ABSTRACT
Production of the translation apparatus of E. coli is carefully matched to the demand for protein synthesis posed by a given growth condition. For example, the fraction of RNA polymerases that transcribe rRNA and tRNA drops from 80% during rapid growth to 24% within minutes of a sudden amino acid starvation. We recently reported in Nucleic Acids Research that the tRNA pool is more dynamically regulated than previously thought. In addition to the regulation at the level of synthesis, we found that tRNAs are subject to demand-based regulation at the level of their degradation. In this point-of-view article we address the question of why this phenomenon has not previously been described. We also present data that expands on the mechanism of tRNA degradation, and we discuss the possible implications of tRNA instability for the ability of E. coli to cope with stresses that affect the translation process.
Introduction
Transfer RNAs (tRNAs) are one of the most abundant cellular species, comprising approximately 15% of total RNA in rapidly growing Escherichia coli cells. As the critical adaptors that pair the codons of mRNA with their cognate amino acids, tRNAs are essential for protein synthesis and thereby for cellular growth. In addition, tRNAs and tRNA-derived fragments also serve regulatory roles in E. coli Citation1,Citation2 and other bacteria (recently reviewed in refs. Citation3,Citation4). For example, in E. coli, uncharged tRNA at the ribosomal A-site is the signal that stimulates RelA to produce guanosine tetraphosphate (ppGpp), which initiates the stress response to amino acid starvation.Citation2 With such key functions, tRNA levels may be expected to be tightly and dynamically regulated to match the changing needs of the cell. In support of this expectation, Kurland and colleagues measured tRNA concentrations under different growth conditions and found that, indeed, individual tRNA abundances vary with growth rate, albeit modestly, in a manner that reflects the codon frequencies in the corresponding mRNA pools.Citation5 In addition, there is evidence that an imbalance in the levels of competing charged tRNAs can compromise the fidelity of protein synthesis.Citation6-Citation8
To the extent that the regulation of tRNA levels has been described, it relies on transcriptional control of tRNA synthesis.Citation9-Citation12 However, while transcriptional activation could rapidly upregulate tRNA levels, transcriptional repression could not, on its own, mediate rapid downregulation of tRNA levels, because tRNA reportedly has a very long half-life in E. coli.Citation13 Thus, in order to quickly downregulate tRNA levels, existing mature tRNAs would need to be inactivated. We recently reported in Nucleic Acids Research that the tRNA pool is rapidly and substantially reduced under several growth conditions that reduce the availability of substrate for protein synthesis. This observation gave rise to a working model, in which a tRNA becomes substrate for degradation whenever the tRNA supply exceeds the demand for tRNAs in protein synthesis.Citation14 We begin this point-of-view article by underlining the importance of careful normalization of gene expression data obtained from stress conditions that disrupt steady-state growth, which may explain why tRNA degradation has not been described earlier.
The important choice of a standard for normalization
For experiments that aim to compare the levels of a specific RNA species among samples of cells experiencing different growth conditions, an appropriate method for normalization of the raw data must be carefully chosen. In the case of northern blot experiments, it is custom to report the signal from the probe that hybridizes to the RNA of interest relative to the signal in the same lane from RNA expressed from a reference gene, often referred to as a “household” gene. Typical reference choices for northern blot, or quantitative PCR techniques mutatis mutandis, include a ribosomal RNA (rRNA) or another RNA (rssA,Citation15 idnT,Citation16 cysGCitation16) which is not expected to vary in cellular level from sample to sample. However, amino acid starvation triggers a major re-orchestration of the genome-wide transcription patternCitation9,Citation17,Citation18 which results in a change in the cellular concentration of most RNA species shortly after the onset of starvation (refs. Citation18-Citation20 and our manuscript in preparation). Therefore, there is no endogenous reference transcript which is known to be suitable for comparing the cellular levels of tRNA (or any other RNA species) during amino acid starvation to the levels during steady-state growth. An alternative approach is to report the signal from the RNA of interest relative to the total amount of RNA loaded in the same lane. Similarly, for quantification of RNA-seq data, reporting the reads mapping to the gene of interest relative to the total mapped reads per sample is a common approach. This method of normalization is only reasonable if the total RNA content per cell is constant across the tested conditions. For experiments such as ours that are specifically designed to investigate whether the amounts of a major component of total RNA differ upon the change in growth conditions, this option is naturally not justifiable.
To circumvent these issues, we make use of spike-in cells from a separate culture, which is added to the experimental samples prior to RNA extraction.Citation14,Citation21 By quantifying a reference RNA that is highly expressed in the spike-in cells and either absent or present at very low levels in the experimental samples, it is possible to obtain accurate normalization by adding only a small aliquot of spike-in cells to the experimental samples. We typically add 1–5% spike-in cells to each sample (based on optical density), and load a control sample containing only spike-in cells to quantify the contribution of the spike-in cells to the signal from the RNA of interest.Citation21 The spike-in-cell approach has the clear advantage that no assumptions about constancy of the levels of the reference RNA across the tested conditions are necessary, because the spike-in cells are added after harvest of the experimental samples. Addition of the spike-in cells prior to RNA extraction (as opposed to the addition of purified spike-in transcripts after RNA purification) ensures that the reference RNA also reflects any sample-to-sample variation in RNA recovery. Using the spike-in-cell approach, we showed that the majority of cellular tRNA is degraded within twenty minutes of the onset of a sudden amino acid starvation.Citation14
Recently, we have also applied the spike-in-cell approach to quantify the kinetics of the transcriptome-wide changes that occur in response to amino acid starvation (manuscript in preparation). Importantly, we find that all three species of rRNA (5S, 16S and 23S rRNA) decrease within minutes of the onset of starvation, albeit more modestly than the tRNAs (rRNA was reduced ∼2-fold one hour into starvation, data not shown). It has been known for a long time that rRNA becomes unstable upon starvation,Citation22 and the degradation pathway has been described by the group of M. P. Deutscher,Citation23-Citation25 but to our knowledge, the rapid onset of rRNA degradation has not previously been reported. Since tRNA and rRNA together make up more than 95% of the total RNA,Citation26,Citation27 this finding highlights why normalizing to total RNA (or total reads in the case of RNA-seq) cannot be recommended for this type of experiment. Without calling attention to specific articles, we remark that this new insight warrants a revisitation of previous work, in which data was interpreted under the assumption of relatively constant cellular levels of tRNA, rRNA or total RNA under different growth-compromising conditions.
A demand-based model for tRNA degradation
In the very beginning of a sudden amino acid starvation, translation is directly limited by the availability of the cognate charged tRNA. Later, when the ppGpp concentration begins to drop, translation is limited by the availability of mRNA substrate.Citation28,Citation29 We found that reduced mRNA availability, induced by treatment with rifampicin, which does not evoke the stringent response, also led to rapid tRNA degradation. This finding, together with the finding that the kinetics of tRNA decay upon amino acid starvation are similar in wild-type E. coli and a relA mutant, suggests that tRNA instability is not a unique consequence of the stringent response, but may occur as a general response to stresses that reduce translation.Citation14 The argument is supported by a recent report of extensive tRNA degradation during oxidative stress,Citation30 which causes stalling of ribosomes at 8-oxoG residues in the mRNACitation31 and thereby also limits the pool of functionally intact mRNA available for translation. Since the capacity to synthesize new protein is severely reduced during amino acid starvation, a tRNA-degradation mechanism that depends on synthesis of new protein may not be feasible. Consistently, we found that global tRNA degradation occurs even in cultures that are pre-treated with chloramphenicol, revealing that a large capacity for tRNA degradation exists, which does not require de novo protein synthesis.Citation14 The simplest model to explain these observations is that there is no molecular signal per se that triggers tRNA degradation. Instead, we propose a demand-based model for tRNA degradation, in which tRNAs are protected from degradation when they are occupied by binding to members of the translational machinery (the aminoacyl-tRNA-synthetases, elongation factor Tu, and the ribosomes), but they become accessible for degradation once they are not engaged in charging and translation. This model is in essence the same scenario as suggested by M.P. Deutscher for rRNA degradation,Citation23 the major difference being that the degradation pathway for tRNA during starvation is undescribed.
Effectors of tRNA degradation
The recent studies mentioned aboveCitation14,Citation30 suggest that tRNA degradation could play an important, but hitherto overlooked, role in the bacterial stress response to amino acid starvation as well as other stresses that affect the availability of substrates for translation. Many questions about the process and the importance of tRNA degradation during stress conditions remain to be answered. The implications of tRNA degradation for bacterial fitness and survival during stress, and the pathway for tRNA degradation are among the most fundamental questions. We are particularly pursuing the latter answer, because we expect that knowledge about the pathway of tRNA degradation can be applied to guide the construction of a mutant that is impaired in tRNA degradation, thereby facilitating the investigation of the effects of tRNA degradation on bacterial fitness and stress survival.
Given the demand-based working model described above, we assume that one or more “household” ribonucleases (RNases) are responsible for tRNA degradation during amino acid starvation, maybe aided by a factor that stimulates ribonucleolytic attack on the tRNA either by modification of the tRNA substrate or by up-regulation of RNase activity. The E. coli genome encodes more than 20 ribonucleases, including a number that participate in the processing of precursor transcripts to generate mature tRNAs, such as RNase E, RNase P, RNase PH and RNase T (reviewed in refs. Citation32,Citation33). However, mature functional tRNAs are normally not considered substrates for any of the cytoplasmic RNases, due to the protective qualities of their compact tertiary structure, their aminoacylated 3′-ends, and their associations with cellular protein partners (reviewed in refs. Citation34,Citation35). The exception to this rule is RNase I, a nonspecific endoribonuclease that is confined to the periplasm.Citation36,Citation37 Upon treatment with agents that damage the cell membrane, RNase I is known to cause massive degradation of cellular RNA, including tRNA.Citation38-Citation40 As pointed out by M. P. Deutscher,Citation34 however, uncontrolled release of active RNase I into the cytoplasm would effectively kill the cell. We have observed that cell survival is unaffected for more than three hours after the sudden onset of amino acid starvation.Citation14 Therefore, we favor the model that tRNA becomes a substrate for one or more of the cytoplasmic RNases upon amino starvation either due to a starvation-induced alteration of the tRNAs that make them more accessible, or simply due to the increased proportion of tRNAs that lose the protection from their protein partners as a consequence of reduced protein synthesis activity during starvation. In the following sections, we show the results of a set of experiments designed to test whether either of two likely candidates, poly(A) polymerase and RNase E, could be the rate-limiting enzyme involved in tRNA degradation.
Poly(A) polymerase I
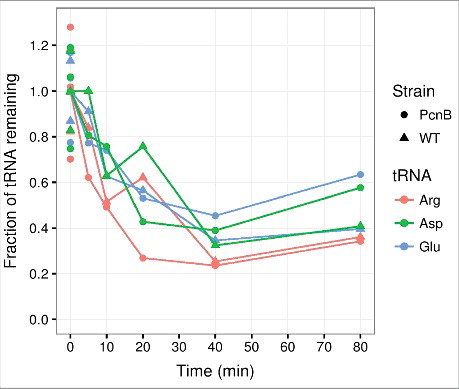
One modification that could target tRNAs for degradation is polyadenylation. Poly(A) polymerase I (PAP I) is the primary enzyme in E. coli that catalyzes the formation of 3′-poly(A) tails on mature RNA transcripts or degradation products, often formed by RNase E, in which strong secondary structures reduce the accessibility of exonucleases at the 3′-end.Citation41 Extension with poly(A) forms a loading platform that facilitates 3′-end processing and degradation of such molecules by 3′-exonucleases.Citation42,Citation43 It has been shown that polyadenylation occurs prior to the degradation of misfolded tRNACitation44 and unprocessed tRNA precursors.Citation45,Citation46 To assess whether PAP I is necessary for the general tRNA decay in the early response to amino acid starvation, we measured tRNA levels in a rph+ derivative of MG1655 and the isogenic ΔpcnB strain, using the method described previously.Citation14,Citation21 shows that all measured tRNA half-lives during starvation for isoleucine were unaffected by the absence of PAP I. Thus, the degradation of mature, functional tRNA seems to occur independently of PAP I activity. We remark that the polynucleotide phosphorylase (PNPase), mainly known for its exonuclease activity, can also generate heteropolymeric tails in vivo, and is responsible for the residual polyadenylation observed in a pcnB mutant.Citation47 Therefore, we cannot exclude that polyadenylation plays a role in tRNA degradation.
Figure 1. PAP I is not required for tRNA degradation at the onset of amino acid starvation. Transfer RNA counts quantified from a northern blot as described in ref. Citation14, Citation21. In brief, cultures of MG1655 rph+ ΔpcnB::cat-sacB (PcnB) and MG1655 rph+ (WT) were grown in MOPS minimal mediaCitation98 supplemented with 0.2% glucose for at least ten generations in exponential phase prior to isoleucine starvation, which was induced by the addition of 400 μg/ml valine.Citation99 At the indicated time points, aliquots of the cultures were harvested into 10% trichloroacetic acid before suspension on ice.Citation100 Spike-in E. coli cells over-expressing the seleno-cysteine tRNA (tRNAsec) were added to constitute 5% of each sample based on OD436 units. RNA was extracted with cold phenol.Citation101 Northern blots were performed on 6% polyacrylamide gels, and probed for tRNAsec, tRNAargVYZQ, tRNAaspTUV, tRNAgltTUVW. Spike-in-normalized counts are shown relative to the average of three measurements from steady state growth (shown at 0 minutes), which is set to unity. The generation times in steady state were 57 min for ΔpcnB and 52 min for the isogenic wild type. The experiment was repeated two times with similar results.
Ribonuclease E
RNase E is considered the rate-limiting enzyme for the degradation of many mRNAsCitation48 and for rRNA decay during carbon starvation and quality control.Citation24 It is also a major endoribonuclease in tRNACitation49 and rRNACitation50 maturation. Furthermore, RNase E is a core member of the degradosome which is a multiprotein complex involved in RNA degradationCitation48 assembled on RNase E through PNPase, RhlB, and enolase binding to the C-terminal half of RNase E.Citation51-Citation53 The degradosome is effective in structured RNA decay from duplex unwinding by the helicase RhIB.Citation54 For these reasons, we investigated whether RNase E may be involved in tRNA degradation after amino acid starvation.
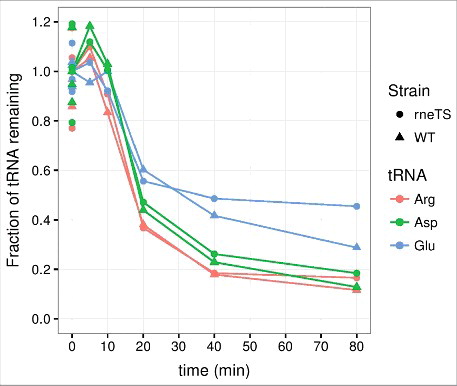
Since RNase E function is essential for E. coli, we employed two different experimental designs to evaluate the effects of RNase E on tRNA half-life. In one setup we employed the temperature-sensitive allele rne-3071 from strain EM1277.Citation55 The timing of the shift to the non-permissive temperature relative to the induction of starvation was an important consideration for this experiment, because the activation of one stress response may alter the cellular response to a second stressor. Namely, cells experiencing a heat shock at the time of amino acid deprivation might alter their response to amino acid starvation and consequently reduce or increase the degree of tRNA decay. In the experiment shown in , the temperature was shifted from 30°C to 43°C five minutes after induction of amino acid starvation. These results revealed that tRNA is effectively degraded under these conditions both in the wildtype and the isogenic rne-3071 strain. Another experiment was conducted with the same strains where the temperature shift was introduced five minutes prior to amino acid starvation to ensure RNase E inactivation at the onset of starvation (data not shown). Again, the two strains behaved similarly, indicating that tRNA turnover is not affected by inactivation of RNase E.
Figure 2. tRNA degradation at the onset of amino acid starvation occurs with similar kinetics in rneTS and WT strains at the nonpermissive temperature. Transfer RNA counts were quantified from a northern blot as described for , except that MG1655 rph+ rne-3071 zce-726::Tn10 (rneTS) and MG1655 rph+ zce-726::Tn10 (WT) were grown at 30°C and shifted to 43°C five minutes after the induction of starvation. The generation times in steady state at 30°C were 75 min for rneTS and 80 min for WT.
In a parallel approach, we avoided the temperature shift altogether by instead inhibiting RNase E via expression of the phage protein Dip/GP37, which binds RNase E and reduces its affinity for RNA.Citation56 Induction of plasmid-encoded dip, or a non-functional variant of dip (GP37_C_his), for one hour prior to amino acid starvation did not result in any inhibition of tRNA turnover (data not shown).
In summary, two key enzymes in bacterial RNA metabolism, poly(A) polymerase and RNase E, are here both ruled out as being the rate-limiting enzyme for tRNA degradation during amino acid starvation. Importantly, we remark that it may not be possible to substantially impair tRNA degradation with a single mutation, since many of the RNases in E. coli possess overlapping activities.Citation33,Citation57
Why degrade tRNA?
E. coli has adapted to live a feast-or-famine existence where nutrients are sometimes plentiful and other times limited, and the durations of individual starvation periods are variable. Nutrient limitation triggers alterations in the activity and promoter preferences of RNA polymerase, and major changes in the cell's gene expression program prime the cell for maintenance and survival rather than growth.Citation58 Several starvation-regulated factors can directly influence transcription by RNA polymerase, including the alarmone ppGpp (binding at the β′-ω and the β′-DksA interfaces of RNA polymerase),Citation59,Citation60 the small RNA 6S (binding the σ70-RNA polymerase holoenzyme)Citation61,Citation62 and a number of protein factors (DksA,Citation63 alternative sigma factors,Citation64 and anti-sigma factorsCitation65) (reviewed in ref. Citation66). The specific nature of the starvation (i.e., the type of nutrient that has become limiting for growth) and the time spent under nutrient limiting conditions affect the accumulation of these modulators of transcription, presumably allowing cells to target their stress response to best compensate for the shortage of particular metabolites.Citation58,Citation67-Citation70 Similarly, we reckon that tRNA degradation may only be beneficial during exposure to a subset of the possible stressors a bacterium may encounter. In our studyCitation14 (as in most other studies on the stringent response, reviewed in refs. Citation71,Citation72) we have used auxotrophic mutants to introduce and maintain amino acid starvation. This has been convenient and necessary for describing molecular processes and dissecting regulatory pathways. However, such experiments most likely represent an unnatural situation. First, because an amino acid auxotroph E. coli cell would most likely quickly be outcompeted in a natural environment where amino acids are scarce,Citation73 and second because, in a prototrophic E. coli cell, amino acid starvation stress is expected to be a transient phenomenon that is generally resolved once the enzymes necessary for biosynthesis of the relevant amino acid(s) have been produced. It would therefore be of great relevance to test the effects of tRNA degradation on bacterial fitness and survival in prototrophic strains exposed to transient amino acid starvation, or even better, on environmental E. coli isolates growing in surroundings that mimic their natural environment.
In the following discussion of the possible effects of tRNA degradation as a stress response, we will concentrate on the well-studied response to amino acid starvation in E. coli K-12. In this case, the stringent response is initiated by ppGpp-signalling, which causes a dramatic reduction in rRNA and tRNA synthesis within minutes.Citation74-Citation77 This initial response is followed by down-regulation of many growth-related genes and up-regulation of the general stress response genes by σS-bound RNA polymerase.Citation18,Citation19
tRNAs and translation accuracy
Given that tRNA synthesis is already strongly reduced in the amino-acid-starved cell, is it likely that degradation of existing tRNA provides any additional benefits? The extensive breakdown of rRNA that is observed on the time-scale of hours to days of starvation for carbon,Citation78 phosphate,Citation13,Citation79 or nitrogenCitation80 is understood as a survival strategy, where the building blocks of the ribosomes are released to provide nutrients for maintenance and biosynthesis in the stressed cells.Citation22 A similar argument can be made for the breakdown of tRNA. Additionally, tRNA degradation may be beneficial for the amino-acid-starved cell because it contributes to optimizing protein synthesis during starvation. One well documented consequence of amino acid starvation is an increase in the rate at which erroneous amino acids are inserted into the nascent polypeptides. Depending on the severity of the starvation, the error rate may increase by an order of magnitude in stringent cells, and two orders of magnitude in relaxed (relA−) cells.Citation7,Citation81 The increased error rate occurs mainly because competing near-cognate tRNAs get access to deliver their amino acids to ribosomes stalled at the “hungry” codons, due to a shortage of cognate tRNA charged with the amino acid starved for.Citation7,Citation8,Citation81 We consider it highly plausible that degradation of the vacant tRNA pool is important for reducing the translational error-frequency because it would reduce the amount of competing near-cognate charged tRNA. The reduction in tRNA levels upon amino acid starvation is a combined effect of the ppGpp-mediated halt in tRNA synthesis and the degradation of already existing tRNA molecules. Which is more important? A conclusive answer requires the experimental comparison of mutants impaired in one or the other pathway. However, at this point it is clear that degradation can overwhelm increased synthesis because tRNA levels initially drop several fold upon amino acid starvation even in a relA− mutant,Citation14 which actually increases the rate of tRNA synthesis upon amino acid starvation, rather than decreasing it.Citation9,Citation82 Therefore, E. coli possesses a very large capacity for tRNA degradation. The most prominent phenotype of a relA− strain is the elevated level of mistranslation during amino acid starvation,Citation83 and we would anticipate that this rate would increase even further if the levels of competing near-cognate tRNAs were not reduced by degradation.
Substrates for translation under nutrient limitation
Another feature of translation that appears important for bacterial growth physiology is the maintenance of an appropriate translation elongation rate.Citation84-Citation87 A recent publication from the laboratories of T. Hwa and collaborators shows convincingly that the translation elongation rate is kept remarkably constant over a wide range of growth rates, and drops less than two-fold in E. coli (from 16 aa s−1 to 9 aa s−1) when the steady-state generation time is increased by 60 fold (from 20 min to 20 hours).Citation88 The authors present compelling evidence that the maintenance of a high translation elongation rate is enabled by a reduction in the fraction of actively translating ribosomes during slow growth.Citation88 Ribosomal inactivation is attributed to factors such as ribosome modulation factor (RMF), ribosome-associated inhibitor A (RaiA), and ribosomal silencing factor (RsfA), which are induced by the elevated level of ppGpp present during slow growth.Citation89-Citation91 T. Hwa and collaboratorsCitation88 argue that ribosomal inactivation occurs without a concomitant decrease in the concentration of available ternary complexes (comprising aminoacylated tRNA, elongation factor TU and GTP), which would ensure the maintenance of a reasonable translation elongation rate. There is an apparent conflict between the latter argument and our demand-based model of tRNA degradation. As a first approximation, we would predict that inactivation of a fraction of the total ribosomes would leave behind unprotected tRNAs, which would be targets for degradation, resulting in a concurrent decrease in the concentration of ternary complexes. However, existing data suggests the contrary, namely that if anything, there are more tRNAs per ribosome at the very slow growth rates (<0.4 doublings h−1) where a large fraction of the ribosomes are inactivated than at higher growth rates (ref. Citation92 and references within ref. Citation93). We hypothesize that the resolution of this apparent conflict lies in the realization that tRNA would not only be protected by active ribosomes, but most likely, to some degree, also by its other protein interaction partners, in particular elongation factor Tu. In support of this hypothesis, the number of EF-Tu's per ribosome were shown to increase from ∼6 during rapid growth to ∼9 at very slow growth rates, thereby reaching a 1:1 stoichiometry with tRNA (ref. Citation93 and references therein). As we have demonstrated, a sudden amino acid starvation causes a substantial drop in tRNA concentrations,Citation14 and this situation differs from the growth at very low growth rates where a steady state has been reached over generations. During the stress induced by sudden starvation, maintenance of an appropriate translation elongation rate seems particularly important because the protein synthesis capacity is low and the need for synthesis of proteins with functions that can help the cell overcome the starvation period is high.Citation94,Citation95 In this case, the overall translation elongation rate must be limited mainly by the concentration of the particular ternary complex that contains tRNA charged with the amino acid starved for. Degradation of the unemployed tRNAs could therefore increase the accuracy of protein translation without negatively affecting the translation elongation rate, thus optimising the quality of the protein products.
Concluding remarks
In our point of view, a demand-based model of tRNA degradation, in which the tRNAs that are engaged with components of the translational apparatus are generally protected, whereas surplus tRNAs are continually subject to degradation, provides a satisfactory explanation for how quality in protein synthesis is ensured during amino acid starvation. Thus, we propose that the levels of components of the translational apparatus are not only co-regulated at the level of their synthesis,Citation5,Citation9,Citation93,Citation96,Citation97 but that the level of actively translating ribosomes indirectly regulates the tRNA pool, via degradation of the unengaged tRNAs. Importantly, to maintain a reasonable translation elongation rate even at very low concentrations of active ribosomes, we propose that their association with other protein factors, in particular the abundant elongation factor Tu, would protect a sizable fraction of the tRNA and thereby set a lower bound on the cellular concentration of ternary complexes.
The predicted beneficial effect of tRNA degradation on the quality of protein synthesis during starvation, and therefore on the bacteria's ability to cope with starvation stress, needs to be addressed experimentally. The identification of a mutant of E. coli that shows impaired or abolished tRNA degradation activity could greatly facilitate these efforts, and is an area of active investigation in our laboratory. Given the pleiotropic phenotypes of many RNase mutants, however, it will likely be very challenging to identify a tRNA-degradation-deficient mutant, which isn't also defective in other ribonucleolytic events, such as rRNA and tRNA maturation or mRNA decay.
We speculate that the demand-based model for tRNA stability can also be extended to other growth conditions, including rapid steady-state growth, where it could serve to dynamically fine-tune the supply of tRNA to the demand in protein synthesis. To this end, measurements of tRNA stability during other growth conditions, using appropriate normalization, are pertinent. Experiments designed to more directly test features of the demand-based model should also be carried out. For example, tRNA stability would be predicted to increase in experiments where the protein synthesis activity is artificially increased during amino acid starvation.
Acknowledgements and Author contributions
Plasmids pGP37 and pGP37_C_his were a gift from Tom Dendooven and Rob Lavigne, strain EM1277 was a gift from Eric Massé, and the MG1655 strain lysogenized with λDE3 was a gift from Anders Løbner-Olesen. Funding of the work in our laboratories is provided by the Danish National Research Foundation under Grant number DNRF120; The Danish Council for Independent Research | Natural Sciences under Grant number 1323–00343B; and The Lundbeck Foundation under Grant number R108-A10583. SLS, MAS, and AOF designed experiments and wrote the manuscript; AOF performed the experiments.
References
- Lalaouna, D, Carrier MC, Semsey S, Brouard JS, Wang J, Wade JT, Massé E. A 3′ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol Cell. 2015;58:393–405. doi:10.1016/j.molcel.2015.03.013.
- Haseltine, WA, Block, R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci U S A. 1973;70:1564–8. doi:10.1073/pnas.70.5.1564.
- Henkin, TM. The T box riboswitch: A novel regulatory RNA that utilizes tRNA as its ligand. Biochim Biophys Acta. 2014;1839:959–963. doi:10.1016/j.bbagrm.2014.04.022.
- Katz, A, Elgamal, S, Rajkovic, A, Ibba, M. Non-canonical roles of tRNAs and tRNA mimics in bacterial cell biology. Mol Microbiol. 2016;101:545–58. doi:10.1111/mmi.13419.
- Dong, H, Nilsson, L, Kurland, CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–63. doi:10.1006/jmbi.1996.0428.
- Kramer, EB, Farabaugh, PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi:10.1261/rna.294907.
- Parker, J. Errors and alternatives in reading the universal genetic code. Microbiol Rev. 1989;53:273–98.
- Sorensen, MA. Charging levels of four tRNA species in Escherichia coli Rel(+) and Rel(−) strains during amino acid starvation: a simple model for the effect of ppGpp on translational accuracy. J Mol Biol. 2001;307:785–98. doi:10.1006/jmbi.2001.4525.
- Ryals, J, Little, R, Bremer, H. Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J Bacteriol. 1982;151:1261–8.
- Lazzarini, RA., Dahlberg, AE. The Control of Ribonucleic Acid Synthesis during Amino Acid Deprivation in Escherichia coli. J Biol Chem. 1971;246:420–429.
- Ikemura, T, Dahlberg, JE. Small Ribonucleic Acids of Escherichia coli. II. Noncoordinate accumulation during stringent control. J Biol Chem. 1973;248:5033–5041.
- Norris, TE, Koch, AL. Effect of growth rate on the relative rates of synthesis of messenger, ribosomal and transfer RNA in Escherichia coli. J Mol Biol. 1972;64:633–49. doi:10.1016/0022-2836(72)90088-5.
- Davis, BD, Luger, SM, Tai, PC. Role of ribosome degradation in the death of starved Escherichia coli cells. J Bacteriol. 1986;166:439–45. doi:10.1128/jb.166.2.439-445.1986.
- Svenningsen, SL, Kongstad, M, Stenum, TS, Munoz-Gomez, AJ, Sorensen, MA. Transfer RNA is highly unstable during early amino acid starvation in Escherichia coli. Nucleic Acids Res. 2017;45:793–804. doi:10.1093/nar/gkw1169.
- Peng, S, Stephan, R, Hummerjohann, J, Tasara, T. Evaluation of three reference genes of Escherichia coli for mRNA expression level normalization in view of salt and organic acid stress exposure in food. FEMS Microbiol Lett. 2014;355:78–82. doi:10.1111/1574-6968.12447.
- Zhou, K, Zhou L, Lim Q', Zou R, Stephanopoulos G, Too HP. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol Biol. 2011;12:18. doi:10.1186/1471-2199-12-18.
- Ryals, J, Bremer, H. relA-dependent RNA polymerase activity in Escherichia coli. J Bacteriol. 1982;150:168–79.
- Traxler, MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–48. doi:10.1111/j.1365-2958.2008.06229.x.
- Traxler, MF, Zacharia VM, Marquardt S, Summers SM, Nguyen HT, Stark SE, Conway T. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli. Mol Microbiol. 2011;79:830–45. doi:10.1111/j.1365-2958.2010.07498.x.
- Durfee, T, Hansen, AM, Zhi, H, Blattner, FR, Jin, DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–96. doi:10.1128/JB.01092-07.
- Stenum, TS, Sørensen, MA, Svenningsen, SL. Quantification of the Abundance and Charging Levels of Transfer RNAs in Escherichia coli. J. Vis. Exp. 2017;126:e56212.
- Mandelstam, J, Halvorson, H Turnover of protein and nucleic acid in soluble and ribosome fractions of non-growing Escherichia coli. Biochim Biophys Acta. 1960;40:43–9. doi:10.1016/0006-3002(60)91313-5.
- Zundel, MA, Basturea, GN, Deutscher, MP. Initiation of ribosome degradation during starvation in Escherichia coli. RNA. 2009;15:977–83. doi:10.1261/rna.1381309.
- Sulthana, S, Basturea, GN, Deutscher, MP. Elucidation of pathways of ribosomal RNA degradation: an essential role for RNase E. RNA. 2016;22:1163–71. doi:10.1261/rna.056275.116.
- Basturea, GN, Zundel, MA, Deutscher, MP. Degradation of ribosomal RNA during starvation: comparison to quality control during steady-state growth and a role for RNase PH. RNA. 2011;17:338–45. doi:10.1261/rna.2448911.
- Baracchini, E, Bremer, H. Determination of synthesis rate and lifetime of bacterial mRNAs. Anal Biochem. 1987;167:245–60. doi:10.1016/0003-2697(87)90160-6.
- Kennel, D. Titration of the gene sites on DNA by DNA-RNA hybridization. II. The Escherichia coli chromosome. J Mol Biol. 1968;34:85–103. doi:10.1016/0022-2836(68)90236-2.
- Sørensen, MA, Jensen, KF, Pedersen, S. High concentrations of ppGpp decrease the RNA chain growth rate: Implications for protein synthesis and translation fidelity during amino acid starvation in Escherichia coli. J. Mol. Biol. 1994;236:441–454. doi:10.1006/jmbi.1994.1156.
- Tian, C, Roghanian M, Jørgensen MG, Sneppen K, Sørensen MA, Gerdes K, Mitarai N. Rapid Curtailing of the Stringent Response by Toxin-Antitoxin Encoded mRNases. J Bacteriol. 2016;198:1918–26. doi:10.1128/JB.00062-16.
- Zhong, J, Xiao C, Gu W, Du G, Sun X, He QY, Zhang G. Transfer RNAs Mediate the Rapid Adaptation of Escherichia coli to Oxidative Stress. PLoS Genet. 2015;11:e1005302. doi:10.1371/journal.pgen.1005302.
- Simms, CL, Hudson, BH, Mosior, JW, Rangwala, AS, Zaher, HS. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 2014;9:1256–64. doi:10.1016/j.celrep.2014.10.042.
- Hartmann, RK, Gößringer, M, Späth, B, Fischer, S, Marchfelder, A. Chapter 8 The Making of tRNAs and More – RNase P and tRNase Z. Progress in Molecular Biology and Translational Science. 2009;85:319–368. doi:10.1016/S0079-6603(08)00808-8.
- Deutscher, MP, Li, Z. Exoribonucleases and their multiple roles in RNA metabolism. Prog Nucleic Acid Res Mol Biol. 2001;66:67–105. doi:10.1016/S0079-6603(00)66027-0.
- Deutscher, MP. Degradation of stable RNA in bacteria. J Biol Chem. 2003;278:45041–4. doi:10.1074/jbc.R300031200.
- Deutscher, MP. How bacterial cells keep ribonucleases under control. FEMS Microbiol Rev. 2015;39:350–61. doi:10.1093/femsre/fuv012.
- Neu, HC, Heppel, LA. The Release of Ribonuclease into the Medium When Escherichia coli Cells Are Converted to Spheroplasts. J Biol Chem. 1964;239:3893–3900.
- Zhu, LQ, Gangopadhyay, T, Padmanabha, KP, Deutscher, MP. Escherichia coli rna gene encoding RNase I: cloning, overexpression, subcellular distribution of the enzyme, and use of an rna deletion to identify additional RNases. J Bacteriol. 1990;172:3146–3151. doi:10.1128/jb.172.6.3146-3151.1990.
- Beppu, T, Arima, K. Induction by Mercuric Ion of Extensive Degradation of Cellular Ribonucleic Acid in Escherichia coli. J Bacteriol. 1969;98:888–897.
- Lambert, PA, Smith, AR. Antimicrobial action of dodecyldiethanolamine: activation of ribonuclease I in Escherichia coli. Microbios. 1976;17:35–49.
- Nakajima, K, Kawamata, J. Studies on the mechanism of action of colistin. IV. Activation of “latent” ribonuclease in Escherichia coli by colistin. Biken J. 1966;9:115–23.
- Haugel-Nielsen, J, Hajnsdorf, E, Regnier, P. The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleolytic processing and exonucleolytic degradation. EMBO J. 1996;15:3144–52.
- Hajnsdorf, E, Braun, F, Haugel-Nielsen, J, Regnier, P. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:3973–7. doi:10.1073/pnas.92.9.3973.
- O'Hara, EB, Chekanova JA, Ingle CA, Kushner ZR, Peters E, Kushner SR. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:1807–11. doi:10.1073/pnas.92.6.1807.
- Li, Z, Reimers, S, Pandit, S, Deutscher, MP. RNA quality control: degradation of defective transfer RNA. EMBO J. 2002;21:1132–8. doi:10.1093/emboj/21.5.1132.
- Mohanty, BK, Maples, VF, Kushner, SR. Polyadenylation helps regulate functional tRNA levels in Escherichia coli. Nucleic Acids Res. 2012;40:4589–603. doi:10.1093/nar/gks006.
- Maes, A, Gracia, C, Hajnsdorf, E, Regnier, P. Search for poly(A) polymerase targets in E. coli reveals its implication in surveillance of Glu tRNA processing and degradation of stable RNAs. Mol Microbiol. 2012;83:436–51. doi:10.1111/j.1365-2958.2011.07943.x.
- Mohanty, BK, Kushner, SR. Polynucleotide phosphorylase functions both as a 3′ right-arrow 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:11966–71. doi:10.1073/pnas.220295997.
- Carpousis, AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi:10.1146/annurev.micro.61.080706.093440.
- Li, Z, Deutscher, MP. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi:10.1017/S1355838202014929.
- Li, Z, Pandit, S, Deutscher, MP. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. The EMBO J. 1999;18:2878–2885. doi:10.1093/emboj/18.10.2878.
- Miczak, A, Kaberdin, VR, Wei, CL, Lin-Chao, S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci U S A. 1996;93:3865–9. doi:10.1073/pnas.93.9.3865.
- Py, B, Causton, H, Mudd, EA, Higgins, CF. A protein complex mediating mRNA degradation in Escherichia coli. Mol Microbiol. 1994;14:717–29. doi:10.1111/j.1365-2958.1994.tb01309.x.
- Vanzo, NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, Krisch HM, Carpousis AJ. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998;12:2770–81. doi:10.1101/gad.12.17.2770.
- Py, B, Higgins, CF, Krisch, HM, Carpousis, AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–72. doi:10.1038/381169a0.
- Masse, E, Escorcia, FE, Gottesman, S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–83. doi:10.1101/gad.1127103.
- Van den Bossche, A, Hardwick SW, Ceyssens PJ, Hendrix H, Voet M, Dendooven T, Bandyra KJ, De Maeyer M, Aertsen A, Noben JP, et al. Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome. eLife. 2016;5:e16413. doi:10.7554/eLife.16413.
- Deutscher, MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–66. doi:10.1093/nar/gkj472.
- Nyström, T. MicroReview: Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol. 2004;54:855–62. doi:10.1111/j.1365-2958.2004.04342.x.
- Ross, W, Sanchez-Vazquez P, Chen AY, Lee JH, Burgos HL, Gourse RL. ppGpp Binding to a Site at the RNAP-DksA Interface Accounts for Its Dramatic Effects on Transcription Initiation during the Stringent Response. Mol Cell. 2016;62:811–823. doi:10.1016/j.molcel.2016.04.029.
- Ross, W, Vrentas, CE, Sanchez-Vazquez, P, Gaal, T, Gourse, RL. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50:420–9. doi:10.1016/j.molcel.2013.03.021.
- Wassarman, KM, Storz, G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–23. doi:10.1016/S0092-8674(00)80873-9.
- Trotochaud, AE, Wassarman, KM. A highly conserved 6S RNA structure is required for regulation of transcription. Nat Struct Mol Biol. 2005;12:313–9. doi:10.1038/nsmb917.
- Rutherford, ST, Villers, CL, Lee, JH, Ross, W, Gourse, RL. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–48. doi:10.1101/gad.1745409.
- Gruber, TM, Gross, CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–66. doi:10.1146/annurev.micro.57.030502.090913.
- Jishage, M, Ishihama, A. A stationary phase protein in Escherichia coli with binding activity to the major σ subunit of RNA polymerase. Proceedings of the National Academy of Sciences to Proc Natl Acad Sci USA. 1998;95:4953–4958. doi:10.1073/pnas.95.9.4953.
- Sharma, UK, Chatterji, D. Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol Rev. 2010;34:646–57. doi:10.1111/j.1574-6976.2010.00223.x.
- Wong, GT, Bonocora RP, Schep AN, Beeler SM, Lee Fong AJ, Shull LM, Batachari LE, Dillon M, Evans C, Becker CJ,. Genome-Wide Transcriptional Response to Varying RpoS Levels in Escherichia coli K-12. J Bacteriol. 2017;199:e00755–16. doi:10.1128/JB.00755-16.
- Hengge, R. Proteolysis of σS (RpoS) and the general stress response in Escherichia coli. Research in Microbiology. 2009;160:667–676. doi:10.1016/j.resmic.2009.08.014.
- Mandel, MJ, Silhavy, TJ. Starvation for Different Nutrients in Escherichia coli Results in Differential Modulation of RpoS Levels and Stability. J Bacteriol. 2005;187:434–442. doi:10.1128/JB.187.2.434-442.2005.
- Hauryliuk, V, Atkinson, GC, Murakami, KS, Tenson, T, Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Micro. 2015;13:298–309. doi:10.1038/nrmicro3448.
- Potrykus, K, Cashel, M. (p)ppGpp: Still Magical? Annu Rev Microbiol. 2008;62:35–51. doi:10.1146/annurev.micro.62.081307.162903.
- Cashel, M, Gentry, D, Hernandez, V, Vinella, D. in Esherichia coli and Salmonella; Cellular and Molecular Biology 1458–1496. Washington (DC): ASM Press; 1996.
- Mason, TG, Slater, JH. Competition between an Escherichia coli tyrosine auxotroph and a prototrophic revertant in glucose- and tyrosine-limited chemostats. Antonie Van Leeuwenhoek. 1979;45:253–63. doi:10.1007/BF00418588.
- Sands, MK, Roberts, RB. The effects of a tryptophan-histidine deficiency in a mutant of escherichia coli. J Bacteriol. 1952;63:505–511.
- Pardee, AB, Prestidge, LS. The dependence of nucleic acid syntheses on the presence of amino acids in escherichia coli. J Bacteriol. 1956;71:677–683.
- Gros, F, Gros, F. Rôle des acides amines dans la synthèse des acides nucléiques chez Escherichia coli. Experimental Cell Research. 1958;14:104–131. doi:10.1016/0014-4827(58)90218-0.
- Cashel, M, Kalbacher, B. The control of ribonucleic acid synthesis in Escherichia coli. V. Characterization of a nucleotide associated with the stringent response. J Biol Chem. 1970;245:2309–18.
- Jacobson, A, Gillespie, D. Metabolic events occurring during recovery from prolonged glucose starvation in Escherichia coli. J Bacteriol. 1968;95:1030–9.
- Maruyama, H, Mizuno, D. Ribosome degradation and the degradation products in starved Escherichia coli. I. Comparison of the degradation rate and of the nucleotide pool between Escherichia coli B and Q-13 strains in phosphate deficiency. Biochim Biophys Acta. 1970;199:159–65. doi:10.1016/0005-2787(70)90704-5.
- Ben-Hamida, F, Schlessinger, D. Synthesis and breakdown of ribonucleic acid in Escherichia coli starving for nitrogen. Biochim Biophys Acta. 1966;119:183–91. doi:10.1016/0005-2787(66)90049-9.
- Johnston, TC, Borgia, PT, Parker, J. Codon specificity of starvation induced misreading. Molecular and General Genetics MGG. 1984;195:459–465. doi:10.1007/BF00341447.
- Lagosky, PA, Chang, FN. Correlation between RNA synthesis and basal level guanosine 5′-diphosphate 3′-diphosphate in relaxed mutants of Escherichia coli. J Biol Chem. 1981;256:11651–6.
- O'Farrell, PH The suppression of defective translation by ppGpp and its role in the stringent response. Cell. 1978;14:545–57. doi:10.1016/0092-8674(78)90241-6.
- Nedialkova, Danny D, Leidel, Sebastian A. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell. 2015;161:1606–1618. doi:10.1016/j.cell.2015.05.022.
- Zhang, G, Hubalewska, M, Ignatova, Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–80. doi:10.1038/nsmb.1554.
- Komar, AA, Lesnik, T, Reiss, C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Letters. 1999;462:387–391. doi:10.1016/S0014-5793(99)01566-5.
- Jacobson, GN, Clark, PL. Quality over quantity: optimizing co-translational protein folding with non-‘optimal’ synonymous codons. Current Opinion in Structural Biology. 2016;38:102–110. doi:10.1016/j.sbi.2016.06.002.
- Dai, X, Zhu M, Warren M, Balakrishnan R, Patsalo V, Okano H, Williamson JR, Fredrick K, Wang YP, Hwa T. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat Microbiol. 2016;2:16231. doi:10.1038/nmicrobiol.2016.231.
- Izutsu, K, Wada, A, Wada, C. Expression of ribosome modulation factor (RMF) in Escherichia coli requires ppGpp. Genes Cells. 2001;6:665–76. doi:10.1046/j.1365-2443.2001.00457.x.
- Agafonov, DE, Kolb, VA, Spirin, AS. A novel stress-response protein that binds at the ribosomal subunit interface and arrests translation. Cold Spring Harb Symp Quant Biol. 2001;66:509–14. doi:10.1101/sqb.2001.66.509.
- Hauser, R, Pech M, Kijek J, Yamamoto H, Titz B, Naeve F, Tovchigrechko A, Yamamoto K, Szaflarski W, Takeuchi N, et al. RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 2012;8:e1002815. doi:10.1371/journal.pgen.1002815.
- Rosset, R, Julien, J, Monier, R. Ribonucleic acid composition of bacteria as a function of growth rate. J Mol Biol. 1966;18:308–20. doi:10.1016/S0022-2836(66)80248-6.
- Klumpp, S, Scott, M, Pedersen, S, Hwa, T. Molecular crowding limits translation and cell growth. Proceedings of the National Academy of Sciences of the United States of America to Proc Natl Acad Sci USA. 2013;110:16754–16759.
- Mandelstam, J. Turnover of protein in starved bacteria and its relationship to the induced synthesis of enzyme. Nature. 1957;179:1179–81. doi:10.1038/1791179a0.
- Kolter, R, Siegele, DA, Tormo, A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–74. doi:10.1146/annurev.mi.47.100193.004231.
- Maaløe O. Regulation of the Protein-Synthesizing Machinery—Ribosomes, tRNA, Factors, and So On. In: Goldberger R.F. (eds.). Biological Regulation and Development, Vol 1. Boston, MA, USA: Springer; 1979.
- Pedersen, S, Bloch, PL, Reeh, S, Neidhardt, FC. Patterns of protein synthesis in E. coli: a catalog of the amount of 140 individual proteins at different growth rates. Cell. 1978;14:179–190. doi:10.1016/0092-8674(78)90312-4.
- Neidhardt, FC, Bloch, PL, Smith, DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–47.
- Leavitt, RI, Umbarger, HE. Isoleucine and valine metabolism in Escherichia coli. XI. Valine Inhibition of the Growth of Escherichia coli Strain K-12. J Bacteriol. 1962;83:624–630.
- Krüger, MK, Sørensen, MA. Aminoacylation of hypomodified tRNAGlu in vivo. J. Mol. Biol. 1998;284:609–620. doi:10.1006/jmbi.1998.2197.
- Varshney, U, Lee, CP, RajBhandary, UL. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J Biol Chem. 1991;266:24712–8.
