ABSTRACT
Cells must make careful use of the resources available to them. A key area of cellular regulation involves the biogenesis of ribosomes. Transcriptional regulation of ribosome biogenesis factor genes through alterations in histone acetylation has been well studied. This work identifies a post-transcriptional mechanism of ribosome biogenesis regulation by Puf protein control of mRNA stability. Puf proteins are eukaryotic mRNA binding proteins that play regulatory roles in mRNA degradation and translation via association with specific conserved elements in the 3ʹ untranslated region (UTR) of target mRNAs and with degradation and translation factors. We demonstrate that several ribosome biogenesis factor mRNAs in Saccharomyces cerevisiae containing a canonical Puf4p element in their 3ʹ UTRs are destabilized by Puf2p, Puf4, and Puf5p, yet stabilized by Puf1p and Puf3p. In the absence of all Puf proteins, these ribosome biogenesis mRNAs are destabilized by a secondary mechanism involving the same 3ʹ UTR element. Unlike other targets of Puf4p regulation, the decay of these transcripts is not altered by carbon source. Overexpression of Puf4p results in delayed ribosomal RNA processing and altered ribosomal subunit trafficking. These results represent a novel role for Puf proteins in yeast as regulators of ribosome biogenesis transcript stability.
Introduction
Ribosome biogenesis is a highly complex process requiring efforts from multiple regulatory proteins performing excision, processing, and folding events of the ribosomal RNAs, as well as assimilation of ribosomal proteins into the ribosomal subunits to create the final products. Life as a ribosome begins with transcription of ribosomal RNA from a large tract of DNA known as rDNA. RNA Polymerase I transcribes the large ~ 6.6kb portion of rDNA that later becomes the 25S, 18S, and 5.8S rRNA species, while RNA Polymerase III transcribes the small portion of rDNA in the opposite direction that becomes the 5S rRNA. Within the large rRNA tract transcribed by RNA PolI, sequences are excised from between the three rRNA species and are termed internal transcribed spacers (ITS), while excised sequences that border on the outside of the central tract are referred to as 5ʹ or 3ʹ external transcribed spacers (ETS) [Citation1,Citation2]. Along the path to ribosome biogenesis, multiple ribosomal proteins attach and release both in the nucleus and after export to the cytoplasm to ensure correct structure and function of the final translational machinery. Many proteins involved in ribosome biogenesis are essential for growth [Citation3], and their expression must be carefully regulated to ensure correct levels of protein at all times. The demands for production are high; in rapidly dividing Saccharomyces cerevisiae yeast, a pre-ribosome is released every two to three seconds [Citation4]. Posttranscriptional regulatory mechanisms play a key role in such expression control.
Puf proteins are a eukaryotic family of posttranscriptional regulatory proteins that bind conserved sequences in the 3ʹ untranslated region (3ʹ UTR) of their target mRNAs. The canonical sequence bound by Puf proteins normally includes a UGU trinucleotide sequence followed by an A/U rich downstream region, although some Puf proteins bind to less well-conserved sequences, which serves to broaden their repertoire of targets [Citation5–Citation17]. The RNA-binding domain of Puf proteins, also known as the Pumilio homology domain, is typically composed of eight imperfect repeats of approximately 36 amino acid residues. Structural studies of Puf proteins bound to their target mRNAs have revealed that the complex adopts a crescent shape in which conserved residues of the Puf protein contact consecutive bases in the target in a one base per residue stacking manner [Citation11,Citation18–Citation21]. Some Puf proteins are able to increase the pool of transcripts they bind by forcing one or more bases away from the binding face of the Puf protein such that targets without perfect retention of the binding sequence may still be bound by Puf proteins [Citation11,Citation12,Citation18–Citation20,Citation22]. Upon binding, Puf proteins typically accelerate decay or inhibit translation of their targets by forming protein-protein interactions with decay machinery to stimulate deadenylation and decapping, or with proteins that inhibit cap binding events of translation initiation [Citation23–Citation27]. The Pumilio homology domain, or repeat domain (RD) of yeast Pufs 1, 3, 4, and 5 has been shown to be sufficient for both RNA binding and binding to decay factors for regulation of mRNA decay [Citation6,Citation28,Citation29]. Some Puf proteins have additional motifs, such as RRMs and glutamine-rich motifs, but these are not well conserved, and little is known of their contributions, if any, to mRNA decay regulation [Citation26,Citation30].
Several global-scale studies have identified hundreds of transcripts physically bound by Puf proteins in S. cerevisiae and other eukaryotes [Citation5,Citation16,Citation31,Citation32]. These studies indicate that Puf proteins tend to bind functionally related classes of mRNAs. In S. cerevisiae, Puf3p binds nuclear-encoded transcripts involved in mitochondrial function, Puf4p binds targets involved in ribosome biogenesis, and Puf5p binds targets involved in regulation of gene expression and cell wall maintenance. While these studies highlight the myriad of targets bound by Puf proteins, binding does not always result in regulation [Citation33].
Puf and Puf-like proteins have previously been shown to be involved in various aspects of ribosome biogenesis. Nop9 is a Puf-like protein in S. cerevisiae required for 18S rRNA synthesis and is associated with the pre-40S ribosomal subunit [Citation34]. It bears 11 pumilio-like repeats and binds both in the pseudoknot region of 18S and close to the Nob1 cleavage site at the 5ʹ end of ITS1 (between cleavage sites D and A2). Its presence at the D-A2 site prevents premature cleavage of the 20S rRNA. It is unclear what the function of Nop9 is at the pseudoknot region, but it must be released from the 18S rRNA prior to full maturation so that correct folding may occur [Citation35]. A puf6Δ in S. cerevisiae results in accumulation of 35S, 27S, and 7S precursor rRNA intermediates and causes defects in nuclear and nucleolar trafficking of large and small ribosomal subunits [Citation36]. Mutation of basic residues of Puf6p conserved between Puf6p and human Puf-A results in similar rRNA processing defects and accumulations [Citation17]. Caenorhabditis elegans Puf proteins puf-5, puf-8, and puf-9 act coordinately with ncl-1 and nos-2, the homologues to Drosophila melanogaster’s Brat and Nanos, respectively, to downregulate expression of fib-1 and control nucleolus size and the rRNA pool [Citation37]. Knockdown of PUF7 in Trypanosoma brucei decreases cleavage of the initial 9.2kb rRNA transcript and decreases abundance of the 2.6kb rRNA of the small subunit, indicating a defect in cleavage of the 3.4kb precursor [Citation38]. Knockdown of PUF7 and PUF10 in T. brucei decreases abundance of the 5.8S rRNA and its immediate 0.6kb precursor [Citation39]. In Arabidopsis thaliana, T-DNA insertion mutants of APUM23 show accumulation of 35S pre-rRNA and unprocessed 18S and 5.8S rRNAs, as well as U3 and U14 snoRNAs, which are involved in maturation of the 18S rRNA [Citation40]. In mice, PUM2 was found to bind PRP-2 mRNA, which has one perfect and two imperfect PREs for PUM2. High PRP-2 expression is linked to decreased expression of several ribosomal protein mRNAs, and it is thought that PUM2 may affect global translation through modulation of PRP-2 mRNA stability [Citation41].
In this study, we focused on understanding the role of S. cerevisiae Puf4p in ribosome biogenesis, given that many mRNAs physically bound to Puf4p are involved in this process. We selected four of the top scoring transcripts binding Puf4p with regard to conservation of their consensus Puf4p binding sequence for further analysis [Citation5]. Each of these transcripts is involved in some aspect of ribosome biogenesis. We show that these transcripts are not only regulated by Puf4p, but by a combination of Puf proteins. Pufs 2, 4 and 5 all contribute to destabilization of the mRNAs, while Pufs 1 and 3 act to stabilize the mRNAs. We also show that a single consensus site in the 3ʹ UTR of these transcripts corresponding to the canonical Puf4p binding sequence is critical not only for Puf-mediated decay regulation, but also decay regulation in the absence of Puf proteins. Unlike other targets of Puf4p regulation, decay of these ribosome biogenesis targets is not inhibited by differing carbon sources. Finally, we demonstrate that overexpression of Puf4p delays ribosomal RNA processing and inhibits ribosomal subunit trafficking. This work may provide a basis for understanding the roles of Puf proteins in ribosome biogenesis in higher eukaryotes.
Results
Targets involved in ribosome biogenesis that physically bind Puf4p are regulated redundantly by Puf4p and Puf5p
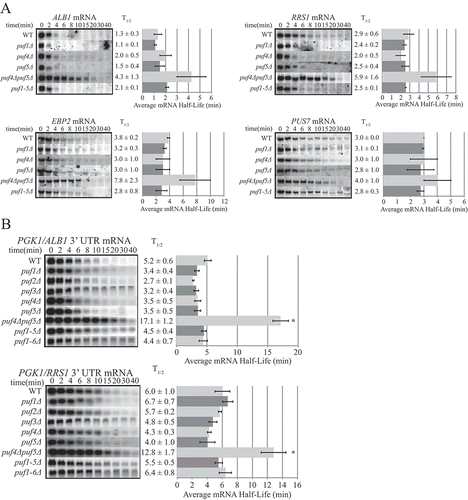
Of the coordinately regulated transcripts that contain Puf4p binding elements in their 3ʹ UTRs as identified in [Citation31], roughly 20% (148 of the 752) were involved in some aspect of ribosomal RNA processing or ribosome maturation and export. Of the transcripts found physically associated with Puf4p as found in [Citation5], one quarter of the 205 genes were involved in such ribosome biogenesis processes, with the rest of the targets falling into other functional categories well below 25% of the total. We therefore selected four of the top ranked targets identified in both studies that were involved in ribosome biogenesis: ALB1, EBP2, PUS7 and RRS1. Each of these targets has one highly conserved Puf4 binding element in its 3ʹ UTR. We began investigating Puf4p’s role in regulating these targets by analyzing their steady-state mRNA levels in a wild-type (WT) and a puf4Δ strain. Unexpectedly, the mRNA levels of these four targets were similar when comparing the two strains (Figure S1). Some Puf proteins, including Puf4p and Puf5p, are known to bind in a promiscuous nature and can flip out nucleotides from the mRNA binding surface to accommodate a larger pool of binding partners [Citation19,Citation20,Citation42–Citation44]. It is also known that multiple Puf proteins can bind and regulate a single target, although usually more than one cognate Puf binding element exists in the 3ʹ UTR [Citation28,Citation29,Citation45]. Specifically, there is significant overlap between mRNAs that are bound and regulated by both Puf4p and Puf5p [Citation5,Citation28,Citation29,Citation32,Citation45]. We therefore tested whether deletion of PUF5 influenced the target mRNA levels. While a single deletion of PUF5 failed to increase the target mRNA pools, a double deletion (puf4Δpuf5Δ) noticeably increased the steady-state levels of all four mRNAs (Figure S1), thus indicating that Puf4p and Puf5p are redundant for regulation of these targets. Previous studies have suggested that the binding pattern of one type of Puf site binds primarily to Puf4p, while another pattern binds both Puf4p and Puf5p [Citation44], while more recent studies suggest that Puf5p is more flexible in its binding, dependent on the curvature of its binding domain [Citation16]. Interestingly, the Puf sites in all four targets tested resemble the Puf4p binding pattern rather than the Puf4p+ Puf5p pattern. Our results therefore demonstrate that the plasticity of Puf regulation is greater than previously described.
Next, we wished to test whether the increased levels of the four transcripts were a result of an altered decay rate mediated by the Puf proteins. We performed transcriptional shutoff assays in WT, puf1Δ, puf4Δ, puf5Δ, and puf4Δpuf5Δ strains that also contain a temperature-sensitive mutation in the RNA polymerase II Subunit B (rpb1-1), which allows mRNA half-life analysis on steady-state mRNA pools following a shift to the non-restrictive temperature to block further transcription. The results mirrored those seen by steady-state analysis in that half-lives of all four transcripts were similar between the WT and single puf deletion strains, but were lengthened in the puf4Δpuf5Δ strain (). Surprisingly, when we tested decay of the target mRNAs in a strain deleted of PUFs 1, 2, 3, 4, and 5 (puf1–5Δ), the half-lives of all four mRNAs resembled the short half-lives seen in the WT strain (). Previous studies have shown that all other Puf target mRNAs tested are stabilized in the puf1–5Δ strain [Citation28,Citation29]. The current findings indicate that the decay rates of these targets are regulated redundantly by Puf4p and Puf5p, and suggest a yet undiscovered secondary mechanism whereby rapid decay of the transcripts is rescued in the absence of Puf proteins 1–5.
Figure 1. mRNAs involved in ribosome biogenesis are regulated redundantly by Puf4p and Puf5p. Decay analyses are shown of endogenous mRNAs (a) or reporter mRNA constructs PGK1-ALB1 3ʹ UTR and PGK1-RRS1 3ʹ UTR (b) in wild-type (WT), single puf deletion, double puf4Δ/puf5Δ deletion, puf1-5Δ deletion, and puf1-6Δ strains. Representative Northern blots are in the left panels, with average half-life (T1/2) listed to the right of each blot, and a graphical representation of the average half-lives in the right panels. Minutes following transcriptional repression at time 0 are indicated above the blots. Error bars represent standard error of the mean (SEM) and are representative of ≥ 3 trials. Asterisks indicate the only significantly different half-life in the group as determined by one-way ANOVA with Tukey’s post-hoc test (p ≤ 0.005).
The 3ʹ UTR of regulated transcripts is sufficient to confer Puf regulation on reporter mRNAs
Unlike the four mRNAs studied here, other mRNAs that are regulated by both Puf4p and Puf5p are at least partially stabilized in single PUF deletion strains [Citation28,Citation29]. One possible explanation for this difference is that the four mRNAs in this study are expressed at lower levels, and thus the amount of either Puf4p or Puf5p in the cell may be sufficient to regulate their decay. To test this hypothesis and also determine if the 3ʹ UTR sequences of these mRNAs are sufficient to confer Puf-mediated decay regulation onto a reporter transcript, 500 nt of the ALB1 or RRS1 3ʹ UTR was cloned onto the PGK1-82Δ open reading frame, which alone is not under Puf regulation. The reporter transcripts were expressed from a GAL-UAS promoter on a high copy vector (2µ), and would thus be expressed at higher levels than the endogenous mRNAs. When decay of these reporter transcripts (PGK1/ALB1 and PGK1/RRS1) was analyzed in the WT, puf1Δ, puf2Δ, puf3Δ, puf4Δ, puf5Δ, puf4Δpuf5Δ, and puf1–5Δ strains (), the patterns of half-lives observed resembled that of the endogenous targets seen in . Deletion of any single PUF failed to stabilize the transcripts beyond WT levels; however, deletion of both puf4Δ and puf5Δ stabilized both transcripts (3.3-fold half-life increase for PGK1/ALB1 and 2.1-fold increase for PGK1/RRS1). Half-lives of the transcripts in the puf1-5Δ strain were again similar to those seen in WT, further supporting a secondary mechanism responsible for stimulating decay in the absence of these Puf proteins. There are six Puf proteins in S. cerevisiae, and while decay stimulation has not been attributed to Puf6p thus far, a role in ribosome biogenesis has been documented [Citation36]. To test whether Puf6p was responsible for the secondary mechanism mediating rapid decay in the absence of Pufs 1-5, the reporter transcripts were analyzed in a puf1–6Δ strain. However, the half-lives of the transcripts were still similar to those in WT, eliminating Puf6 as the secondary decay mechanism (). Together, these results show that the 3ʹ UTRs of these target mRNAs are sufficient to confer Puf regulation, and the redundant regulatory actions of Puf4p and Puf5p are not due to the low level expression of the endogenous mRNA targets.
Reporter mRNAs are not stabilized in galactose
When cells are grown in galactose, global transcript turnover is not significantly altered as compared to in cells grown in dextrose, with a few exceptions [Citation46]. However, prior work from our lab has demonstrated that transcripts whose decay is regulated by Puf proteins in dextrose are stabilized in galactose, frequently over 2-fold [Citation28,Citation33]. Posttranscriptional modifications of Puf proteins such as phosphorylation are known to inhibit or stimulate activity [Citation47,Citation48], and phosphorylation of yeast Puf3p upon glucose depletion has been shown to stimulate translation of its targets [Citation49]. These findings provide an attractive model whereby Puf activity is regulated quickly by the changing carbon source [Citation33]. We therefore wished to determine if the transcripts in this study were also stabilized in galactose. As shown in , the reporter transcripts PGK1/ALB1 and PGK1/RRS1 were not substantially stabilized in galactose. For PGK1/ALB1, the half-life decreased 1.5-fold in galactose versus dextrose, while the half-life of PGK1/RRS1 increased only 1.4-fold. Since the decay stimulation by Puf4p and Puf5p on other mRNAs is inhibited by galactose [Citation28], these results suggest that the secondary mechanism acting to regulate decay of these transcripts in the absence of Puf proteins may also be acting here in a carbon source independent manner.
Figure 2. Reporter mRNAs are not stabilized in galactose, while Puf protein overexpression rescues rapid decay. Decay analyses are shown of reporter constructs PGK1-ALB1 3ʹ UTR and PGK1-RRS1 3ʹ UTR in (a) the WT strain in the presence of dextrose or galactose, or (b) in the double puf4Δ/puf5Δ deletion strain containing an empty vector (EV) or a vector overexpressing Puf4p or Puf5p. Representative Northern blots are in the left panels, with average half-life (T1/2) listed to the right of each blot, and a graphical representation of the average half-lives in the right panels. Minutes following transcriptional repression at time 0 are indicated above the blots. Error bars represent SEM (n ≥ 3). The asterisk in (a) indicates a significant difference as determined by Student’s t-Test (p ≤ 0.05). The letters next to the bar graph in (b) indicate significant differences as determined by one-way ANOVA with Tukey’s post-hoc test (PGK1-ALB1, p ≤ 0.005; PGK1-RRS1, p ≤ 0.05).
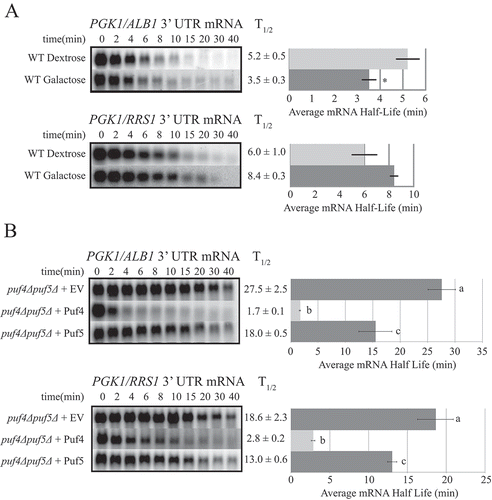
Puf4p expression in a puf4Δpuf5Δ strain rescues rapid decay
We next sought to determine if exogenous expression of either Puf4p or Puf5p individually in the puf4Δpuf5Δ strain could rescue rapid decay of the target mRNAs. As shown in , complementation of the puf4Δpuf5Δ strain with Puf4p expressed from a CEN vector dramatically shortened the half-lives of the mRNAs as compared to the puf4Δpuf5Δ strain with an empty vector. The PGK1/ALB1 half-life decreased 16.2-fold, while the PGK1/RRS1 half-life decreased 6.6-fold. In fact, these half-lives are 3.1-fold and 2.1-fold shorter than those of the respective reporter mRNAs in the WT strain. Since PUF4 expression from the CEN vector was 20-fold higher than endogenous PUF4 levels as measured by qPCR (Figure S2), these results suggest that endogenous levels of Puf4p are limited in the cell, as higher exogenous levels promoted increased decay stimulation. Exogenous Puf5p expression from a CEN vector displayed partial rescue of decay, decreasing the PGK1/ALB1 half-life 1.5-fold and decreasing the PGK1/RRS1 half-life 1.4-fold versus the empty vector strain (). PUF5 expression from the CEN vector was 15-fold higher than endogenous PUF5 levels as measured by qPCR (Figure S2). Thus, given equally increased expression of both exogenously-expressed PUF4 and PUF5, exogenous Puf4p appears to be much more active in stimulating decay of the mRNA targets.
Other Puf proteins play contrasting roles in decay regulation
Given our data showing that the target reporter transcripts were stabilized in a puf4Δpuf5Δ strain but destabilized in a puf1-5Δ strain back to a half-life mirroring WT, it is possible that the loss of Puf1p, Puf2p, Puf3p, and/or Puf6p could be responsible for this destabilization. Specifically, one or more of these proteins may have a stabilizing role, unique to these targets. While the vast majority of Puf proteins stimulate decay upon binding to their targets, some Puf proteins under certain circumstances act to stabilize target transcripts [Citation50,Citation51]. To investigate this possibility, we deleted PUF1, PUF2, PUF3, or PUF6 in the puf4Δpuf5Δ background and analyzed the decay of PGK1/ALB1 and PGK1/RRS1. The deletion of PUF2 in addition to PUF4 and PUF5 further stabilized both mRNAs 1.6-fold over the respective half-lives seen in the puf4Δpuf5Δ strain (). This result identifies Puf2p as a third Puf protein that acts to promote decay of these targets. Like Puf4p and Puf5p, the individual removal of Puf2p had no effect on target decay ()), but all three appear to play redundant roles in stimulating decay, with combinations of their loss additive. In contrast, deletion of PUF1, PUF3, or PUF6 individually in the puf4Δpuf5Δ background had little effect on decay of the targets, as the half-lives of the triple mutants were similar to the puf4Δpuf5Δ double mutant ()).
Figure 3. Puf2p acts with Puf4p and Puf5p to stimulate mRNA decay, while Puf1p and Puf3p stabilize mRNA targets. Decay analyses are shown of reporter constructs PGK1-ALB1 3ʹUTR and PGK1-RRS1 3ʹ UTR in (a) triple mutant strains or (b) in the puf1-5Δ strain containing an empty vector (EV) or a vector overexpressing the Puf1p repeat domain (Puf1RD) or full length Puf1p (Puf1FL) or the Puf3p repeat domain (Puf3RD) or full length Puf3p (Puf3FL). Representative Northern blots are in the left panels, with average half-life (T1/2) listed to the right of each blot, and a graphical representation of the average half-lives in the right panels. Minutes following transcriptional repression at time 0 are indicated above the blots. Error bars represent SEM (n ≥ 3). Asterisks in (a) represent the only significantly different half-life as compared to the double puf4Δpuf5Δ half-life as determined by one-way ANOVA with Tukey’s post-hoc test (p ≤ 0.05).
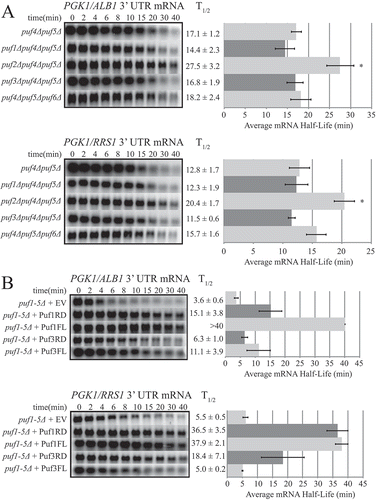
Although the individual removal of PUF1 or PUF3 in the puf4Δpuf5Δ background did not alter target decay, it is possible that one of the remaining Pufs in the triple mutant is sufficient to stabilize the mRNAs as compared to the rapid decay in the quintuple PUF mutant. To test whether the presence of one of these Pufs alone can stabilize the target transcripts, we expressed either full-length or the Puf repeat domain of Puf1p or Puf3p from a 2µ vector in the puf1-5Δ strain. As shown in ), expression of either Puf1p or Puf3p was able to stabilize the reporter transcripts. Of the two, Puf1p acted more strongly to stabilize the transcripts, with half-lives increased 4- to 10- fold. The full-length Puf1p was the most potent stabilizer, though the Puf1p repeat domain alone was sufficient for stabilization. The repeat domain of Puf3p stabilized the transcripts 2- to 3- fold, while the full-length Puf3p stabilized only the PGK1/ALB1 transcript 3-fold. Both Puf1p and Puf3p have previously been shown to stimulate decay of their target mRNAs [Citation29,Citation30,Citation33]. Therefore, these results demonstrate a novel role for Puf1p and Puf3p in stabilizing these particular transcripts, and the first evidence of a stabilizing role for Puf proteins in the yeast S. cerevisiae. Moreover, the secondary mechanism that may be responsible for stimulating decay in the absence of Puf proteins may be inhibited by the presence of either Puf1p or Puf3p.
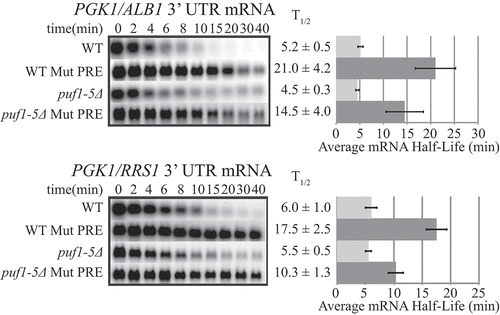
The PREs in the target mRNAs are necessary for decay regulation
Each of the four endogenous mRNA targets contains a single Puf binding site, also termed a Pumilio Response Element (PRE), shown in Figure S3, which matches the canonical Puf4p PRE [Citation5,Citation44]. To investigate the necessity of these sites for mRNA decay regulation, the conserved UGUA sequences within the PREs of the PGK1/ALB1 and PGK1/RRS1 3ʹ UTRs were mutated to ACAC, then the half-lives of the transcripts examined. As seen in ), the PGK1/ALB1 mutant PRE transcript was stabilized 4-fold over its respective native PRE transcript, and the PGK1/RRS1 mutant PRE transcript was stabilized 3-fold over its respective native PRE transcript in the WT strain containing all Puf proteins. In addition, the PGK1/ALB1 and PGK1/RRS1 mutant PRE transcripts were stabilized 3-fold and 2-fold, respectively, in the puf1-5Δ strain. Thus, the canonical PREs are necessary not only for decay regulation by Puf proteins, as expected, but also for decay regulation by the secondary mechanism that is acting in the absence of Puf proteins.
Figure 4. A single Puf Response Element (PRE) in the 3ʹ UTR is critical for decay regulation. Decay analyses are shown of native PRE reporter constructs PGK1-ALB1 3ʹ UTR and PGK1-RRS1 3ʹ UTR and respective mutant PRE reporter constructs (Mut PRE) in the WT and puf1-5Δ strains. In the mutant PRE constructs, the canonical UGUA portion of the PRE was mutated to ACAC. Representative Northern blots are in the left panels, with average half-life (T1/2) listed to the right of each blot, and a graphical representation of the average half-lives in the right panels. Minutes following transcriptional repression at time 0 are indicated above the blots. Error bars represent SEM (n ≥ 3).
Overexpression of Puf4p causes ribosomal RNA processing defects
The four mRNA targets examined in this study are all involved in various aspects of ribosome biogenesis. Ebp2p is involved in some of the initial rRNA excision events, as its removal causes accumulation of the 35S transcript [Citation52]. Rrs1p is also involved in rRNA excision events, as its deletion results in kinetic defects not only in 35S processing, but also in processing 27S to 25S and 20S to 18S [Citation53]. Given our data showing that overexpression of Puf4p shortens the half-lives of target transcripts below those seen in WT cells, we hypothesized that such alterations in half-lives would negatively impact the production of protein from those transcripts. Decreased levels of ribosome biogenesis regulatory factors might then result in slower rRNA processing. To examine this possibility, steady-state levels of rRNA processing intermediates were analyzed from WT cells containing either an empty CEN vector or a CEN vector expressing PUF4. As shown in ,b), overexpression of Puf4p caused a greater than 2-fold accumulation of the initial 35S precursor rRNA transcript. In accordance with a slowed processing of 35S, overexpression of Puf4p caused a significant decrease in the levels of 20S, the immediate precursor to the final 18S rRNA, and also a decrease in 7S, the precursor to the final 5.8S rRNA (see Figure S4 for the rRNA cleavage and excision pathway). These alterations are very similar to those found in prior work analyzing mutations in ribosome processing factors [Citation54,Citation55]. Moreover, like our results, these prior studies typically did not detect differences in the levels of the large, stable pools of 18S and 5.8S final rRNA products due to limitations in the assay and the fact that the kinetics of processing are slowed, but not blocked. Overexpression of Puf4p also caused a moderate increase in the levels of the 23S precursor. The 23S variant is an aberrant excision product that occurs when the cell fails to excise the 5ʹ ETS at sites A0 and A1, and uses alternative cleavage at site A3. Increased 35S and 23S precursor and decreased 20S precursor are consistent with other mutations that result in inhibition of cleavage at site A2. Together, these results demonstrate that Puf4p overexpression causes rRNA processing defects, including slowed 5ʹ ETS excision and A2 cleavage inhibition, likely due to the destabilization of mRNAs coding for ribosome biogenesis factors.
Figure 5. Puf4p overexpression causes accumulation of rRNA processing intermediates. (a) Representative gel images are shown of the steady state levels of rRNA processing intermediates/precursors in the WT strain in the presence of a CEN empty vector (CEN EV) or a CEN vector overexpressing Puf4p (Puf4 OE). The scRI loading controls for each set of rRNA species is shown directly underneath the corresponding images. The diagrams to the right of each gel image are visual representations of the sequence regions within the rRNA intermediates/precursors. The three final species of rRNA originating from the PolI transcript (18S, 25S and 5.8S) are labeled in white; sites of processing are labeled in black letters A-D. (b) Graphical representation of the rRNA intermediate/precursor band intensities in (a). Error bars represent SEM (n = 3). Asterisks indicate significant differences between reported levels for EV and 4OE as determined by Student’s t-Test (p ≤ 0.01) (c) Representative gel images of rRNA processing intermediates/precursors following an L-[Methyl-3H]-Methionine pulse-chase labeling assay in a WT strain in the presence of a CEN empty vector (CEN EV) or a CEN vector overexpressing Puf4p (Puf4 OE). Minutes following the addition of unlabeled methionine at time 0 are indicated above the gels (n = 3).
![Figure 5. Puf4p overexpression causes accumulation of rRNA processing intermediates. (a) Representative gel images are shown of the steady state levels of rRNA processing intermediates/precursors in the WT strain in the presence of a CEN empty vector (CEN EV) or a CEN vector overexpressing Puf4p (Puf4 OE). The scRI loading controls for each set of rRNA species is shown directly underneath the corresponding images. The diagrams to the right of each gel image are visual representations of the sequence regions within the rRNA intermediates/precursors. The three final species of rRNA originating from the PolI transcript (18S, 25S and 5.8S) are labeled in white; sites of processing are labeled in black letters A-D. (b) Graphical representation of the rRNA intermediate/precursor band intensities in (a). Error bars represent SEM (n = 3). Asterisks indicate significant differences between reported levels for EV and 4OE as determined by Student’s t-Test (p ≤ 0.01) (c) Representative gel images of rRNA processing intermediates/precursors following an L-[Methyl-3H]-Methionine pulse-chase labeling assay in a WT strain in the presence of a CEN empty vector (CEN EV) or a CEN vector overexpressing Puf4p (Puf4 OE). Minutes following the addition of unlabeled methionine at time 0 are indicated above the gels (n = 3).](/cms/asset/91b5d13e-f4d9-45fc-ba73-d9cc5cdb46cc/krnb_a_1521211_f0005_b.gif)
To further analyze the effect of Puf4p overexpression on the kinetics of rRNA processing, a pulse-chase rRNA labeling assay was performed. Ribosomal RNA is methylated on the 2ʹ-O-ribose group of many of its bases [Citation56]. Using L-[Methyl-3H]-Methionine as a tritiated methyl donor, the rRNA was radiolabeled, then samples were collected at increasing times after a chase with unlabeled methionine. As seen in ), the kinetics of processing 27S into 25S rRNA and 20S into 18S rRNA were similar when comparing strains with the empty vector versus Puf4p overexpression. However, the original 35S transcript is barely detectable even at the beginning of the chase in the empty vector strain due to rapid processing, while in the strain overexpressing Puf4p, the 35S precursor has accumulated and persists two minutes into the chase. This data provides additional support that Puf4p overexpression slows the kinetics of rRNA processing at the earliest cleavage events, such as 5ʹ ETS excision, as a result of destabilization of the mRNAs coding for factors that are involved in such events.
Overexpression of Puf4p causes mislocalization of ribosomal subunits
In addition to factors involved in rRNA cleavage events, some mRNAs targeted for destabilization by Puf4p code for proteins involved in ribosome subunit trafficking. Alb1p is vital to the export of ribosomal subunits, functioning along with Arx1p to recycle the anti-association factor Tif6 to the nucleus [Citation57]. Rrs1p also has a role in the export of ribosomal subunits; rrs1 mutants display 60S export defects [Citation58]. We therefore investigated whether overexpression of Puf4p disrupts export of ribosomal subunits. To this end, confocal microscopy was used to visualize the localization of the ribosomal subunits in a WT strain ± an empty CEN vector, or a WT strain overexpressing Puf4p. GFP-tagged versions of the large ribosomal subunit protein RPL11B or small ribosomal subunit protein RPS2 were used as markers for the large and small ribosomal subunits, and SIK1-mRFP was used as a nucleolar marker. As shown in ), RPL11B (large subunit) was largely diffuse between the nucleus and cytoplasm in WT cells with or without the empty CEN vector. However, overexpression of Puf4p caused punctate nuclear foci of RPL11B to appear in many cells (white arrows) that were largely absent in the WT strain ± empty CEN vector. Overexpression of Puf4p also caused more nuclear accumulation of RPS2 (small subunit). In WT cells ± empty CEN vector, there is a noticeable spatial deficit of RPS2 that coincides with the position of the nucleus (white perpendicular symbols). Upon overexpression of Puf4p, the incidence of these areas decreased, indicating more RPS2 was residing in the nucleus.
Figure 6. Puf4p overexpression results in nuclear accumulation of ribosomal subunits. (a) Representative panels are shown from confocal microscopy of cells from the WT strain, or the WT strain containing an empty vector (EV) or a vector overexpressing Puf4p (4OE). Labels above the panels indicate the stain/protein being observed. Arrows on the upper set of panels indicate the position of nuclear foci as detected by RPL11B-eGFP. Arrows on the lower set panels indicate the position of nuclei in the cells as detected by a decreased signal of RPS2-eGFP. (b) Graphical representation of the nuclear to cytoplasmic ratio of GFP fluorescence intensity in WT cells ± empty vector or vector overexpressing Puf4p. Letters a and b indicate significant differences using one-way ANOVA (p < 0.005) and Tukey’s post-hoc test. Error bars represent SEM and are representative of ≥ 250 cells spread across 3 individual growth trials.
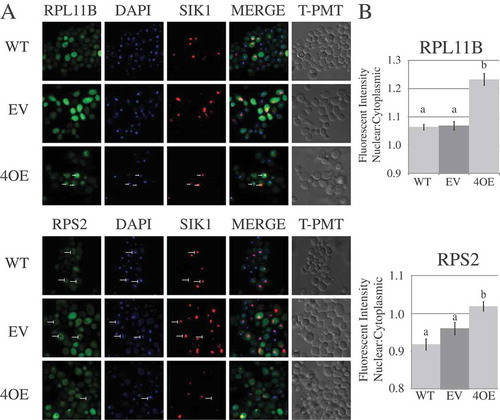
To quantitate the nuclear retention of large and small ribosomal subunits upon Puf4p overexpression, GFP signal was analyzed in the microscopy images using CellProfiler software [Citation59]. Briefly, the software identified the position of the nucleus from the DAPI channel, and the intensity of the GFP signal corresponding to either RPL11B-eGFP or RPS2-eGFP was calculated for the nuclear and cytoplasmic areas of the cell. As shown in ), there was a statistically significant increase in the mean fluorescent intensity ratio of nuclear to cytoplasmic signal for both RPL11B and RPS2. The results indicate that overexpression of Puf4p causes mislocalization of large and small ribosomal subunits, likely a result of the inhibition of their export from the nucleus due to destabilization of the mRNAs coding for ribosome export factors.
Discussion
Cell growth is heavily dependent on ribosome production, and changing growth conditions necessitates strict control of ribosome biogenesis through both transcriptional and post-transcriptional regulation. This work focused on the role of Puf proteins in the post-transcriptional regulation of ribosome biogenesis factors, and thereby ribosome production in Saccharomyces cerevisiae. Previous work found that Puf4p physically bound multiple mRNAs involved in various aspects of ribosome biogenesis, both in the maturation of pre-ribosomal RNA and the export of pre-ribosomes to the cytoplasm. Our analyses of the top-scoring mRNAs bound by Puf4p demonstrate that these transcripts are uniquely destabilized by a combination of Puf proteins 2, 4 and 5. Our results also provide the first evidence of the role of Puf proteins 1 and 3 in stabilizing transcripts in S. cerevisiae. Decay regulation of each transcript appears to occur through a single canonical Puf4p binding site in the 3ʹ UTR, indicating competition between Puf proteins for binding access. We also show evidence for a Puf-independent secondary mechanism of decay regulation that uses the same binding site. The importance of Puf-mediated decay regulation of ribosome biogenesis factor transcripts was further established by demonstrating that Puf4p overexpression causes ribosomal RNA processing delays and inhibits nuclear export of pre-ribosomal subunits.
The data presented suggest the following model for Puf-mediated regulation of the ribosome biogenesis transcripts and the resulting cellular implications (). In a normal cell with all Puf proteins present, there is likely a competition between different Puf proteins for binding the single consensus Puf Response Element (PRE) in the 3ʹ UTR, though it is possible that Pufs may be binding non-canonical elements. Binding of Puf2p, Puf4p, or Puf5p stimulates decay, while binding of Puf1p or Puf3p results in stabilization. Since the half-lives of the target transcripts are short in a wild-type cell expressing all Puf proteins, the actions of Puf2p, Puf4p, or Puf5p to stimulate decay supersede the stabilizing activities of Puf1p or Puf3p. Furthermore, since the presence of Puf1p or Puf3p can stabilize the transcripts in the absence of the other Pufs, the stabilizing activities of Puf1p or Puf3p supersede the secondary Puf-independent mechanism of decay stimulation.
Figure 7. Puf proteins regulate mRNA decay to ensure proper ribosome biogenesis. (a) Puf2p, Puf4p, and Puf5p can each act to stimulate mRNA decay of ribosome biogenesis factors through binding to a single PRE site (UGU). Puf1p and Puf3p act through the same PRE to stabilize these mRNAs, potentially by blocking access of another decay factor to the PRE. In the absence of Puf proteins, a secondary mechanism that also requires the PRE site acts to stimulate decay. (b) At physiological levels of Puf proteins, processing of rRNA transcripts occurs normally. When Puf4p is overexpressed, processing is slowed, resulting in higher levels of the initial 35S and aberrant 23S transcripts, and lower levels of 20S and 7s intermediates. (c) At physiological levels of Puf proteins, ribosomal subunits are shuttled from the nucleus at a normal rate. When Puf4p is overexpressed, trafficking of ribosomal subunits is inhibited, resulting in abnormal nuclear retention of ribosomal subunits.
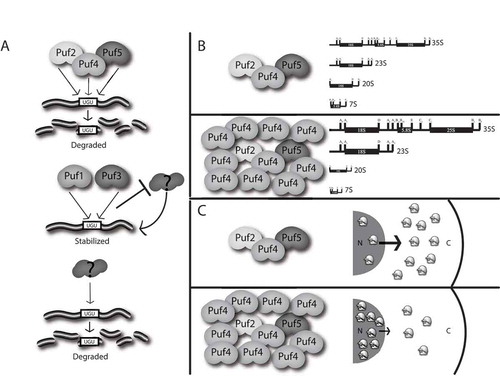
The mechanism of action by which Puf1p and Puf3p stabilize these transcripts is unclear. Some possibilities include stabilization through stimulation of translation, the blocking of access of the PRE to decay stimulatory factors, or the inability to modify the mRNP to stimulate decay. We do not believe that Puf1p and Puf3p are stimulating translation under the glucose replete conditions used in this study, as all previous work has demonstrated the roles of these Puf proteins in decay stimulation under such conditions [Citation28,Citation33], and only in glucose depleted conditions has Puf3p been shown to stimulate translation [Citation49]. It is possible that Puf1p and Puf3p actively stabilize these transcripts by recruiting stabilizing factors. However, we think it more likely that Puf1p and Puf3p simply occupy the PRE and prevent other non-Puf decay stimulating factors from binding. It is possible that the overexpression of Puf1p and Puf3p in these studies are exaggerating the effect that these Pufs may have at endogenous levels. However, endogenous Puf3p has been shown to physically bind ALB1 [Citation60], EBP2, and PUS7 [Citation61] mRNAs, although the PREs do not represent canonical Puf3p PREs. Puf proteins that bind non-canonical PREs may be in a non-functional conformation, preventing them from recruiting decay factors, while still occupying the site. Another possibility is that the local mRNP structure around non-canonical targets prevents these Pufs from activating decay. For example, Pub1p, another 3ʹ UTR binding protein, promotes decay of some transcripts while stabilizing others, dependent on the sequence of the mRNA. Remodeling of the mRNP structure by Pub1p has been suggested to be at least partly responsible for this alteration in mRNA stability [Citation62]. Puf3p has also been shown to require the Poly(A)-Binding protein Pab1p to promote proper activities of poly(A)-tail deadenylation by Pan proteins and Ccr4p, suggesting that perturbations to overall mRNP structure could inhibit Puf stimulation of decay [Citation25].
The data also suggest that Puf expression levels are important to maintaining appropriate levels of proteins involved in ribosome biogenesis. Overexpression of Puf4p inhibited both pre-rRNA processing events and trafficking of ribosomal subunits from the nucleus to the cytoplasm (). The decreased lifespan of target mRNAs upon Puf4p overexpression is concomitant with this phenotype, suggesting that decreased protein production caused by increased decay stimulation of these and likely other targets is responsible for these defects.
A recent study to globally analyze yeast Puf binding affinity to mRNAs utilized a novel tagging system in which Puf3p, Puf4p, and Puf5p were fused to the Polyuridine-Polymerase (PUP) gene from C. elegans, and all mRNAs that bore poly-U tails as a result were analyzed [Citation32]. The mRNAs were scored for the length of the poly-U tails, which is an indirect measurement of their association time with the Puf protein. All four targets studied here are canonical Puf4p targets with 9-residue binding elements; ALB1, RRS1, and EBP2 were top scorers bound by Puf4p, while PUS7 scored lower, which may be why in our studies it did not respond as dramatically to the double puf4Δpuf5Δ deletion. While none of the targets contain the canonical 10-residue Puf5p binding element, most were found bound by Puf5p, albeit in a lower scoring group than found with Puf4p; ALB1 and RRS2 scored above EBP2, while PUS7 was not identified by the Puf5p analysis. These results support our data showing that Puf4p overexpression in a puf4Δpuf5Δ strain is better at rescuing rapid decay of target mRNAs than overexpression of Puf5p.
Puf2p’s discovered role in regulation of these transcripts was unexpected. A previous study suggested the consensus Puf2p binding sequence is UAAU-XXX-UAAU, where X is usually an A or U [Citation63]. This sequence is unlike those of other PUF proteins, which necessarily include the UGU trinucleotide. The ALB1 3ʹUTR does not contain this sequence, while the RRS1 3ʹ UTR contains dual UAAU motifs separated by a less conserved 9 nucleotide linker, yet both seem to be negatively regulated by Puf2p. While studies have analyzed Puf2p binding [Citation63], no study has yet implicated Puf2p in decay stimulation of a specific mRNA target, and deletion of puf2 does not alter the lifespans of other tested mRNAs [Citation28,Citation29]. Thus, the current findings represent the first bona fide targets of Puf2p regulation of decay.
Puf6p has previously been shown to play a role in ribosome biogenesis, as its deletion causes rRNA processing and ribosomal subunit trafficking defects [Citation36]. In the absence of Pufs 1-5, rapid decay of the target mRNAs was rescued, suggesting that either Puf6p stimulates decay in the absence of other Pufs, or there exists a non-Puf secondary mechanism of decay stimulation. Since in the deletion of Pufs 1-6, mRNAs were still rapidly decayed, Puf6p is not involved in this secondary mechanism. The current understanding of Puf6p activity is that it inhibits translation of ASH1 mRNA [Citation47,Citation64]. It is also thought to be required, along with Loc1p, for efficient loading of Rpl43p onto pre-60S particles; Puf6p and Loc1p are released upon Rpl43p loading [Citation65]. Deletion of Puf6p results in 7s precursor rRNA accumulation, although the mechanism by which Puf6p ensures proper 7s to 5.8S processing is still unclear [Citation17]. Thus, Puf6p’s mechanism of action in ribosome biogenesis appears to be quite different than the mRNA decay regulatory activities of Pufs 1-5.
The nature of the non-Puf secondary mechanism acting to promote decay of the target mRNAs in the absence of Pufs is unclear. We show that the mechanism acts through the Puf Response Element, as mutation of the conserved UGUA to ACAC results in stabilization of the mRNA in the absence of Pufs. There are other proteins known to bind similar AU-rich elements (AREs) in 3ʹ UTRs for decay regulation, such as Vts1, Cth1/2p, Whi3p, and Pub1p [Citation62,Citation66–Citation71]. There are numerous other uncharacterized RNA-binding proteins that may also be responsible for the non-Puf secondary mechanism of decay of the target mRNAs. The fact that there are multiple mechanisms all acting to carefully control the lifespans of ribosome biogenesis factor transcripts demonstrates the importance of this regulation for cell fitness in varying environments.
Most studies of ribosome biogenesis gene expression focus on the transcriptional control of these genes, including through the actions of the TORC1 complex in response to nutrient availability [Citation72,Citation73]. There is also some evidence for translational control of ribosome-associated transcripts [Citation74,Citation75]. This work demonstrates a new layer of regulation of ribosome biogenesis factors through mRNA decay control by both Puf-mediated and non-Puf mechanisms. Further studies will elucidate any cooperation between these two mechanisms and dissect the combinatorial activities of Puf proteins on this regulation.
Materials and methods
Plasmids, oligonucleotides, and yeast strains
All plasmids, oligonucleotides, and yeast strains used in this study are listed in Supplementary Tables S1-S3, respectively. Strain yWO260 was created by mating yWO3 and yWO7, and strain yWO289 was created by mating yWO260 and yWO261 as described [Citation76]. The diploids were sporulated, the resulting tetrads dissected, and the spores genotyped. Strain yWO261 was creating by transforming yWO39 with BamHI digested pWO183 to replace the URA3 gene with the KanMX3 G418 resistance cassette and plating the transformants on yeast extract/peptone/dextrose (YEPD) containing 300µg/ml Geneticin (Life Technologies). Transformations were performed via LiOAc-mediated high efficiency transformation as described [Citation77]. Strains yWO306, yWO307, and yWO308 were created by PCR amplification of the nourseothricin resistance cassette (NAT1) contained in pWO241 [Citation78] using primers oWO869/870, oWO918/919, and oWO911/912, respectively, and the resultant products were transformed into yWO289 as previously described [Citation77]. yWO317 and yWO318 were created in a similar manner by PCR amplification of the nourseothricin resistance cassette with oWO857/858 and transformation into yWO289 and yWO205, respectively. Transformants were allowed to recover overnight in YEPD media, then plated on YEPD containing 100µg/ml nourseothricin (Jena Bioscience) to select for replacement of PUF genes by the nourseothricin resistance cassette. Gene replacement was verified by PCR amplifying a region spanning upstream of the insertion site to inside the resistance cassette using primers oWO901/833, oWO919/833, oWO913/833, and oWO863/833.
Creation of pWO226 was achieved by digesting pWO165 with XhoI/EcoRI, then transforming the resultant fragment and pWO13 into yWO3. Transformants were selected on media lacking uracil. Plasmids were recovered from yeast cells by digestion with Zymolyase (MP Biomedicals) at 37°C for 30 minutes, harvesting the cells, and proceeding with alkaline lysis using the Zyppy plasmid miniprep kit (Zymo Research) per the manufacturer’s instructions. Creation of the PGK1-ALB1 3ʹ UTR (pWO231) and PGK1-RRS1 3ʹ UTR (pWO232) reporter constructs was performed by PCR amplifying ~ 500nt of the 3ʹ UTRs of the respective genes while introducing a 5ʹ ClaI site and a 3ʹ HindIII site for ALB1, and a 5ʹ EcoRV and 3ʹ HindIII site for RRS1. The digested PCR products were ligated into the PGK1Δ82 ORF vector (pWO102) and the resulting constructs sequence verified. Creation of the PGK1-ALB1 3ʹ UTR (pWO233) and PGK1-RRS1 3ʹ UTR (pWO234) reporter constructs was performed by digesting pWO167 with SmaI and transforming the resultant linear DNA with pWO231 or pWO232 into yWO3. Cells with recombinant plasmids were selected on media lacking leucine. Plasmids were recovered from yeast cells by digestion and alkaline lysis as described above. The PGK1-ABL1 and PGK1-RRS1 mutant PRE reporter constructs were created by site directed mutagenesis using the Quikchange II-XL site directed kit (Agilent Technologies) per manufacturer’s instructions with primers oWO786/oWO787 and oWO788/oWO789, respectively. Creation of pWO238 was performed by digesting pWO166 with SmaI and transforming the resultant linear DNA and pWO237 into yWO236. Cells with recombinant plasmids were selected on media lacking histidine, and plasmids recovered as described above. Creation of pWO253 was performed by amplifying PUF5 from pWO196 using primers oWO936 and oWO938, then transforming the resultant PCR product and pWO116 linearized by HindIII/XhoI into yWO3. Cells with recombinant plasmids were selected on media lacking leucine.
Steady state analysis of endogenous mRNAs
WT (yWO3), puf4Δ (yWO22), puf5Δ (yWO17), or puf4Δpuf5Δ (ywo39) cells were grown in 20 ml YEP media containing 2% dextrose at 30°C until an OD600 of 0.4 was reached. Cells were then harvested and total RNA was isolated as previously described [Citation79]. 40μg of total RNA was loaded onto a 1.25% agarose gel (containing formaldehyde and MOPS) and electrophoresed at 70V for 4 hours. RNA was transferred to a membrane (Biobond Plus Nylon Membrane, Sigma-Aldrich), UV crosslinked, and probed with [Citation32]P 5ʹ-end labeled oligonucleotides complementary to ALB1 (oWO318), PUS7 (oWO319), EBP2 (oWO320) or RRS1 (oWO321) mRNA, and with an oligonucleotide complementary to scRI RNA (oWO21) as a loading control. Membranes were exposed to phosphorimager screens, and imaging and quantification of RNA levels were performed using ImageQuant Software (Molecular Dynamics).
In vivo decay analysis of endogenous mRNAs
WT (yWO7), puf4Δ (yWO106), puf5Δ (yWO49), puf4Δpuf5Δ (yWO289), or puf1-5Δ (yWO268) cells were grown in 200 ml YEP media plus 2% dextrose at 24°C until an OD600 of 0.4 was reached. These strains harbor a mutation in RNA polymerase II subunit b (rpb1-1), which causes temperature-sensitive inactivation of the polymerase at 37°C. Cells were harvested and resuspended in 10 ml YEP media at 24°C. 10 ml of YEP media plus 8% dextrose at 50°C was added to raise the temperature to 37°C and effectively stop transcription. 2 ml aliquots of cells were collected at time points up to 40 minutes, and total RNA was isolated as previously described [Citation79]. Northern analysis, detection and quantification proceeded as described above.
In Vivo decay analysis of reporter mRNAs
Decay analysis of reporter mRNAs from pWO231, 232, 233, 234, 235, and 236 was performed in the same manner as decay analysis of endogenous mRNAs. Briefly, indicated strains were transformed with the reporter construct and any other indicated plasmids and grown in media lacking uracil, leucine, or both uracil and leucine as noted. The temperature of the culture was shifted to 37°C to stop new transcription, cell aliquots at the indicated time points were collected, and total RNA was extracted. Northern analysis, detection and quantification proceeded as described above. PGK1-ALB1 and PGK1-RRS1 reporter mRNAs were detected with oWO521 and oWO522, respectively, which are complementary to the junction between the end of the PGK1Δ82 ORF and beginning of the ALB1 or RRS1 3ʹ UTR.
Steady state analysis of rRNA
20 ml cultures of yWO3 containing pWO58 or pWO116 were grown at 30°C to an OD600 of 0.4, then the cells harvested and frozen on dry ice. Total RNA was extracted via a hot phenol method previously described [Citation80], and 10μg of RNA was loaded on a 1.25% agarose gel (containing formaldehyde and MOPS) for large rRNAs and on a 6% polyacrylamide (19:1 acryl:bis) 8M urea TBE for the 7S and 5.8S rRNAs. The agarose gel was electrophoresed for 8 hours at 70V and the polyacrylamide for 6 hours at 300V. The RNA was transferred to nylon membrane as previously described for the agarose gel and electroblotted for polyacrylamide gel in 0.5X TBE at 10.5V for hours. The membraneswereUV crosslinked, and probed with [32P] radiolabeled oligonucleotides oWO714-724, and scRI loading control as previously described. Membranes were exposed to phosphorimager screens, and imaging and quantification of RNA levels were performed using ImageQuant Software (Molecular Dynamics).
Pulse chase labeling of rRNA
Pulse chase labeling of rRNA was performed essentially as described [Citation54,Citation55]. Briefly, 20 ml cultures of yWO3 containing pWO58 or pWO116 were grown at 30°C in SC-Met-Leu media containing 2% dextrose to an OD600 of 0.4. 10 ml of cells were harvested then resuspended in 3 ml SC-Met-Leu media containing 2% dextrose. Cells were allowed to grow at 30°C for 25 min. 250μCi of L-[Methyl-3H]-Methionine (Perkin-Elmer) was added and cells were allowed to grow two minutes. L-Methionine was then added to a final concentration of 0.6 mM. A 600μl cell aliquot was immediately collected, spun down for 15 sec at 3,000 rpm, supernatant removed, and placed in a dry ice/methanol bath. Time points were collected in the same manner at 1, 2, 4, and 8 minutes after methionine chase. Total RNA was extracted via hot phenol method as previously described. 20,000 cpm from each time point was loaded on a 1.25% agarose gel (containing formaldehyde and MOPS) and electrophoresed for 8 hours at 70V. The RNA was then transferred to a nylon membrane, UV crosslinked, and the membrane allowed to dry. The membrane was then coated four times with ENH3ANCE (Perkin-Elmer), allowing 15 minutes for drying in between each coat and after the final coat, as per manufacturer’s instructions. Autoradiography film was exposed to the membrane for 96 hr at −80°C prior to development.
Confocal fluorescent microscopy analysis of subcellular localization of RPL11Bp and RPS2p
A 10 ml culture of yWO236 containing pWO238 (RPL11B-GFP) or pWO239 (RPS2-GFP) and pWO240 (SIK1-mRFP) and either pWO58, pWO116, or no plasmid were grown in SC-His-Met-Leu or SC-His-Met, respectively, containing 2% dextrose to an OD600 of 0.4. 1 ml of cell suspension was collected and 122 μl of 37% formaldehyde was added to bring the concentration to 4%. Cells were fixed at room temp with end-over-end rocking for 20 minutes. Cells were then washed with 1X PBS three times and resuspended in 30 μl 1X PBS. Cells were mounted onto a cleaned glass slide essentially as described [Citation81]. Briefly, a 9 mm circle was etched into the center of a glass slide. Glass chips were removed and the slide was cleaned in 95% ethanol and allowed to air dry. 30 μl of 1% polyethyleneimine (Sigma-Aldrich) was pipetted onto the etched circle. The slide was allowed to sit for 5 minutes, at which point the polyethyleneimine was removed by aspiration and the remaining solution was allowed to air dry. The 30 μl of cells were then pipetted onto the polyethyleneimine and allowed to settle for 30 minutes. Nonadherent cells were removed by two washes with 1X PBS by covering the slides with the PBS and shaking on an orbital shaker for five minutes. The cells were allowed to dry briefly before 50 μl of mounting media (1X PBS, 50% glycerol, 100ng/ml DAPI, and 2.73mg/ml p-phenylenediamine) was placed onto a glass coverslip and the coverslip placed over the cells. Excess mounting media was removed by blotting with absorbent paper and the coverslip was sealed with clear acrylic nail polish. Cells were viewed for GFP, mRFP, and DAPI fluorescence within 48 hr using a Zeiss LSM-700 confocal microscope. A single plane view comprising 4 μm was captured for each fluorophore. Overlays of sample cells were created using Photoshop image software.
Subcellular nuclear/cytoplasmic localization ratios for RPL11Bp and RPS2p were calculated using Cellprofiler analysis software [Citation59]. Briefly, to assist the software in identifying nuclei, DAPI channels were adjusted for signal threshold by creating a custom pipeline using the auto threshold method. A second custom pipeline was then created wherein signal from the DAPI channel was used to identify nuclei, and signal from the GFP channel was used to identify the cytoplasm (extranuclear space). Mean GFP fluorescent intensity (MFI) was calculated for each compartment and was expressed as a ratio of nuclear MFI/cytoplasmic MFI. Averages and SEM were calculated for each data set and ANOVA with Tukey’s Honestly Significant Differences posthoc test statistics were calculated using the R statistical software package. All Cellprofiler pipelines and the R code for the ANOVA statistics can be found at http://openwetware.org/wiki/OlivasLab.
PUF overexpression qPCR
Relative levels of PUF4 or PUF5 mRNA expression were analyzed by qPCR via the following method. A 20 ml culture of yWO3 containing pWO58, pWO116, or pWO253 was grown to an OD600 of 0.4 in SC-leucine containing 2% dextrose. RNA was extracted via the hot phenol method described previously. Equivalent masses of RNA were DNAse treated with the Turbo DNA-Free Kit (ThermoFisher) per manufacturer’s instructions. Equivalent masses of DNAse-free RNA were reverse transcribed into cDNA using iScript Reverse Transcription Supermix for RT-qPCR (BioRad) per manufacturer’s instructions. Equivalent volumes of equivalent dilutions of resultant cDNA were loaded into 15 µl qPCR reactions containing PowerUP SYBR Green Master Mix (Applied Biosystems) with oWO939 and oWO940 for PUF4 amplification, oWO941 and oWO942 for PUF5 amplification, and oWO632 and oWO652 for TDH1 amplification. PCR cycling and fluorescence analysis took place on the CFX96 Real Time System (BioRad) per manufacturer’s instructions. Relative expression levels were calculated with TDH1 expression as the reference gene using CFX Manager (BioRad).
Supplemental Material
Download Zip (1.1 MB)Acknowledgments
We would like to thank Dr. David Stillman for pWO183 within the set of marker swap vectors [Citation82], Dr. Pamela Silver for the RPL11B-GFP expression vector, Dr. Marvin Wickens for the Puf4FL/CEN plasmid, and Drs. Arlen Johnson and Edward Marcotte for the RPS2-GFP and SIK1-mRFP expression vectors. We would also like to thank members of the Olivas Lab for helpful discussions and reviews of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Woolford JL Jr., Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013 Nov;195(3):643–681. PubMed PMID: 24190922; PubMed Central PMCID: PMC3813855.
- Osheim YN, French SL, Keck KM, et al. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell. 2004 Dec 22;16(6):943–954. PubMed PMID: 15610737.
- Steffen KK, McCormick MA, Pham KM, et al. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics. 2012 May;191(1):107–118. PubMed PMID: 22377630; PubMed Central PMCID: PMC3338253.
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999 Nov;24(11):437–440. PubMed PMID: 10542411.
- Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004 Mar;2(3):E79. PubMed PMID: 15024427; PubMed Central PMCID: PMCPmc368173. Eng.
- Jackson JS Jr., Houshmandi SS, Lopez Leban F, et al. Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast. Rna. 2004 Oct;10(10):1625–1636. PubMed PMID: 15337848; PubMed Central PMCID: PMC1370648.
- Murata Y, Wharton RP. Binding of pumilio to maternal hunchback messenger-Rna is required for posterior patterning in drosophila embryos. Cell. 1995 Mar 10;80(5):747–756. PubMed PMID: WOS:A1995QM39900011; English.
- Nakahata S, Katsu Y, Mita K, et al. Biochemical identification of Xenopus pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J Biol Chem. 2001 Jun 15;276(24):20945–20953. PubMed PMID: WOS:000169297900021; English.
- Souza GM, Da Silva AM, Kuspa A. Starvation promotes dictyostelium development by relieving PufA inhibition of PKA translation through the YakA kinase pathway. Development. 1999 Jul;126(14):3263–3274. PubMed PMID: WOS:000081818800020; English.
- Tadauchi T, Matsumoto K, Herskowitz I, et al. Post-transcriptional regulation through the HO 3 ‘-UTR by Mpt5, a yeast homolog of Pumilio and FBF. Embo J. 2001 Feb 1;20(3):552–561. PubMed PMID: WOS:000166848400024; English.
- Wang X, McLachlan J, Zamore PD, et al. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002 Aug 23;110(4):501–512. PubMed PMID: 12202039.
- Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001 Apr;7(4):855–865. PubMed PMID: 11336708.
- Wreden C, Verrotti AC, Schisa JA, et al. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997 Aug;124(15):3015–3023. PubMed PMID: WOS:A1997XP68800015; English.
- Zamore PD, Bartel DP, Lehmann R, et al. The PUMILIO-RNA interaction: a single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry. 1999 Jan 12;38(2):596–604. PubMed PMID: 9888799.
- Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. Rna-A Publ Rna Soc. 1997 Dec;3(12):1421–1433. PubMed PMID: WOS:000071255700006; English.
- Wilinski D, Qiu C, Lapointe CP, et al. RNA regulatory networks diversified through curvature of the PUF protein scaffold. Nat Commun. 2015;6:8213. PubMed PMID: 26364903; PubMed Central PMCID: PMC4570272.
- Qiu C, McCann KL, Wine RN, et al. A divergent Pumilio repeat protein family for pre-rRNA processing and mRNA localization. Proc Natl Acad Sci USA. 2014 Dec 30;111(52):18554–18559. PubMed PMID: 25512524; PubMed Central PMCID: PMC4284587.
- Zhu D, Stumpf CR, Krahn JM, et al. A 5ʹ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc Natl Acad Sci USA. 2009 Dec 1;106(48):20192–20197. PubMed PMID: 19918084; PubMed Central PMCID: PMC2787145.
- Wang Y, Opperman L, Wickens M, et al. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc Natl Acad Sci USA. 2009 Dec 01;106(48):20186–20191. PubMed PMID: 19901328; PubMed Central PMCID: PMC2787170.
- Miller MT, Higgin JJ, Hall TM. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat Struct Mol Biol. 2008 Apr;15(4):397–402. PubMed PMID: 18327269; PubMed Central PMCID: PMC2802072.
- Koh YY, Wang Y, Qiu C, et al. Stacking interactions in PUF-RNA complexes. Rna. 2011 Apr;17(4):718–727. PubMed PMID: 21372189; PubMed Central PMCID: PMC3062182.
- Edwards TA, Pyle SE, Wharton RP, et al. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001 Apr 20;105(2):281–289. PubMed PMID: 11336677.
- Goldstrohm AC, Hook BA, Seay DJ, et al. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006 Jun;13(6):533–539. PubMed PMID: WOS:000238122500018; English.
- Goldstrohm AC, Seay DJ, Hook BA, et al. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem. 2007 Jan 5;282(1):109–114. PubMed PMID: WOS:000243166500013; English.
- Lee D, Ohn T, Chiang YC, et al. PUF3 acceleration of deadenylation in vivo can operate independently of CCR4 activity, possibly involving effects on the PAB1-mRNP structure. J Mol Biol. 2010 Jun 18;399(4):562–575. PubMed PMID: WOS:000279138700003; English.
- Miller MA, Olivas WM. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip Rev RNA. 2011 Jul-Aug;2(4):471–492. PubMed PMID: 21957038.
- Blewett NH, Goldstrohm AC. A eukaryotic translation initiation factor 4E-binding protein promotes mRNA decapping and is required for PUF repression. Mol Cell Biol. 2012 Oct;32(20):4181–4194. PubMed PMID: 22890846; PubMed Central PMCID: PMC3457345.
- Russo J, Olivas WM. Conditional regulation of Puf1p, Puf4p, and Puf5p activity alters YHB1 mRNA stability for a rapid response to toxic nitric oxide stress in yeast. Mol Biol Cell. 2015 Mar 15;26(6):1015–1029. PubMed PMID: 25631823; PubMed Central PMCID: PMC4357503.
- Ulbricht RJ, Olivas WM. Puf1p acts in combination with other yeast Puf proteins to control mRNA stability. Rna. 2008 Feb;14(2):246–262. PubMed PMID: 18094119; PubMed Central PMCID: PMC2212245.
- Olivas W, Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. Embo J. 2000 Dec 1;19(23):6602–6611. PubMed PMID: 11101532; PubMed Central PMCID: PMC305854.
- Foat BC, Houshmandi SS, Olivas WM, et al. Profiling condition-specific, genome-wide regulation of mRNA stability in yeast. Proc Natl Acad Sci USA. 2005 Dec 6;102(49):17675–17680. PubMed PMID: 16317069; PubMed Central PMCID: PMC1295595.
- Lapointe CP, Preston MA, Wilinski D, et al. Architecture and dynamics of overlapped RNA regulatory networks. Rna. 2017 Aug 02;23:1636–1647. PubMed PMID: 28768715.
- Miller MA, Russo J, Fischer AD, et al. Carbon source-dependent alteration of Puf3p activity mediates rapid changes in the stabilities of mRNAs involved in mitochondrial function. Nucleic Acids Res. 2014 Apr;42(6):3954–3970. PubMed PMID: 24371272; PubMed Central PMCID: PMC3973295.
- Thomson E, Rappsilber J, Tollervey D. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. Rna. 2007 Dec;13(12):2165–2174. PubMed PMID: 17956976; PubMed Central PMCID: PMC2080597.
- Wang B, Ye K. Nop9 binds the central pseudoknot region of 18S rRNA. Nucleic Acids Res. 2017 Apr 07;45(6):3559–3567. PubMed PMID: 28053123; PubMed Central PMCID: PMC5389560.
- Li Z, Lee I, Moradi E, et al. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009 Oct;7(10):e1000213. PubMed PMID: 19806183; PubMed Central PMCID: PMC2749941.
- Yi YH, Ma TH, Lee LW, et al. A genetic cascade of let-7-ncl-1-fib-1 modulates nucleolar size and rRNA pool in caenorhabditis elegans. PLoS Genet. 2015 Oct;11(10):e1005580. PubMed PMID: 26492166; PubMed Central PMCID: PMC4619655.
- Droll D, Archer S, Fenn K, et al. The trypanosome Pumilio-domain protein PUF7 associates with a nuclear cyclophilin and is involved in ribosomal RNA maturation. FEBS Lett. 2010 Mar 19;584(6):1156–1162. PubMed PMID: 20153321; PubMed Central PMCID: PMC2855960.
- Schumann Burkard G, Kaser S, de Araujo PR, et al. Nucleolar proteins regulate stage-specific gene expression and ribosomal RNA maturation in Trypanosoma brucei. Mol Microbiol. 2013 May;88(4):827–840. PubMed PMID: 23617823.
- Abbasi N, Kim HB, Park NI, et al. APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010 Dec;64(6):960–976. PubMed PMID: 21143677.
- Scott RE, White-Grindley E, Ruley HE, et al. P2P-R expression is genetically coregulated with components of the translation machinery and with PUM2, a translational repressor that associates with the P2P-R mRNA. J Cell Physiol. 2005 Jul;204(1):99–105. PubMed PMID: 15617101.
- Opperman L, Hook B, DeFino M, et al. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat Struct Mol Biol. 2005 Nov;12(11):945–951. PubMed PMID: 16244662.
- Koh YY, Opperman L, Stumpf C, et al. A single C. elegans PUF protein binds RNA in multiple modes. Rna. 2009 Jun;15(6):1090–1099. PubMed PMID: 19369425; PubMed Central PMCID: PMC2685523.
- Valley CT, Porter DF, Qiu C, et al. Patterns and plasticity in RNA-protein interactions enable recruitment of multiple proteins through a single site. Proc Natl Acad Sci USA. 2012 Apr 17;109(16):6054–6059. PubMed PMID: 22467831; PubMed Central PMCID: PMC3341033.
- Hook BA, Goldstrohm AC, Seay DJ, et al. Two yeast PUF proteins negatively regulate a single mRNA. J Biol Chem. 2007 May 25;282(21):15430–15438. PubMed PMID: 17389596.
- Munchel SE, Shultzaberger RK, Takizawa N, et al. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol Biol Cell. 2011 Aug 1;22(15):2787–2795. PubMed PMID: 21680716; PubMed Central PMCID: PMC3145553.
- Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 2008 Apr 15;22(8):1037–1050. PubMed PMID: 18413716; PubMed Central PMCID: PMC2335325.
- Kedde M, van Kouwenhove M, Zwart W, et al. A Pumilio-induced RNA structure switch in p27-3ʹ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010 Oct;12(10):1014–1020. PubMed PMID: 20818387.
- Lee CD, Tu BP. Glucose-regulated phosphorylation of the PUF protein Puf3 regulates the translational fate of its bound mRNAs and association with RNA granules. Cell Rep. 2015 Jun 16;11(10):1638–1650. PubMed PMID: 26051939; PubMed Central PMCID: PMC4472502.
- Archer SK, Luu VD, de Queiroz RA, et al. Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle. PLoS Pathog. 2009 Aug;5(8):e1000565. PubMed PMID: 19714224; PubMed Central PMCID: PMC2727004.
- Suh N, Crittenden SL, Goldstrohm A, et al. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics. 2009 Apr;181(4):1249–1260. PubMed PMID: 19221201; PubMed Central PMCID: PMC2666496.
- Huber MD, Dworet JH, Shire K, et al. The budding yeast homolog of the human EBNA1-binding protein 2 (Ebp2p) is an essential nucleolar protein required for pre-rRNA processing. J Biol Chem. 2000 Sep 15;275(37):28764–28773. PubMed PMID: WOS:000089330700058; English.
- Tsuno A, Miyoshi K, Tsujii R, et al. RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 2000 Mar;20(6):2066–2074. PubMed PMID: 10688653; PubMed Central PMCID: PMC110823.
- de la Cruz J, Kressler D, Tollervey D, et al. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3ʹ end formation of 5.8S rRNA in Saccharomyces cerevisiae. Embo J. 1998 Feb 16;17(4):1128–1140. PubMed PMID: 9463390; PubMed Central PMCID: PMC1170461.
- White J, Li Z, Sardana R, et al. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol Cell Biol. 2008 May;28(10):3151–3161. PubMed PMID: 18332120; PubMed Central PMCID: PMC2423152.
- Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002 Jul;27(7):344–351. PubMed PMID: 12114023.
- Lebreton A, Saveanu C, Decourty L, et al. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J Cell Biol. 2006 May 8;173(3):349–360. PubMed PMID: 16651379; PubMed Central PMCID: PMC2063836.
- Miyoshi K, Shirai C, Horigome C, et al. Rrs1p, a ribosomal protein L11-binding protein, is required for nuclear export of the 60S pre-ribosomal subunit in Saccharomyces cerevisiae. FEBS Lett. 2004 May 7;565(1–3):106–110. PubMed PMID: 15135061.
- Kamentsky L, Jones TR, Fraser A, et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011 Apr 15;27(8):1179–1180. PubMed PMID: WOS:000289301600028; English.
- Freeberg MA, Han T, Moresco JJ, et al. Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae. Genome Biol. 2013 Feb 14;14;14(2):R13. PubMed PMID: 23409723; PubMed Central PMCID: PMC4053964.
- Kershaw CJ, Costello JL, Talavera D, et al. Integrated multi-omics analyses reveal the pleiotropic nature of the control of gene expression by Puf3p. Sci Rep. 2015 Oct 23;5:15518. PubMed PMID: 26493364; PubMed Central PMCID: PMC4616039.
- Duttagupta R, Tian B, Wilusz CJ, et al. Global analysis of Pub1p targets reveals a coordinate control of gene expression through modulation of binding and stability. Mol Cell Biol. 2005 Jul;25(13):5499–5513. PubMed PMID: 15964806; PubMed Central PMCID: PMC1156976.
- Yosefzon Y, Koh YY, Chritton JJ, et al. Divergent RNA binding specificity of yeast Puf2p. Rna. 2011 Aug;17(8):1479–1488. PubMed PMID: 21685478; PubMed Central PMCID: PMC3153972.
- Gu W, Deng Y, Zenklusen D, et al. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004 Jun 15;18(12):1452–1465. PubMed PMID: 15198983; PubMed Central PMCID: PMC423195.
- Yang YT, Ting YH, Liang KJ, et al. The roles of Puf6 and Loc1 in 60S biogenesis are interdependent, and both are required for efficient accommodation of Rpl43. J Biol Chem. 2016 Sep 9;291(37):19312–19323. PubMed PMID: 27458021; PubMed Central PMCID: PMC5016672.
- Aviv T, Lin Z, Ben-Ari G, et al. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat Struct Mol Biol. 2006 Feb;13(2):168–176. PubMed PMID: 16429151.
- Rendl LM, Bieman MA, Smibert CA. S. cerevisiae Vts1p induces deadenylation-dependent transcript degradation and interacts with the Ccr4p-Pop2p-Not deadenylase complex. Rna. 2008 Jul;14(7):1328–1336. PubMed PMID: 18469165; PubMed Central PMCID: PMC2441989.
- Pedro-Segura E, Vergara SV, Rodriguez-Navarro S, et al. The Cth2 ARE-binding protein recruits the Dhh1 helicase to promote the decay of succinate dehydrogenase SDH4 mRNA in response to iron deficiency. J Biol Chem. 2008 Oct 17;283(42):28527–28535. PubMed PMID: 18715869; PubMed Central PMCID: PMC2568921.
- Cai Y, Futcher B. Effects of the yeast RNA-binding protein Whi3 on the half-life and abundance of CLN3 mRNA and other targets. PLoS One. 2013;8(12):e84630. PubMed PMID: 24386402; PubMed Central PMCID: PMC3875557.
- Holmes KJ, Klass DM, Guiney EL, et al. Whi3, an S. cerevisiae RNA-binding protein, is a component of stress granules that regulates levels of its target mRNAs. PLoS One. 2013;8(12):e84060. PubMed PMID: 24386330; PubMed Central PMCID: PMC3873981.
- Vasudevan S, Peltz SW. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol Cell. 2001 Jun;7(6):1191–1200. PubMed PMID: 11430822.
- Huber A, French SL, Tekotte H, et al. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. Embo J. 2011 Jul 05;30(15):3052–3064. PubMed PMID: 21730963; PubMed Central PMCID: PMC3160192.
- Urban J, Soulard A, Huber A, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007 Jun 8;26(5):663–674. PubMed PMID: 17560372.
- Lahr RM, Fonseca BD, Ciotti GE, et al. La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. Elife. 2017 Apr 7;6. PubMed PMID: 28379136; PubMed Central PMCID: PMC5419741. DOI:10.7554/eLife.24146
- Hong S, Freeberg MA, Han T, et al. LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. Elife. 2017 Jun 26;6. PubMed PMID: 28650797; PubMed Central PMCID: PMC5484620. DOI:10.7554/eLife.25237
- Codon AC, Gasent-Ramirez JM, Benitez T. Factors Which Affect the Frequency of Sporulation and Tetrad Formation in Saccharomyces cerevisiae Baker’s Yeasts. Appl Environ Microbiol. 1995 Apr;61(4):1677. PubMed PMID: 16535013; PubMed Central PMCID: PMC1388431.
- Gietz RD, Schiestl RH, Willems AR, et al. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995 Apr 15;11(4):355–360. PubMed PMID: 7785336.
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999 Oct;15(14):1541–1553. PubMed PMID: 10514571.
- Caponigro G, Muhlrad D, Parker R. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol Cell Biol. 1993 Sep;13(9):5141–5148. PubMed PMID: 8355674; PubMed Central PMCID: PMC360202.
- Wei Y, Tsang CK, Zheng XF. Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. Embo J. 2009 Aug 5;28(15):2220–2230. PubMed PMID: 19574957; PubMed Central PMCID: PMC2726700.
- Atkin AL. Preparation of yeast cells for confocal microscopy. Methods Mol Biol. 1999;122:131–139. PubMed PMID: 10231788.
- Voth WP, Jiang YW, Stillman DJ. New ‘marker swap’ plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast. 2003 Aug;20(11):985–993. PubMed PMID: 12898713.
