ABSTRACT
Riboswitches are dynamic RNA motifs that are mostly embedded in the 5ʹ-untranslated regions of bacterial mRNAs, where they regulate gene expression transcriptionally or translationally by undergoing conformational changes upon binding of a small metabolite or ion. Due to the small size of typical ligands, relatively little free energy is available from ligand binding to overcome the often high energetic barrier of reshaping RNA structure. Instead, most riboswitches appear to take advantage of the directional and hierarchical folding of RNA by employing the ligand as a structural ‘linchpin’ to adjust the kinetic partitioning between alternate folds. In this model, even small, local structural and kinetic effects of ligand binding can cascade into global RNA conformational changes affecting gene expression. Single-molecule (SM) microscopy tools are uniquely suited to study such kinetically controlled RNA folding since they avoid the ensemble averaging of bulk techniques that loses sight of unsynchronized, transient, and/or multi-state kinetic behavior. This review summarizes how SM methods have begun to unravel riboswitch-mediated gene regulation.
Riboswitches, an ideal target for single molecule microscopy
Bacteria synthesize or import the small organic molecules and inorganic ions needed for their metabolism and survival, whereas they destroy or export toxic substances. However, in a constantly changing environment with many emerging threats and opportunities, feedback mechanisms are often crucial for nimbly adjusting the intracellular concentrations of metabolites and cofactors. Riboswitches are noncoding, cis-acting RNA motifs that undergo structural changes to regulate gene expression upon binding specific cellular metabolites or ions (henceforth referred to as ligands) [Citation1–Citation6]. These RNA elements modulate gene expression by either regulating mRNA synthesis (transcriptional riboswitches) or by adjusting protein expression from an existing mRNA (translational riboswitches) (). To this end, riboswitches are composed of two distinct domains that act synergistically to accomplish gene regulation. The first domain, the aptamer, binds the ligand and defines the class of the riboswitch. Upon ligand binding, the aptamer transmits the on or off signal to the second domain, called the expression platform. Conformational changes in this domain modulate the expression of the downstream gene(s). Compared to the aptamer domain, the expression platform tends to be significantly less conserved [Citation3]. This is consistent with the fact that some classes of riboswitches regulate gene expression via multiple, distinct mechanisms. Transcriptional modulation is the most common regulatory action of riboswitches in Gram-positive bacteria, where ligand binding to the aptamer typically induces the formation of a terminator structure in the expression platform. This leads to premature dislodging of the RNA polymerase (RNAP) during mRNA synthesis and, thus, aborted expression of the downstream gene [Citation7,Citation8] (). The other common mechanism, mostly found in Gram-negative bacteria, regulates the translation initiation process. Here, ligand binding typically leads to the formation of a stem-loop in the expression platform that sequesters the Shine-Dalgarno (SD) sequence [Citation4,Citation9]. Consequently, ribosome binding is hindered, and protein synthesis inhibited ().
Figure 1. Riboswitch-mediated gene regulation.
(A) A simplified overview of gene regulation upon ligand binding to a typical riboswitch. Pausing of RNAP during transcription allows the time for ligand to bind the riboswitch in concentration dependent manner. ‘High’ ligand concentrations lead to downregulation of gene expression, whereas ‘low’ ligand concentrations allow gene expression to go forward. In the case of transcriptional riboswitches, ligand binding induces the formation of a typical terminator stem-loop followed by a poly-uridine stretch that together dislodge the RNAP and thus terminate transcription. In the case of translational riboswitches, ligand binding promotes the formation of a sequester stem that prevents ribosome binding to the SD sequence. RNAP: RNA polymerase; SD: Shine-Dalgarno sequence; AUG: start codon. (B) The kinetic partitioning mechanism of mRNA selection by the ribosome. Cofactors including protein S1, the three initiation factors (IF1-3) and initiator fMet-tRNAfMet assist in loading an mRNA embedding a preQ1-sensing riboswitch onto the 30S ribosomal subunit (left). As the 30S and 70S initiation complexes (IC) form, commitment to translation increases while the vulnerability to mRNA decay decreases, as mediated by the bacterial degradosome, Hfq and Rho (bottom).
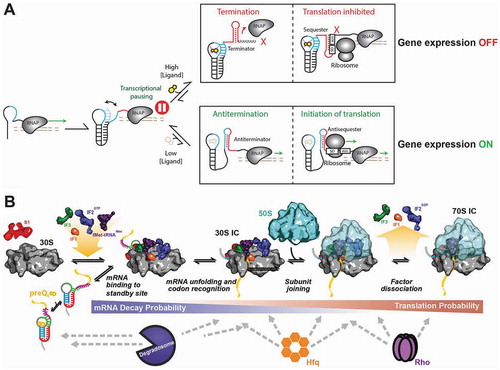
Recent work has shed light on additional variations of these mechanisms (). For example, a ligand-bound translational riboswitch that impairs translation of an mRNA also subjects it more likely to degradation by ribonucleases or transcription termination by the Rho protein [Citation10,Citation11]. Similarly, ligand binding to the Escherichia coli lysC riboswitch has been shown to induce access of the degradosome to the RNA, acting in concert with translational control to prevent expression of the downstream gene [Citation12]. Other examples implicate the ligand in modulating access by the Rho termination factor [Citation13–Citation15], or additional accessory proteins to their respective RNA binding sites [Citation16,Citation17], showing that riboswitches play a central role in the modulation of gene expression and synergistically exploit the available cellular machinery to potentiate their regulatory effectiveness, often through kinetic partitioning between alternate routes ().
Riboswitches typically undergo allosteric structural rearrangements in response to the binding of a metabolite or ion that is small in size compared to the riboswitch and especially the gene expression machinery. This is true even for larger riboswitches such as the T-box systems, where the riboswitch binds an entire tRNA, but the allosteric change originates only from discrimination of the presence or absence of a small aminoacyl modification on the 3ʹ-end of the tRNA [Citation18–Citation21]. Consequently, the amount of binding energy liberated via the net gain of just a few kcal/mol of hydrogen bonds, ion-ion and/or stacking interactions [Citation22–Citation27] may seem insufficient to overcome the potentially large (tens to hundreds of kcal/mol) free energy barriers to interconvert alternative RNA secondary structures [Citation28,Citation29]. Notably, the ligand often binds as a structural ‘linchpin’ to topologically close distal RNA segments and compact the aptamer [Citation30]. In turn, this may empower riboswitches to shift the kinetic partitioning between alternative RNA folds to bring distal sequence elements together on the timescale of transcription, taking advantage of the directional and hierarchical co-transcriptional folding of RNA [Citation28,Citation31–Citation36]. Kinetic partitioning may leverage a branch point in the folding landscape at which a small free energy bias can have a significant effect on the ultimate secondary structure within a reasonable amount of time (i.e., seconds rather than days). In this way, the ligand ‘steers’ the folding before the entire sequence has been transcribed, when the free energy wells are not quite as deep as in the final transcript; this kinetically favored fold then becomes trapped as more of the RNA is transcribed. Such hair-trigger kinetic control through dynamic, low-barrier conversion of alternate riboswitch folds may also be a general and effective way to ‘proofread’ against closely related, near-cognate ligands present in the cell [Citation37,Citation38].
Over the years, traditional bulk assays from biochemistry and molecular biology have been employed to validate novel riboswitches and identify the ligands of orphan riboswitches [Citation8,Citation39–Citation41]. In addition, the field has learned about regulatory mechanisms and differentiated between so far at least 40 classes of riboswitches [Citation1,Citation41,Citation42]; yet, bulk assays often lose important information through ensemble averaging over typically billions of molecules. In particular, it is not possible to characterize kinetic processes that cannot be sufficiently synchronized across a population of molecules, such as equilibrium kinetics, the partitioning between distinct reaction pathways, transient and rare states, and the inherent diversity of molecular behaviors. Notably, a seemingly homogeneous population of biomolecules often displays heterogeneous behavior where the ergodic assumption – that each molecule over time visits the same set of states to the same extent – does not hold. Such behavior is known as either ‘dynamic’ or ‘static’ heterogeneity depending on the observed greater or lesser extent, respectively, of interconversion between states [Citation43–Citation45]. Observing one molecule at a time is the only way to fully assess such heterogeneity.
For dynamic structural motifs such as riboswitches, single-molecule (SM) techniques are often essential for a quantitative description of their folding and unfolding [Citation46]. More specifically, the kinetic differences of riboswitch folding in the absence and presence of ligand relative to the kinetics of the cellular gene expression machinery are key determinants of whether the downstream gene(s) are expressed. Consequently, their ability to uniquely detect and quantify rare molecular events, transiently visited states and diverse kinetic behaviors have led to a quick adoption of SM methods for probing riboswitch mechanisms. In this article, we survey the utility of SM techniques based on fluorescence and force spectroscopy in revealing riboswitch function.
Leading SM tools used to study riboswitches
The two main SM techniques so far employed for studying riboswitches are fluorescence and force based. Due to their access to molecular detail, inherent sensitivity due to probe locations specified by Stokes shift [Citation47] between absorption and emission, and ability to probe specified locations by site-specific fluorophore modification, SM fluorescence techniques have quickly gained momentum for riboswitch studies. In addition, SM fluorescence detection via area detectors (i.e., CCD cameras) allows for super-resolved localization (upto ~1 nm), whereas the characteristic absorption and emission spectra of distinct fluorophores allow for multiplexing [Citation48–Citation52]. Complementarily, if two fluorophores with overlapping emission and absorption spectra come to within 2–8 nm of each other, their transition dipole-dipole interaction results in non-radiative energy transfer from the donor to the acceptor. This phenomenon is known as Förster (or fluorescence) resonance energy transfer (FRET). Since a dipole-dipole interaction has a well-defined inverse-sixth-power dependence on the inter-fluorophore distance, FRET is a very powerful method to measure intra- and intermolecular motions of biomolecules. Strategic placement of the two fluorophores results in real-time observation of specific conformational changes of the host molecule or relative motions between two molecules in a complex. A detailed description of FRET as a technique and its adaptation for different SM experiments is found in previous reviews [Citation46,Citation53–Citation55]. In short, the dually labeled RNA is typically immobilized on a surface and the dynamics between the folded and the unfolded states detected upon observing the evolution of FRET states over time (). A typical FRET time trajectory shows dynamics between alternate conformations. Hidden Markov model (HMM) idealization is often used to analyze these dynamic FRET trajectories and provide rates of folding/binding and unfolding/dissociation (). Additional information can be derived from FRET histograms that bin all FRET values arising from a certain time segment (for example, the initial 10 seconds) of all molecule trajectories observed, or sometimes from a long-lived single trajectory; in addition, different forms of transition density plots help visualize the results of HMM fitting as two-dimensional heat maps summarizing the frequency of FRET transitions observed in a population of single molecules (for more information, we refer the reader to the review by Blanco et al. [Citation56]).
Figure 2. Single molecule methods have enriched the study of riboswitches.
(A) Experimental setup (top) and typical single molecule time traces (bottom) obtained from an smFRET experiment monitoring riboswitch folding dynamics in either the absence or presence of ligand. A streptavidin-coated microscope slide allows the immobilization of single biotinylated riboswitch molecules that are monitored by total internal reflection fluorescence microscopy (TIRFM). Direct excitation of the donor dye leads to a FRET signal from the energy transfer between donor and acceptor. Close proximity between the donor and acceptor dyes leads to high FRET [Citation26]. (B) Experimental setup (top) and typical single molecule time traces (bottom) obtained from a SIM-KARTS experiment that uses TIRFM to monitor binding of an excess of a fluorophore-labeled DNA probe from solution to an immobilized riboswitch in either the absence or presence of ligand. Individual probe binding and dissociation cycles are detectable as fluorescence spikes (black idealized HMM trace) that exhibit periods of burst and non-burst behavior (green and red periods, respectively, on top of the trace, identified by spike train analysis) [Citation57]. (C) Experimental setup (top) and representative force-extension curves and FRET traces (bottom) from a combined optical tweezers/smFRET experiment unfolding a riboswitch in either the absence or presence of ligand. This combined ‘force-FRET’ approach directly correlates local and global structural changes upon folding, including events associated with ligand binding. Adapted with permission from Duesterberg et al. [Citation66].
![Figure 2. Single molecule methods have enriched the study of riboswitches.(A) Experimental setup (top) and typical single molecule time traces (bottom) obtained from an smFRET experiment monitoring riboswitch folding dynamics in either the absence or presence of ligand. A streptavidin-coated microscope slide allows the immobilization of single biotinylated riboswitch molecules that are monitored by total internal reflection fluorescence microscopy (TIRFM). Direct excitation of the donor dye leads to a FRET signal from the energy transfer between donor and acceptor. Close proximity between the donor and acceptor dyes leads to high FRET [Citation26]. (B) Experimental setup (top) and typical single molecule time traces (bottom) obtained from a SIM-KARTS experiment that uses TIRFM to monitor binding of an excess of a fluorophore-labeled DNA probe from solution to an immobilized riboswitch in either the absence or presence of ligand. Individual probe binding and dissociation cycles are detectable as fluorescence spikes (black idealized HMM trace) that exhibit periods of burst and non-burst behavior (green and red periods, respectively, on top of the trace, identified by spike train analysis) [Citation57]. (C) Experimental setup (top) and representative force-extension curves and FRET traces (bottom) from a combined optical tweezers/smFRET experiment unfolding a riboswitch in either the absence or presence of ligand. This combined ‘force-FRET’ approach directly correlates local and global structural changes upon folding, including events associated with ligand binding. Adapted with permission from Duesterberg et al. [Citation66].](/cms/asset/2f7b3a2e-d804-45aa-a0e9-c4ee0d9fb9a8/krnb_a_1536594_f0002_oc.jpg)
Although SM fluorescence techniques are highly valued for their broad applicability, they have to contend with the concern that attachment of fluorescent labels could potentially perturb folding of the RNA. This concern can be addressed by comparing the behaviors of the labeled RNA and a dye-free control. In addition, photobleaching of the fluorescent dyes limits the range of timescales that can be monitored, although it can be extended employing oxygen scavenger systems and through photon budgeting (i.e., intermittent illumination and detection). Alternatively, Single Molecule Kinetic Analysis of RNA Transient Structure (SiM-KARTS) has been developed to investigate RNA structure at the single molecule level without the need for any direct labeling. Instead, the repeated binding of a short, fluorophore labeled, complementary oligonucleotide probe against a region of interest can be used to interrogate the solvent accessibility of a short stretch of RNA (), in concept similar to solution-based RNA footprinting with a chemical or enzymatic reagent, but with the added advantage of the temporal signature of repeat binding providing for high assignment accuracy and information on structural changes over time at the single molecule level [Citation57]. For example, the ligand-dependent accessibility of the SD sequence of an mRNA with an embedded preQ1-sensing translational riboswitch was probed by SiM-KARTS using a short anti-SD oligonucleotide derived from the sequence of the corresponding bacterial 16S ribosomal RNA, thus quantitatively reporting on changes in SD sequence accessibility over time as the RNA folds and unfolds [Citation57] (). Spike train analysis of individual fluorescent probe binding trajectories revealed that single mRNAs interconvert between periods of more and less frequent binding capacity, interpreted as periods of high and low SD accessibility, respectively (). Such measurements provide mechanistic clues as to how riboswitches regulate gene expression through the modulation of inherently bursty translation initiation events.
Complementarily, the most prominent force-based SM techniques employ optical tweezers to manipulate microbeads with a focused infrared laser beam and impose pico-Newton (pN) forces on a biomolecule tethered between two beads or a bead and a surface [Citation58–Citation61] (). SM-force techniques generally perturb the biological system under investigation by an external force along a specified axis and record its mechanical response in real-time. Given their high spatial (0.1 to 10 nm) and temporal (milliseconds to microseconds) resolution, force spectroscopy assays are well suited to study the dynamic folding pathways of riboswitches upon perturbation; they require handles for bead and surface attachment instead of fluorophore labeling of the RNA [Citation59,Citation62]. Potential caveats are that they may impose unnatural, directed mechanical stress on a biomolecule or damage it through use of high laser power. Conversely, by applying a specific force trajectory on an RNA molecule, it is possible to mimic the folding free energy landscape of an RNA as it emerges from biological motors such as RNAP or the ribosome. For example, force-extension curves using constant force have identified discrete states that can be adopted by riboswitches during their directional folding [Citation63,Citation64], and optical tweezers can be combined with an SM fluorescence readout to monitor the local response to a mechanical force [Citation65,Citation66] ().
Unveiling the kinetic mechanisms of riboswitches
The earliest SM studies of riboswitches were devoted to deciphering the kinetic mechanism of ligand-mediated riboswitch folding. One of the initial smFRET studies of riboswitch dynamics by Lemay et al. investigated the folding of an adenine riboswitch as a function of magnesium ion (Mg2+) concentration and detected discrete folding intermediates between undocked and docked conformations [Citation67]. Detection of such intermediate states is critical in order to interpret the stepwise mechanism of riboswitch action [Citation68]. Similarly, studies on various riboswitches subsequently revealed crucial kinetic parameters related to their folding. For example, in a study of a lysine-dependent translational riboswitch, Fiegland et al. employed smFRET to measure the opening and closing rates of its aptamer domain and predicted an apparent dissociation equilibrium constant for lysine binding [Citation69]. The authors further discussed the physiological viability of ligand recognition and hinted that co-transcriptional folding might allow the aptamer domain to quickly sense its environment and achieve equilibrium between bound and unbound populations. The short time windows for ligand sensing imposed by the rapid progress of RNAP imply that a binding equilibrium is rarely established during transcription, but that the response is rather governed by the ligand binding kinetics rather its the equilibrium dissociation constant (KD).
The observation that riboswitches often employ ligands as structural linchpins that are positioned such that they can adjust the kinetic partitioning between possible alternate folds naturally raises the question of ligand binding pathways. In most cases, the binding of ligand and RNA conformational changes go hand-in-hand, and hence there is considerable interest in the field to assess their coupling mechanism. Classically, the two extreme models of molecular recognition are the induced fit (IF) and the conformational selection (CS) models. A comprehensive discussion of these two mechanisms is described by Hammes et al. [Citation70]. Briefly, in the IF mechanism the ligand binds to an unfolded state of the aptamer and promotes conformational changes into the folded state (i.e., binding precedes folding). In contrast, in the CS mechanism the riboswitch samples, in the absence of ligand, both unfolded and folded states. The ligand specifically binds the folded conformation, by Le Chatelier [Citation71] shifting the conformational equilibrium towards the folded state (i.e., folding precedes binding). Suddala et al. employed smFRET to specifically probe which pathway a preQ1 class-I riboswitch follows [Citation72]. The authors demonstrated that the presence of Mg2+ ions shifts the ligand binding mechanism from IF to CS. In other words, Mg2+ promotes folding of the aptamer so that it can sample both folded and unfolded conformations even in the absence of ligand. Thus, kinetic partitioning is increasingly shifted by Mg2+ towards preQ1 binding the pre-folded conformation. In the absence of Mg2+, by contrast, this kinetic path is less taken and riboswitch folding needs to be stabilized first by ligand, instead favoring the IF mechanism. To further validate this conclusion, the authors employed transition state analysis with Φ-values that quantify the fractional abundance of specific folded state contacts in the folding transition state, using ligand ‘mutations’ to preQ0 and guanine as ligands. Due to their loss of functional groups, these ligands have a progressively lower affinity for binding to the aptamer compared to ‘wild-type’ preQ1. In the presence of Mg2+, the transition state energies for all ligands shifted similarly to those of their respective docked state energies, indicating that the majority of ligand-RNA interactions are already established in the transition state as expected for the CS mechanism. By contrast, in the absence of Mg2+ the preQ0 ligand shows a relatively minor change in transition state energy, suggesting that the docking transition states involving the two ligands (preQ1 and preQ0) are nearly identical and specific ligand-RNA contacts are not yet formed in this early transition state, supporting an IF mechanism of folding. These data dissect the strong interdependence of ligand- and Mg2+-mediated folding mechanisms of riboswitches and underscore the detailed information needed to reliably distinguish CS and IF folding mechanisms. Notably, many naturally occurring riboswitches completely enclose their ligand to reach maximal binding specificity, and such occlusion from solvent will require some level of induced fit for full closure. CS and IF therefore should be viewed not as mutually exclusive, but rather as idealizations of opposites on a sliding scale.
Other SM studies have provided additional elucidation of ligand-controlled riboswitch folding pathways. For example, Haller et al. reported that in the larger TPP riboswitch Mg2+ first stabilizes the base of the aptamer [Citation73]. The rest of the aptamer, consisting of two dynamic forearms, creates a flexible cavity that can accommodate the ligand. Thus, the ligand acts as a chaperone to facilitate further folding of the riboswitch by closing the forearms through an IF mechanism, eventually determining the outcome of gene expression. SAM riboswitches follow similar two-step hierarchical folding mechanisms selectively induced by metal ions and ligand binding. However, the folding of the riboswitch is characterized by stacking and rotation of helical segments upon ligand binding through a CS mechanism [Citation74–Citation76]. Even more complex, a cyclic diguanylate (c-di-GMP)-dependent riboswitch exists in four distinct ligand-free sub-populations that differ in dynamics between their undocked and docked conformations. In the presence of c-di-GMP and Mg2+, the docked state becomes stabilized. Furthermore, analysis of mutants demonstrated that tertiary interactions distal to the ligand binding site help pre-organize the RNA for accelerated ligand recognition and binding [Citation77]. Further insight into how secondary structure units impact tertiary structure formation and dynamics was provided by Soulière et al. using a preQ1 class II riboswitch. The authors discovered that the RNA employs a stem-loop insertion into a classical pseudoknots as a tool to fine-tune the ligand response [Citation78]. In a different study, Holmstrom et al. used SM fluorescence assays for a hydroxocobalamin (HyCbl) riboswitch where the ligand itself can, conveniently, act as a quencher of the fluorophore [Citation79]. This allowed the authors to monitor the kinetics of ligand binding directly, independently of the conformational changes of the aptamer. Their data revealed that the undocking rate constant associated with the disruption of a long-range kissing loop interaction is substantially decreased by ligand binding. Continuing in a similar vein, a recent study by Warhaut et al. showed that an adenine binding riboswitch adopts two pre-formed, ligand-free secondary structure isomers termed apoA and apoB [Citation80]. In the absence of ligand, the riboswitch alternates between these two competing conformers. The ligand only binds to the apoA state, leading to a shift in equilibrium towards the ligand-bound, or holo, state. Using smFRET, the authors detected docking dynamics between stable undocked and docked sub-states of the apoA and holo conformations, while complementary NMR techniques revealed an unstable kissing-loop fold in the apoA and holo states [Citation80]. This study further substantiates a model of kinetic partitioning that, via a cascade of structural rearrangements, eventually leads to a global change in gene expression.
Although a majority of single molecule studies of riboswitches were performed on isolated aptamer domains, several recent studies have started to consider both the aptamer and the expression platform as a unit [Citation58,Citation62,Citation81–Citation83]. In fact, oftentimes a 3ʹ segment of a helix is shared by the aptamer and the expression platform, indicating that structural competition for this segment can be a direct link between ligand binding and gene expression. For example, Manz et al. recently studied the dynamics of the SAM-I riboswitch aptamer and its terminator by smFRET [Citation82]. They found that the anti-terminator and terminator structures coexist, and that SAM binding only gradually shifts the equilibrium toward the terminator.
Finally, RNA-ligand interaction analyses, different chemical denaturants, and other perturbation methods have also been employed to understand the folding landscapes of additional riboswitches [Citation84,Citation85], together demonstrating how SM fluorescence imaging can provide powerful insights into the ligand-guided folding mechanisms of riboswitches.
Following the folding of riboswitches co-transcriptionally
Folding of nascent transcripts can be modulated by the molecular properties of the RNAP that carries out their transcription. For example, the site-specific pausing of RNAP together with co-transcriptional RNA-protein interactions have been shown in several cases to be important for co-transcriptional RNA folding [Citation86]. In this context, it has been shown that the transcription rate of the transcriptional flavin mononucleotide (FMN)-sensing riboswitch from Bacillus subtilis has a profound impact on its ability to sense the ligand and regulate gene expression [Citation87]. That is, ligand binding and the kinetics of transcription are intimately linked to allow fine-tuning of riboswitch folding [Citation88]. During the past decade, the emergence of new biochemical and SM techniques has allowed researchers to isolate the transcription elongation complex (EC) and to analyze it in ever greater detail [Citation89].
In one of the first applications of optical tweezers to the riboswitch field, Greenleaf et al. investigated the folding landscape of the pBuE adenine riboswitch and uncovered multiple states in the hierarchical folding of this RNA [Citation59]. Following this study, similar folding pathways were uncovered for other riboswitches such as those for TPP and guanine [Citation58,Citation60,Citation66,Citation90]. These studies helped to identify discrete states adopted by the aptamer in the bound and unbound conformations, showing that riboswitch folding often is not confined to two mutually exclusive conformations. For example, the guanine riboswitch can adopt six different states with three of these dependent on the presence of ligand [Citation54]. Very fast multi-state folding of the aptamer (in under 1 s) allows the ligand to efficiently modulate the conformation of the RNA in a very short time window as necessary for regulating transcription.
One of the main technical challenges in the transcriptional riboswitch field is the ability to follow the folding pathway of the RNA as the RNAP transcribes it. Indeed, during RNA biosynthesis, the nascent RNA can adopt intermediate conformers that determine the final output of gene expression [Citation31,Citation34]. This behavior is exemplified by riboswitches that have to fold, bind their cognate ligand, and regulate gene expression by partitioning from an antiterminator to a terminator stem-loop that dislodges the RNAP during transcription. As a result, dissecting the folding landscape of riboswitches as they are transcribed is needed to fully understand how the binding of the ligand triggers conformational changes that modulate the stability of a complex as stable as the EC.
Frieda and Block developed an SM force microscopy assay to address this question [Citation58]. Specifically, they investigated the hierarchical folding of the pbuE riboswitch as it is transcribed by RNAP. By suspending a short initial transcription complex via the DNA template and the RNAP between dual beams of optical tweezers, they were able to follow RNA co-transcriptional folding in real-time. Using this dumbbell configuration, extension between the two microbeads and spikes in force-extension curves permitted characterization of the folding landscape of the RNA as it emerges from the RNAP. By applying different forces to the EC, the researchers defined typical SM signatures of RNA folding in the absence and presence of the adenine ligand. They found that, at high force, the aptamer does not fold properly even in the presence of adenine. As the applied force is decreased, the folding pathway of the aptamer can be observed with the P2 stem folding first, followed by the P3 stem. Notably, the researchers were able to identify an adenine-dependent termination event in real-time. Based on the short time window for ligand binding (2 s) compared to the lifetime of the aptamer in the bound conformation (10 s), they were able to determine that the riboswitch is kinetically driven, revealing evidence of such a mechanism at the SM level and underscoring the significance of co-transcriptional folding for understanding riboswitch mechanisms.
While experiments with optical tweezers have improved our understanding of global riboswitch folding, a comprehensive understanding further requires a correlation with local structural rearrangements. Uhm et al. pursued such studies by analyzing the co-transcriptional folding of a TPP-sensing riboswitch using smFRET [Citation91]. In this study, while using fluorescently labeled RNA incorporated into an EC of a bacteriophage RNAP, the researchers were able to restart transcription upon addition of NTPs allowing them to survey the real-time folding of the riboswitch as it emerges from the RNAP. By altering the rate of the transcription process with varying NTP concentrations, the researchers found that ligand binding is temporally coordinated, defining a time window during which TPP can bind the aptamer co-transcriptionally. This study demonstrated how the transcription process can be dynamically coordinated with ligand binding.
More recently, Widom et al. uncovered a direct role of RNAP in riboswitch folding by analyzing a paused elongation complex (PEC) formed with a preQ1-responsive riboswitch and bacterial RNAP [Citation83]. The authors found that the unbound state of the riboswitch promotes RNAP pausing within the expression platform, whereas the bound state acts as a ‘release factor’ for this transcriptional pausing. In addition, by studying this particular PEC by smFRET they found that RNAP conversely impacts riboswitch folding via electrostatic shielding from the DNA template to stabilize the docked conformation. In addition to studying a native PEC at the SM level, this work provides new evidence into riboswitch dynamics in which the transcriptional machinery cross-couples with ligand-dependent riboswitch folding.
Future outlook: riboswitch folding under cellular conditions
SM techniques have advanced the study of isolated riboswitches to a new level in terms of identifying intermediate folding states, measuring quantitative kinetics, and determining folding mechanisms. In the complex environment of a living cell, however, a riboswitch-embedding mRNA can potentially interact with many additional RNA-binding proteins during gene expression (), a challenge that is only beginning to be addressed.
As one mechanism that has been largely underappreciated, the detailed process of translation initiation is of particular interest. Indeed, while it is known that ‘simple’ RNA secondary structures already can prevent ribosome binding, much less information is available concerning the converse impact of the large pre-initiation complex (PIC) on riboswitch folding and dynamics (). Moreover, it is important to ask whether a riboswitch can impact only the pioneering round of translation, upon which it becomes unfolded, or whether it can (re)bind ligand and refold to impact subsequent translation initiation events. More studies in the context of the bacterial translation initiation factors and both subunits of the ribosome are required since all of these factors can potentially impact the folding pattern of a given riboswitch ().
Furthermore, in bacteria transcription and translation are intimately coupled, that is, once the SD sequence and start codon have been transcribed, the ribosome can initiate translation and begin generating protein in parallel to further RNA synthesis by RNAP [Citation92]. In fact, it has been shown that the ribosome can physically push into the RNAP so that the two processes become coupled [Citation93]. The recent cryo-electron microscopy (cryo-EM) characterization of an ‘expressome’, containing an mRNA transcribed by RNAP while being translated by the ribosome (), opens the gate to new studies probing the effects of this larger context on riboswitch folding and function [Citation94].
Figure 3. The ‘Goliath’ of the gene expression machinery is overpowered by local structural changes upon binding of a small ligand ‘David’ to a riboswitch.
The ligand, riboswitch and expressome are drawn to scale, with insets using expanded scale bars to show more detail for ligand and riboswitch. A ‘wave of kinetic selection’ exploits the directional and hierarchical folding of RNA upon ligand binding as a structural ‘linchpin’ to allow for discrimination (‘proofreading’) of even subtle differences between near-cognate ligands and efficient regulation of the gene expression machinery. Adapted from pdb-5MY1 by Kohler et al. [94] (for the expressosome) and pdb-3Q50 by Jenkins et al. [25] (for the riboswitch and ligand).
![Figure 3. The ‘Goliath’ of the gene expression machinery is overpowered by local structural changes upon binding of a small ligand ‘David’ to a riboswitch.The ligand, riboswitch and expressome are drawn to scale, with insets using expanded scale bars to show more detail for ligand and riboswitch. A ‘wave of kinetic selection’ exploits the directional and hierarchical folding of RNA upon ligand binding as a structural ‘linchpin’ to allow for discrimination (‘proofreading’) of even subtle differences between near-cognate ligands and efficient regulation of the gene expression machinery. Adapted from pdb-5MY1 by Kohler et al. [94] (for the expressosome) and pdb-3Q50 by Jenkins et al. [25] (for the riboswitch and ligand).](/cms/asset/3ef9ed88-53b0-4775-aafa-1181ac096cc6/krnb_a_1536594_f0003_oc.jpg)
In conclusion, with the constant discovery of new riboswitches in increasingly exotic bacteria, and the assignment of ligands to orphan riboswitches [Citation41], one may expect a parallel rise in the complexity of the mechanisms and model systems amenable to SM techniques. As one example, a recently discovered new class of riboswitches uses a B12-responsive riboswitch as part of a trans-acting RNA that modulates accessibility by an RNA-binding protein [Citation17,Citation95]. Another new dimension currently arising – after extensive studies of the folding and dynamics of isolated riboswitches in vitro – is the realization that RNAs may behave quite differently in the crowded molecular environment of the cell, where transient encounters with a plethora of non-canonical binding partners may dominate RNA-guided gene expression in ways that we are just beginning to understand [Citation96]. This may help the ‘David’ of a small ligand overpower the much larger ‘Goliath’ of the gene expression machinery in ways both unexpected and profound, fine-tuning gene regulation by waves of kinetic selection yet awaiting to be discovered ().
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Alexander Johnson-Buck for a careful reading of the manuscript, Dr. Paul Lund for , and all members of the Walter laboratory and the reviewers for valuable feedback.
Additional information
Funding
References
- McCown PJ, Corbino KA, Stav S, et al. Riboswitch diversity and distribution. RNA. 2017;23(7):995–1011.
- Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15(3):342–348.
- Barrick JE, Breaker RR. The power of riboswitches. Sci Am. 2007;296(1):50–57.
- Sherwood AV, Henkin TM. Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu Rev Microbiol. 2016;70:361–374.
- Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152(1–2):17–24.
- Garst AD, Edwards AL, Batey RT. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol. 2011;3(6). DOI:10.1101/cshperspect.a003533
- Barrick JE, Corbino KA, Winkler WC, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101(17):6421–6426.
- Dar D, Shamir M, Mellin JR, et al. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science (New York, NY). 2016;352(6282):aad9822.
- Nou X, Kadner RJ. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci USA. 2000;97(13):7190–7195.
- Bastet L, Chauvier A, Singh N, et al. Translational control and Rho-dependent transcription termination are intimately linked in riboswitch regulation. Nucleic Acids Res. 2017;45(12):7474–7486.
- Takemoto N, Tanaka Y, Inui M. Rho and RNase play a central role in FMN riboswitch regulation in Corynebacterium glutamicum. Nucleic Acids Res. 2015;43(1):520–529.
- Caron M-P, Bastet L, Lussier A, et al. Dual-acting riboswitch control of translation initiation and mRNA decay. Proc Natl Acad Sci USA. 2012;109(50):E3444–53.
- Chauvier A, Picard-Jean F, Berger-Dancause J-C, et al. Transcriptional pausing at the translation start site operates as a critical checkpoint for riboswitch regulation. Nat Commun. 2017;8:13892.
- Hollands K, Proshkin S, Sklyarova S, et al. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci USA. 2012;109(14):5376–5381.
- Hollands K, Sevostiyanova A, Groisman EA. Unusually long-lived pause required for regulation of a Rho-dependent transcription terminator. Proc Natl Acad Sci USA. 2014;111(19):E1999–2007.
- DebRoy S, Gebbie M, Ramesh A, et al. A riboswitch-containing sRNA controls gene expression by sequestration of a response regulator. Science (New York, NY). 2014;345(6199):937–940.
- Mellin JR, Koutero M, Dar D, et al. Riboswitches. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science (New York, NY). 2014;345(6199):940–943.
- Grundy FJ, Henkin TM. The T box and S box transcription termination control systems. Front Biosci. 2003;8:d20–31.
- Suddala KC, Cabello-Villegas J, Michnicka M, et al. Hierarchical mechanism of amino acid sensing by the T-box riboswitch. Nat Commun. 2018;9(1):1896.
- Zhang J, Ferre-D’Amare AR. Structure and mechanism of the T-box riboswitches. Wiley Interdiscip Rev RNA. 2015;6(4):419–433.
- Sherwood AV, Frandsen JK, Grundy FJ, et al. New tRNA contacts facilitate ligand binding in a Mycobacterium smegmatis T box riboswitch. Proc Natl Acad Sci USA. 2018;115(15):3894–3899.
- Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455(7217):1263–1267.
- Stoddard CD, Montange RK, Hennelly SP, et al. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure (London, England: 1993). 2010;18(7):787–797.
- Huang L, Serganov A, Patel DJ. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell. 2010;40(5):774–786.
- Jenkins JL, Krucinska J, McCarty RM, et al. Comparison of a preQ1 riboswitch aptamer in metabolite-bound and free states with implications for gene regulation. J Biol Chem. 2011;286(28):24626–24637.
- Suddala KC, Rinaldi AJ, Feng J, et al. Single transcriptional and translational preQ1 riboswitches adopt similar pre-folded ensembles that follow distinct folding pathways into the same ligand-bound structure. Nucleic Acids Res. 2013;41(22):10462–10475.
- Liberman JA, Suddala KC, Aytenfisu A, et al. Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics. Proc Natl Acad Sci USA. 2015;112(27):E3485–94.
- Aboul-Ela F, Huang W, Abd Elrahman M, et al. Linking aptamer-ligand binding and expression platform folding in riboswitches: prospects for mechanistic modeling and design. Wiley Interdiscip Rev RNA. 2015;6(6):631–650.
- Fallmann J, Will S, Engelhardt J, et al. Recent advances in RNA folding. J Biotechnol. 2017;261:97–104.
- Jones CP, Ferre-D’Amare AR. Long-range interactions in riboswitch control of gene expression. Annu Rev Biophys. 2017;46:455–481.
- Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–175.
- Zhao B, Guffy SL, Williams B, et al. An excited state underlies gene regulation of a transcriptional riboswitch. Nat Chem Biol. 2017;13(9):968–974.
- Helmling C, Klötzner D-P, Sochor F, et al. Life times of metastable states guide regulatory signaling in transcriptional riboswitches. Nat Commun. 2018;9(1):944.
- Lai D, Proctor JR, Meyer IM. On the importance of cotranscriptional RNA structure formation. RNA. 2013;19(11):1461–1473.
- Gong S, Wang Y, Wang Z, et al. Co-transcriptional folding and regulation mechanisms of riboswitches. Molecules. 2017;22(7). DOI:10.3390/molecules22071169
- Feng J, Walter NG, Brooks CL 3rd. Cooperative and directional folding of the preQ1 riboswitch aptamer domain. J Am Chem Soc. 2011;133(12):4196–4199.
- Wacker A, Buck J, Richter C, et al. Mechanisms for differentiation between cognate and near-cognate ligands by purine riboswitches. RNA Biol. 2012;9(5):672–680.
- Zhang J, Ferre-D’Amare AR. Direct evaluation of tRNA aminoacylation status by the T-box riboswitch using tRNA-mRNA stacking and steric readout. Mol Cell. 2014;55(1):148–155.
- Tapsin S, Sun M, Shen Y, et al. Genome-wide identification of natural RNA aptamers in prokaryotes and eukaryotes. Nat Commun. 2018;9(1):1289.
- Nelson JW, Atilho RM, Sherlock ME, et al. Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol Cell. 2017;65(2):220–230.
- Greenlee EB, Stav S, Atilho RM, et al. Challenges of ligand identification for the second wave of orphan riboswitch candidates. RNA Biol. 2018;15(3):377–390.
- Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67.
- Zhuang X, Kim H, Pereira MJ, et al. Correlating structural dynamics and function in single ribozyme molecules. Science (New York, NY). 2002;296(5572):1473–1476.
- Solomatin SV, Greenfeld M, Herschlag D. Implications of molecular heterogeneity for the cooperativity of biological macromolecules. Nat Struct Mol Biol. 2011;18(6):732–734.
- Solomatin SV, Greenfeld M, Chu S, et al. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463(7281):681–684.
- Suddala KC, Walter NG. Riboswitch structure and dynamics by smFRET microscopy. Methods Enzymol. 2014;549:343–373.
- Lakowicz JR. Principles of fluorescence spectroscopy. New York, NY: Springer; 2006.
- Larson JD, Rodgers ML, Hoskins AA. Visualizing cellular machines with colocalization single molecule microscopy. Chem Soc Rev. 2014;43(4):1189–1200.
- Perez-Gonzalez C, Grondin JP, Lafontaine DA, et al. Biophysical approaches to bacterial gene regulation by riboswitches. Adv Exp Med Biol. 2016;915:157–191.
- St-Pierre P, McCluskey K, Shaw E, et al. Fluorescence tools to investigate riboswitch structural dynamics. Biochim Biophys Acta. 2014;1839(10):1005–1019.
- Walter NG, Huang CY, Manzo AJ, et al. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat Methods. 2008;5(6):475–489.
- Yildiz A, Selvin PR. Fluorescence imaging with one nanometer accuracy: application to molecular motors. Acc Chem Res. 2005;38(7):574–582.
- Joo C, Balci H, Ishitsuka Y, et al. Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem. 2008;77:51–76.
- Ray S, Widom JR, Walter NG. Life under the microscope: single-molecule fluorescence highlights the RNA world. Chem Rev. 2018;118(8):4120–4155.
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5(6):507–516.
- Blanco M, Walter NG. Analysis of complex single-molecule FRET time trajectories. Methods Enzymol. 2010;472:153–178.
- Rinaldi AJ, Lund PE, Blanco MR, et al. The Shine-Dalgarno sequence of riboswitch-regulated single mRNAs shows ligand-dependent accessibility bursts. Nat Commun. 2016;7:8976.
- Frieda KL, Block SM. Direct observation of cotranscriptional folding in an adenine riboswitch. Science (New York, NY). 2012;338(6105):397–400.
- Greenleaf WJ, Frieda KL, Foster DAN, et al. Direct observation of hierarchical folding in single riboswitch aptamers. Science (New York, NY). 2008;319(5863):630–633.
- Chandra V, Hannan Z, Xu H, et al. Single-molecule analysis reveals multi-state folding of a guanine riboswitch. Nat Chem Biol. 2017;13(2):194–201.
- Ritchie DB, Woodside MT. Probing the structural dynamics of proteins and nucleic acids with optical tweezers. Curr Opin Struct Biol. 2015;34:43–51.
- Neupane K, Yu H, Foster DA, et al. Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 2011;39(17):7677–7687.
- Savinov A, Block SM. Folding and catalysis of the glms ribozyme riboswitch studied at the single-molecule level. Biophys J. 2017;112(3):368a.
- Savinov A, Perez CF, Block SM. Single-molecule studies of riboswitch folding. Biochim Biophys Acta. 2014;1839(10):1030–1045.
- Chemla YR. High-resolution, hybrid optical trapping methods, and their application to nucleic acid processing proteins. Biopolymers. 2016;105(10):704–714.
- Duesterberg VK, Fischer-Hwang IT, Perez CF, et al. Observation of long-range tertiary interactions during ligand binding by the TPP riboswitch aptamer. eLife. 2015;4. DOI:10.7554/eLife.12362.
- Lemay JF, Penedo JC, Tremblay R, et al. Folding of the adenine riboswitch. Chem Biol. 2006;13(8):857–868.
- Lemay JF, Penedo JC, Mulhbacher J, et al. Molecular basis of RNA-mediated gene regulation on the adenine riboswitch by single-molecule approaches. Methods Mol Biol. 2009;540:65–76.
- Fiegland LR, Garst AD, Batey RT, et al. Single-molecule studies of the lysine riboswitch reveal effector-dependent conformational dynamics of the aptamer domain. Biochemistry. 2012;51(45):9223–9233.
- Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: a flux description of reaction mechanism. Proc Natl Acad Sci USA. 2009;106(33):13737–13741.
- Le Chatelier H, Boudouard O. Limits of flammability of gaseous mixtures. Bulletin de la Société Chimique de France (Paris). 1898;19:483–488.
- Suddala KC, Wang J, Hou Q, et al. Mg(2+) shifts ligand-mediated folding of a riboswitch from induced-fit to conformational selection. J Amer Chem Soc. 2015;137(44):14075–14083.
- Haller A, Altman RB, Souliere MF, et al. Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proc Natl Acad Sci USA. 2013;110(11):4188–4193.
- Heppell B, Blouin S, Dussault AM, et al. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol. 2011;7(6):384–392.
- Haller A, Rieder U, Aigner M, et al. Conformational capture of the SAM-II riboswitch. Nat Chem Biol. 2011;7(6):393–400.
- Boudreault J, Perez-Gonzalez DC, Penedo JC, et al. Single-molecule approaches for the characterization of riboswitch folding mechanisms. Methods Mol Biol. 2015;1334:101–107.
- Wood S, Ferre-D’Amare AR, Rueda D. Allosteric tertiary interactions preorganize the c-di-GMP riboswitch and accelerate ligand binding. ACS Chem Biol. 2012;7(5):920–927.
- Souliere MF, Altman RB, Schwarz V, et al. Tuning a riboswitch response through structural extension of a pseudoknot. Proc Natl Acad Sci USA. 2013;110(35):E3256–64.
- Holmstrom ED, Polaski JT, Batey RT, et al. Single-molecule conformational dynamics of a biologically functional hydroxocobalamin riboswitch. J Am Chem Soc. 2014;136(48):16832–16843.
- Warhaut S, Mertinkus KR, Hollthaler P, et al. Ligand-modulated folding of the full-length adenine riboswitch probed by NMR and single-molecule FRET spectroscopy. Nucleic Acids Res. 2017;45(9):5512–5522.
- Manz AS, Paeng K, Kaufman LJ. Single molecule studies reveal temperature independence of lifetime of dynamic heterogeneity in polystyrene. J Chem Phys. 2018;148(20):204508.
- Manz C, Kobitski AY, Samanta A, et al. Single-molecule FRET reveals the energy landscape of the full-length SAM-I riboswitch. Nat Chem Biol. 2017;13(11):1172–1178.
- Widom JR, Nedialkov YA, Rai V, et al. Ligand modulates cross-coupling between riboswitch folding and transcriptional pausing. Mol Cell. 2018;in press.
- Shaw E, St-Pierre P, McCluskey K, et al. Using sm-FRET and denaturants to reveal folding landscapes. Methods Enzymol. 2014;549:313–341.
- Baird NJ, Inglese J, Ferre-D’Amare AR. Rapid RNA-ligand interaction analysis through high-information content conformational and stability landscapes. Nat Commun. 2015;6:8898.
- Zhang J, Landick R. A two-way street: regulatory interplay between RNA polymerase and nascent RNA structure. Trends Biochem Sci. 2016;41(4):293–310.
- Wickiser JK, Winkler WC, Breaker RR, et al. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18(1):49–60.
- Lemay JF, Desnoyers G, Blouin S, et al. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7(1):e1001278.
- Dangkulwanich M, Ishibashi T, Bintu L, et al. Molecular mechanisms of transcription through single-molecule experiments. Chem Rev. 2014;114(6):3203–3223.
- Anthony PC, Perez CF, García-García C, et al. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc Natl Acad Sci USA. 2012;109(5):1485–1489.
- Uhm H, Kang W, Ha KS, et al. Single-molecule FRET studies on the cotranscriptional folding of a thiamine pyrophosphate riboswitch. Proc Natl Acad Sci USA. 2017;115(2):331–336.
- Castro-Roa D, Zenkin N. In vitro experimental system for analysis of transcription-translation coupling. Nucleic Acids Res. 2012;40(6):e45.
- Proshkin S, Rahmouni AR, Mironov A, et al. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science (New York, NY). 2010;328(5977):504–508.
- Kohler R, Mooney RA, Mills DJ, et al. Architecture of a transcribing-translating expressome. Science (New York, NY). 2017;356(6334):194–197.
- Moore SJ, Mayer MJ, Biedendieck R, et al. Towards a cell factory for vitamin B12 production in Bacillus megaterium: bypassing of the cobalamin riboswitch control elements. N Biotechnol. 2014;31(6):553–561.
- Daher M, Widom JR, Tay W, et al. Soft interactions with model crowders and non-canonical interactions with cellular proteins stabilize RNA folding. J Mol Biol. 2018;430(4):509–523.
