:10.1074/jbc.M113.467795
- Schimmel P, Guo M. A tipping point for mistranslation and disease. Nat Struct Mol Biol. 2009;16:348–9. doi:10.1038/nsmb0409-348
- Ribas de Pouplana L, Santos MA, Zhu JH, Farabaugh PJ, Javid B. Protein mistranslation: friend or foe? Trends Biochem Sci. 2014;39:355–62. doi:10.1016/j.tibs.2014.06.002
- Lu J, Bergert M, Walther A, Suter B. Double-sieving-defective aminoacyl-tRNA synthetase causes protein mistranslation and affects cellular physiology and development. Nat Commun. 2014;5:5650. doi:10.1038/ncomms6650
- Liu Y, Satz JS, Vo MN, Nangle LA, Schimmel P, Ackerman SL. Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc Natl Acad Sci USA. 2014;111:17570–5. doi:10.1073/pnas.1420196111
- Wang X, Pan T. Methionine Mistranslation Bypasses the Restraint of the Genetic Code to Generate Mutant Proteins with Distinct Activities. PLoS Genet. 2015;11:e1005745. doi:10.1371/journal.pgen.1005745
- Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, Schimmel P, Senju S, Nishimura Y, Chuang JH, Ackerman SL. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–9. doi:10.1126/science.1249749
- Su HW, Zhu JH, Li H, Cai RJ, Ealand C, Wang X, Chen YX, Kayani MU, Zhu TF, Moradigaravand D, et al. The essential mycobacterial amidotransferase GatCAB is a modulator of specific translational fidelity. Nat Microbiol. 2016;1:16147. doi:10.1038/nmicrobiol.2016.147
- Nangle LA, Motta CM, Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem Biol. 2006;13:1091–100. doi:10.1016/j.chembiol.2006.08.011
- Mohler K, Aerni HR, Gassaway B, Ling J, Ibba M, Rinehart J. MS-READ: Quantitative measurement of amino acid incorporation. Biochim Biophys Acta. 2017. doi:10.1016/j.bbagen.2017.01.025.
- Moghal A, Mohler K, Ibba M. Mistranslation of the genetic code. FEBS Lett. 2014;588:4305–10. doi:10.1016/j.febslet.2014.08.035
