ABSTRACT
Type III CRISPR-Cas systems code for a multi-subunit ribonucleoprotein (RNP) complex that mediates DNA cleavage and synthesizes cyclic oligoadenylate (cOA) second messenger to confer anti-viral immunity. Both immune activities are to be activated upon binding to target RNA transcripts by their complementarity to crRNA, and autoimmunity avoidance is determined by extended complementarity between the 5ʹ-repeat tag of crRNA and 3ʹ-flanking sequences of target transcripts (anti-tag). However, as to how the strategy could achieve stringent autoimmunity avoidance remained elusive. In this study, we systematically investigated how the complementarity of the crRNA 5ʹ-tag and anti-tag (i.e., tag complementarity) could affect the interference activities (DNA cleavage activity and cOA synthesis activity) of Cmr-α, a type III-B system in Sulfolobus islandicus Rey15A. The results revealed an increasing suppression on both activities by increasing degrees of tag complementarity and a critical function of the 7th nucleotide of crRNA in avoiding autoimmunity. More importantly, mutagenesis of Cmr3α exerts either positive or negative effects on the cOA synthesis activity depending on the degrees of tag complementarity, suggesting that the subunit, coupling with the interaction between crRNA tag and anti-tag, function in facilitating immunity and avoiding autoimmunity in Type III-B systems.
Introduction
CRISPR (clustered regularly interspaced short palindromic repeats) loci and cas (CRISPR-associated) genes protect bacteria and archaea from invasion of viruses and plasmids [Citation1,Citation2]. The system is able to obtain a short genetic fragment (protospacer) from invading nucleic acid and store the information in CRISPR loci [Citation3]. Then, transcription of CRISPR loci and processing of the resulting transcript generate CRISPR RNA (crRNA) that contains the genetic information of past invasion [Citation4,Citation5]. Upon subsequent invasion, crRNA guides Cas proteins to recognize invading nucleic acid by complementarity between crRNA and protospacer, and destroy the targeted invading nucleic acid [Citation2,Citation4]. Depending on the cas gene content and mechanisms of invading nucleic acid destruction, the CRISPR-Cas systems are classified into six different types [Citation6,Citation7]. Among them, type I, II and V systems target dsDNA [Citation7], although type V systems can also be activated for indiscriminate ssDNA cleavage after cleavage of the dsDNA target [Citation8–Citation10]; type VI system is activated by binding to target RNA for indiscriminate ssRNA cleavage [Citation11]. Type III system exhibits the most complicated activities, including target RNA cleavage [Citation12], target RNA-activated DNA degradation [Citation13–Citation16] and cOA synthesis [Citation17–Citation20], the first of which is carried out by the backbone subunit [Citation21–Citation25], while the active sites of the latter two reside in the largest subunit, Cas10. Further, target RNA cleavage could deactivate the latter two activities, providing a temporal control mechanism to avoid autoimmunity [Citation17–Citation19,Citation26].
Autoimmunity avoidance also requires an effective self vs non-self discrimination strategy. For the dsDNA targeting CRISPR-Cas systems, including type I, II and V, the strategy is to recognize protospacer adjacent motif (PAM), a short motif immediately flanking protospacer in invading DNAs but absent from the CRISPR loci in host genomes [Citation27–Citation30]. By contrast, type III systems employ a different self vs non-self discrimination mechanism; that is, sequence complementarity between the 5ʹ-repeat tag of crRNA and the corresponding 3ʹ-protospacer flanking sequence (i.e. tag complementarity) prevents self-immunity [Citation31]. It has been further shown that self vs non-self discrimination occurs at the RNA level: mismatches between the 5ʹ-tag of crRNA and the anti-tag region of the target RNA activates DNA cleavage and cOA generation by different type III RNPs whereas their complementarity suppresses both activities [Citation13–Citation18]. Further, not full complementarity to the 8-nt tag is required for self-protection. In vivo studies have shown that the minimal complementarity required for protection is base-pairing at the 5th, 6th and 7th nt for the type III-A system in Staphylococcus epidermidis [Citation31], while base-pairing at the 6th and 7th nt is enough for protection in the type III-B system in Pyrococcus furiosus [Citation13]. In agreement with the in vivo studies, structural analysis indicates that only four of the eight nucleotides in the crRNA tag, i.e. from 4th to 7th nt, are accessible for Watson-Crick pairing and should play a role in self vs non-self discrimination [Citation15,Citation32]. The structure prediction is then supported by in vitro studies, showing that base-pairing at the four positions suppresses both DNA cleavage and cOA synthesis activities in a type III-A system [Citation15,Citation18]. Meanwhile, it was inferred that tag complementarity would change the route of 3ʹ-flanking sequence of target RNA and thereby control the activity of Cas10 subunit [Citation15,Citation32,Citation33]. Most recently, structural studies confirmed such a mechanism [Citation34,Citation35], showing that the non-complementary flanking sequence is positioned in the cleft of Cas10 and induces conformational changes of the subunit, while the complementary flanking sequence forms 4 base pairs with crRNA tag and does not interact with Cas10 directly, such that tag complementarity could determine the interaction between 3ʹ-flanking sequence and Cas10 and thus the activation of the latter. Nevertheless, the specific mechanism how a 4-nt motif achieves such a stringent switch is not fully understood, especially for extremely thermophilic organisms, of which the physiological temperature is much higher than the melting temperature of any 4 bp RNA duplex [Citation36].
In this study, we systematically analyzed the effects of varying degrees of tag complementarity on the DNA cleavage and cOA synthesis activities of a type III-B Cmr-α effector from the model archaeon Sulfolobus islandicus Rey15A. We found that each of the four available nucleotides (4th ~ 7th) of crRNA contributes to self vs non-self discrimination, while the 7th nt is more critical than the other three nucleotides. We further demonstrate that Cmr3α functions in shaping Cmr-α activities upon varying degrees of tag complementarity, hinting a mechanism to ensure both high immune activity and effective self-protection.
Results
Parallel inhibition of cOA synthesis and DNA cleavage activities by tag complementarity in a type III-B CRISPR-Cas system
It has been shown that base-pairing of target RNA to the 4 ~ 7th nt of crRNA tag provides self-protection in several different type III systems [Citation13,Citation15,Citation18,Citation31]. To evaluate the specific functions of each nucleotide in the quadruple motif of crRNA tag for the type III-B Cmr-α system of S. islandicus Rey15A, we analyzed the Cmr-α activation ability of all possible target RNA variations that are either complementary or noncomplementary to each nucleotide at the 4-nt region. In addition, although base-pairing at the 8th position of crRNA tag has been shown not to play a role in self-protection [Citation15,Citation18], target RNA variants complementary to this nucleotide are also included in the experiments. To minimize the number of target RNA variants, the noncomplementary nucleotides are set as ‘A’ as the SS1-46 RNA, which we have used previously [Citation16]. Thus, the experiments could only analyze how tag complementarity affect Cmr-α activity but would not reveal possible effects of various sequences of target RNA 3ʹ-flanking region. In total, we designed 23 target RNAs (T1~ T23, ), which, as well as the cognate target RNA (CT) and anti-tag target RNA (ATT, target RNA fully complementary to SS1 crRNA), were synthesized by in vitro transcription assay and purified by denaturing PAGE as described in the Methods and Materials section. Then, the target RNAs were assayed for their ability to activate the cOA synthesis activity and DNA cleavage activity of Cmr-α-RNP (Supplementary figure S1 and S2), and the resulting incorporated ATP and three main DNA cleavage products were quantified, respectively. The values derived from CT RNA were set as ‘1’, with which relative activities of all other target RNAs were calculated.
Figure 1. Relative DNA cleavage activity (white bar) and cOA synthesis activity (grey bar) of Cmr-α RNP in the presence of target RNAs carrying different 3ʹ-flanking sequence. The DNA cleavage activity and cOA synthesis activity were measured as described in the Method and Materials section and the relative activities derived from each target RNA were calculated with the data from CT set as ‘1’. The nucleotides matching the 4th ~ 8th positions from each target RNA are marked in red. Error bar represents SD of three independent experiments
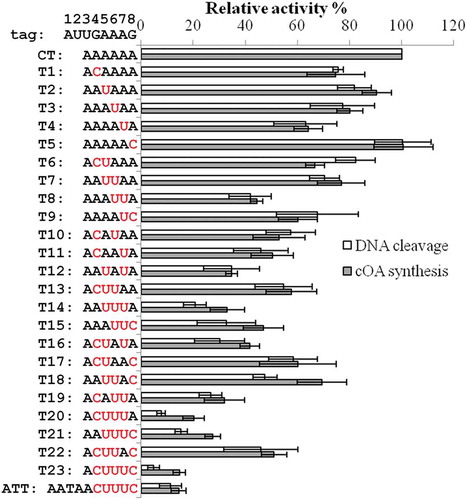
As shown in , varying degrees of tag complementarity suppressed the DNA cleavage activity and cOA synthesis activity in the same pattern, indicating that the two activities are controlled by the same switch. For target RNAs (T1 ~ T5) that only contains one match in the anti-tag region, base-pairing at the 7th position showed the most pronounced influence on both activities (ca. 40% reduction) whereas base-pairing to the 4th, 5th or 6th position reduced the activities by about 20%, and in contrast, base-pairing at the 8th position did not show any detectable influence (). These results indicated that the 7th nucleotide plays a critical role in mediating autoimmunity avoidance, in agreement with previous reports [Citation13,Citation31].
Analysis of target RNAs with double- or triple-matched nucleotides in the tag region revealed () that those contained a paired nucleotide at the 7th position (T8, T11, T12, T14, T16 and T19) showed lower ability to activate Cmr-α than those carrying a mismatch at the position (T6, T7, T10 and T13), further demonstrating the central role of the 7th nucleotide. Combination of base-pairing at the 7th position with one at the 4th, 5th or 6th position further reduced the activation of Cmr-α (comparison of T8, T11, T12 and T4), while introduction of an additional base-pairing at the 8th position did not yield any influence (comparison of T9 and T4), consistent with the results that the 8th nucleotide is not involved in self-protection (comparison of T5 and CT). In addition, T12 and T19 that contain non-consecutive matches showed similar activation ability as T8 and T14 that carry consecutive matches, indicating that non-consecutive complementarity also provides effective self-protection. Moreover, T20 and ATT (full tag complementarity) exhibited the lowest activation ability, indicating that full self-protection requires base-pairing to all the four bases of the quadruple motif as revealed from previous investigations of other type III systems [Citation15,Citation18,Citation34].
Cmr3α shapes the suppression of cOA synthesis in a type III-B CRISPR-Cas system
In the structure of a hybrid CmrΔ1 complex, the 5ʹ-tag of crRNA is mainly grasped by Cmr3 [Citation32]. Thus, we asked whether Cmr3α could modulate the interaction between crRNA tag and target RNA anti-tag region, and contributes to self vs non-self discrimination in S. islandicus Cmr-α system. As a member of the RAMP protein superfamily, Cmr3 shares basic structure with Cmr4, both containing two loops and a β-hairpin thumb towards to the duplex of crRNA and target RNA in Cmr-RNP::target RNA complex [Citation32,Citation33] (Supplementary figure 3). In Cmr4, the thumb separates the duplex of crRNA and target RNA for every 6-nt and employs a conserved tryptophan residue to place the cleavage site close to the active site, which resides in loop 1 (L1) [Citation32]. Similarly, the Cmr3 thumb also contains conserved residues with large side chain (), the function of which remains unknown. Further, the Cmr3 loop 1 is close to the 4 ~ 7th nucleotides of crRNA tag, and sequence alignment shows that it contains a conserved glycine residue ().
Figure 2. Sequence alignment of Cmr3α homologs using ESPript 3.x (http://espript.ibcp.fr) [Citation50]. Secondary structure is shown above the alignment, based on the structure of Pyrococcus furiosus Cmr3 (PDB: 3X1L, Chain B). Sequences for the alignment are from Acidilobus saccharovorans 345–15 (ASAC_0008), Sulfolobus islandicus L.S.2.15 (LS215_0820), Pyrococcus furiosus DSM 3638 (PF1128), Sulfolobus islandicus LAL14/1 (SiL_0600), Sulfolobus islandicus REY15A (SiRe_0597, SiRe_0895) and Sulfolobus islandicus Y.G.57.14 (YG5714_0697). Shown are sequences containing loop 1 and thumb regions, which are highlighted in red box. The residues marked with black arrows are chosen for substitution mutation (M1: I123A-Y124A; M3: G24F; M4: G25F; M7: F13A-K14A-W15A), while those marked with white arrows are chosen for deletion mutation (M2: deletion of I21-L22-L23; M5: deletion of Y18-N19-S20; M6: deletion from Y18 to L23)
![Figure 2. Sequence alignment of Cmr3α homologs using ESPript 3.x (http://espript.ibcp.fr) [Citation50]. Secondary structure is shown above the alignment, based on the structure of Pyrococcus furiosus Cmr3 (PDB: 3X1L, Chain B). Sequences for the alignment are from Acidilobus saccharovorans 345–15 (ASAC_0008), Sulfolobus islandicus L.S.2.15 (LS215_0820), Pyrococcus furiosus DSM 3638 (PF1128), Sulfolobus islandicus LAL14/1 (SiL_0600), Sulfolobus islandicus REY15A (SiRe_0597, SiRe_0895) and Sulfolobus islandicus Y.G.57.14 (YG5714_0697). Shown are sequences containing loop 1 and thumb regions, which are highlighted in red box. The residues marked with black arrows are chosen for substitution mutation (M1: I123A-Y124A; M3: G24F; M4: G25F; M7: F13A-K14A-W15A), while those marked with white arrows are chosen for deletion mutation (M2: deletion of I21-L22-L23; M5: deletion of Y18-N19-S20; M6: deletion from Y18 to L23)](/cms/asset/3aa7ea9c-28a1-4968-9956-c4ef7a8a35f4/krnb_a_1642725_f0002_oc.jpg)
To investigate whether these moieties of Cmr3α function in regulating the Cmr-α activities in response to varying levels of tag complementarity, we designed seven Cmr3α mutants by amino acid substitution at the thumb or the root region of L1 and amino acid deletion at the middle region of L1 (), M1: alanine substitution of Ile123 and Tyr124 in thumb; M2: deletion of Ile21, Leu22 and Leu23; M3: phenylalanine substitution of Gly24; M4: phenylalanine substitution of Gly25; M5: deletion of Tyr18, Asn19 and Ser20; M6: deletion from Tyr18 to Leu23; M7: alanine substitution of Phe13, Lys14 and Trp15. The plasmids expressing the mutated Cmr3α proteins with a 6xHis tag were constructed and the resulting Cmr3α mutant proteins were employed to pull down the corresponding mutated Cmr-α effector complexes as previously described for the His-tagged Cmr6 copurification [Citation16]. SDS-PAGE analysis shows that the mutated Cmr-α complex was obtained for Cmr3α-M1, M2, M3 and M5 but not for M4, M6 and M7 (Supplementary figure 4), indicating that the M4, M6 and M7 mutation could have interfered protein folding of Cmr3α or assembly of the Cmr-α complex. Further, the backbone RNA cleavage assay indicates that the obtained mutated Cmr-α complexes were all active (Supplementary figure 4). Then, the four Cmr3α-mutated and WT Cmr-α complexes (WT, M1, M2, M3 and M5) were tested for their cOA synthesis activity in the presence of each target RNA and the incorporated ATP was quantified. The values obtained from CT were set as ‘1’ with which relative activity was calculated for all other target RNAs (Supplementary figure S5 and S6). The resulting data were further analyzed with t-test to evaluate whether Cmr3α mutation exerts significant effect on the relative cOA synthesis activity for each target RNA (Supplementary table 5), and the data with significant variations (p < 0.05) are shown in .
Figure 3. Comparison of the relative cOA synthesis activity of wild type (WT, white bar) and Cmr3α mutants (grey bar) in the presence of different target RNAs. A: WT and M2; B: WT and M1; C: WT and M5. Only the results with significant differences are shown. Error bar represents SD of three independent experiments
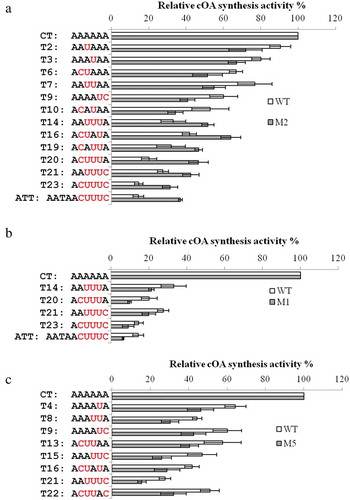
The results show that the M3 mutation did not affect the relative cOA synthesis activity for any target RNA (Supplementary table 5, Supplementary figure S6), while the M1, M2 and M5 mutations at least resulted in alteration of the relative activity for a subgroup of target RNAs (Supplementary table 5, ). Specifically, M2 (deletion of Ile21, Leu22 and Leu23) showed a lower activity than WT in the presence of T2, T3, T6, T7, T9 and T10, but a considerably higher activity in the presence of T14, T16, T19, T20, T21, T23 and AAT (about doubled activity for T20, T23 and AAT). Strikingly, the former group of target RNAs only matches crRNA tag at one or two nucleotides and exerts minor inhibition on Cmr-α activities, while the latter group matches crRNA tag at three or four nucleotides of the quadruple motif, including the 7th nucleotide, and provides efficient self-protection. Together, the data indicate that the three residues within L1 could both facilitate Cmr-α activity upon low level of tag complementarity and suppress Cmr-α activity upon high level of tag complementarity. In contrast to M2, M1 (I123A-Y124A) showed about 30%~60% lower cOA synthesis activity than WT in the presence of the target RNAs that provide efficient self-protection, including T14, T20, T21, T23 and AAT. The data suggest that Ile123 and Tyr124 within thumb function against tag complementarity-dependent self-protection. Similar to M1, M5 mutation resulted in lower cOA synthesis activity in the presence of T4, T8, T9, T13, T15, T16, T21 and T22. These RNAs complement crRNA tag at one or two positions including the 7th nucleotide or three positions either including or not including the 7th nucleotide, representing a group that moderately or efficiently suppresses Cmr-α activity. Taken together, the data demonstrate that the L1 and thumb regions of Cmr3α play an important role in regulating the Cmr-α activity upon varying degrees of crRNA tag complementarity, shedding novel insights into the self vs non-self discrimination mechanism of type III systems.
Discussion
In this study, we show that mutation of Cmr3α loop1 and thumb moieties altered Cmr-α cOA synthesis activity depending on different tag complementarity degrees. In type III CRISPR-Cas systems, tag complementarity suppresses the DNA cleavage and cOA synthesis activity of the Cas10 subunit as a self-immunity avoidance mechanism () [Citation13–Citation18]. Therefore, our data indicate that Cmr3α could regulate the suppression on Cmr-α activity by tag complementarity, hinting a role of the subunit in controlling self-immunity.
We demonstrate that three mutations, including alanine substitution of I123-Y124 in thumb, deletion of Y18-N19-S20 and deletion of I21-L22-L23 in loop 1, reduced cOA synthesis activity in the presence of different subgroups of target RNAs, suggesting that these motifs functions in facilitating Cmr-α-mediated immunity with the corresponding target RNAs (). Among them, I21-L22-L23 could moderately enhance Cmr-α activity upon low tag complementarity, suggesting its function in facilitating immunity against the invaders, the anti-tag region of which accidentally complements crRNA tag at one or two bases. In contrast, the two large hydrophobic residues (I123-Y124) in thumb act upon high tag complementarity, implying that these residues could interfere with the suppression of immune activity, possibly increasing potential autoimmunity risk. Strikingly, deletion of I21-L22-L23 within loop 1 resulted in increased cOA synthesis activity upon high tag complementarity, suggesting that the motif exerts dual effects on Cmr-α activity, i.e. it could both enhance Cmr-α activity upon low tag complementarity and suppress Cmr-α activity upon high tag complementarity. Together, the data suggest dual functions of Cmr3 in both facilitating immunity and preventing autoimmunity in type III systems.
The specific mechanisms how Cmr3α regulates Cmr-α activity remain in mystery. Nevertheless, three evidences support that Cmr3 performs its function by modulating the interaction between crRNA tag and target RNA anti-tag region: first, the interaction between tag and anti-tag region determines the binding site of the latter and only not bound by crRNA tag, the anti-tag region could interact with the Cas10 subunit and activate it to confer immunity [Citation34]; second, the loop 1 and thumb structures of Cmr3 are adjacent to crRNA tag and thumb is believed to separate crRNA tag and anti-tag region at the 8th position [Citation32]; third, we show that the observed effects of Cmr3α mutation is dependent on different tag complementarity degrees (). Together, we infer that Cmr3α could either separate 3ʹ-flanking sequence from crRNA tag to achieve higher Cmr-α activity or tether flanking sequence to crRNA tag to suppress Cmr-α activity. Such mechanism might provide a secondary regulation after tag complementarity to ensure both high immune activity and stringent autoimmunity avoidance.
Currently, two different strategies are employed by CRISPR-Cas systems for self vs non-self discrimination, i.e., PAM-dependent and tag complementarity-dependent strategies. The former is utilized by those targeting dsDNA, including type I, II and V systems [Citation27–Citation30], while the latter is employed by type III, as well as type VI as reported recently [Citation37]. Type III and type VI are activated by target RNA for indiscriminate nucleic acid degradation instead of targeting dsDNA; therefore, antisense transcription of CRISPR loci could provide potential targets for type III and VI systems [Citation38,Citation39], which might be the reason why these systems have evolved tag complementarity-dependent self discrimination mechanism. Apparently, such a mechanism holds advantages over the PAM-dependent strategy, which is likely to result in escapers with PAM mutation [Citation40,Citation41]. On the other hand, the PAM-dependent strategy is able to stringently restrict immune activity to PAM-containing targets by various PAM-readout mechanisms [Citation42]. In this study, our findings hint a role of Cmr3 in controlling the stringency of autoimmunity avoidance based on the tag complementarity-dependent strategy. Conceivably, type VI system might also employ an unknown mechanism to ensure the stringency of autoimmunity avoidance.
In type III CRISPR-Cas systems, the functions of non-catalytic subunits, including Cmr3 (Csm4 in III-A), Cmr5 (Csm2 in III-A), Cmr1 (Csm5 in III-A) and Cmr6, are gradually emerging from structural and biochemical characterization of type III effector complexes. Cmr1 and Csm5 that cap type III RNP at the 3ʹ-end promote target binding to facilitate Cas10 activation [Citation43–Citation45], while Csm2 promotes target RNA cleavage by Csm3 in a type III-A RNP [Citation45]. Structural studies imply that Csm4 could play a role in regulating cOA synthesis [Citation34]. Here, we reveal a dual function for Cmr3 in regulating Cmr activity upon different tag complementarity degrees, and it is expected that the corresponding subunit in Csm systems should perform similar functions. By now, only Cmr6, a unique subunit in type III-B RNP, remains to be further characterized. This subunit also possesses a loop and thumb towards crRNA-target RNA duplex as Cmr4 and Cmr3 [Citation32], and these structural moieties in Cmr6 could play an important yet unknown function, which would be of special interest in future research.
Materials and methods
Construction of pAC-cmr3α plasmid and derivative plasmids
The pAC-cmr3α plasmid was constructed with a ‘two-step’ method as described previously [Citation16]. First, the cmr3α gene (SiRe_0895) fragment was amplified with the primer set of Cmr3-up-NdeI and Cmr3-dw-SalI (Supplementary Table S1), digested with NdeI and SalI and inserted into pSeSD1 [Citation46] between the same sites, giving pCmr3α plasmid (Supplementary Table S3). Second, the cmr3α expression cassette, including the arabinose-inducible promoter araS-SD, the coding region of cmr-3α, coding sequence of 6× His-tag, and the transcriptional terminator, was amplified from the pCmr3α plasmid using the primer pair of MRS-up and MRS-dw (Supplementary table S1). The resulting PCR product was digested with SmalI and XhoI and inserted into pAC-MS1 [Citation16] between the SmaI and SalI sites, yielding pAC-cmr3α (Supplementary table S2).
To generate plasmids expressing mutated Cmr-3α subunit (M1, M2, M3, M4, M5, M6 and M7), splicing, overlapping and extension PCR (SOE-PCR) [Citation47] was employed to amplify the Cmr3α mutant expression cassette, using the primers listed in Supplementary Table S1 (including overlapping primers and the primer pair of MRS-up and MRS-dw as two flanking primers). The resulting SOE-PCR fragments were inserted into pAC-MS1 as described above, yielding plasmids to express mutated Cmr-3α proteins. All mutations were confirmed by determination of DNA sequences of the constructed plasmids (Eurofins Genomics, Germany).
Purification of cmr-α-RNPs from S. islandicus
The pAC-cmr3α plasmid and its variants were electroporated into S. islandicus MF1 [Citation16] as previously described. Then transformants were grown in SCV media (Basal media supplemented with 0.2% sucrose, 0.2% Casamino acids and 1% vitamin solution) at 78°C [Citation48] until the optical density at 600 nm reached about 0.7, when cell mass was harvested and used for Cmr-α-RNP purification as previously described [Citation16,Citation49]. Specifically, cell pellets were resuspended in Buffer A (20 mM HEPES pH 7.5, 30 mM Imidazole, 500 mM NaCl), and the cells were disrupted by French press, followed by centrifuge at 12,000 rpm for 20 min to remove cell debris. The cell extract was loaded onto a 1 ml HisTrap HP column (GE Healthcare, Waukesha, WI, USA) and protein bound by the column were stepwise eluted with buffers containing 30 mM, 70 mM and 200 mM imidazole successively. Fractions containing the target protein were concentrated and further loaded onto a Superdex 200 Hiload column (GE Healthcare, Waukesha, WI, USA) pre-balanced with Buffer C (20 mM Tris-HCl pH 7.5, 250 mM NaCl). Sample fractions from the Superdex column were analyzed by SDS-PAGE and those containing Cmr-α were pooled together and used for further analysis.
Generation of target RNAs by in vitro transcription and their purification
The target RNAs carrying different flanking sequences used in the DNA cleavage assay and cOA synthesis assay were generated by in vitro transcription, followed by PAGE gel purification. Specifically, a forward primer (SS1-46-Fwd) and reverse primers containing sequence variations at the 5ʹ region (Supplementary table 3) were designed and synthesized by IDT, USA. Annealing and extension of the forward primer and each reverse primer, followed by chloroform extraction and isopropanol precipitation, generates dsDNA template for in vitro transcription. The in vitro transcription assay was performed with TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA) in a 20 μl reaction mixture containing 4 μl 5X TranscriptAid Reaction Buffer, 8 μl ATP/CTP/GTP/UTP mix, 2 μg template DNA, 2 μl TranscriptAid Enzyme Mix. Then, the reaction mixture was treated with 2 μl of DNase I (1 U/μl) for 30 min at 37°C and mixed with 20 μl of 2× RNA loading dye. After heating at 95°C for 2 min and cooling down on ice for 5 min, the transcripts were loaded on 10% denaturing polyacrylamide gel and visualized by ethidium bromide staining. The main bands were cut from the gel and the RNA in gel was purified as described previously [Citation16]. At last, the transcripts were resolved in 50 μl RNase-free H2O and the concentration of them was determined with Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA).
Nucleic acid cleavage assay
Radio-labelled SS1-46 RNA and S10 ssDNA (Supplementary Table S4) were used for RNA cleavage assay and DNA cleavage assay, respectively. The substrates were synthesized from Integrated DNA Technologies (IDT, Coralville, IA, USA.), gel-purified and labelled with γ[Citation32]P-ATP using T4 PNK (New England Biolabs, Ipswich, MA, USA) as described previously [Citation16].
RNA cleavage assay was carried out in a 10 µl reaction mixture containing 50 mM Tris-Cl (pH 7.5), 10 mM MgCl2, 5 mM DTT, 20 nM of labelled SS1-46 and 20 nM of the wild-type or mutated Cmr-α-RNP. The mixture was incubated at 70°C for 20 min and then supplemented with 10 µl of 2× RNA loading dye. At last, the sample was denatured by heating at 95°C for 2 min analyzed by denaturing PAGE in an 18% gel with the results recorded by phosphor imaging and read by a Typhoon FLA 7000 (GE Healthcare, Waukesha, WI, USA).
The DNA cleavage assay was conducted in a 10-µl reaction mixture contains 20 mM Mes, pH 6.0, 5 mM MnCl2, 5 mM DTT, 20 nM labelled S10 ssDNA and 5 nM of the wild-type or a mutated Cmr-α-RNP, in the presence of 200 nM one of the target RNAs. The mixtures were incubated at 70°C for 20 min and analyzed as described for the RNA cleavage assay.
The DNA cleavage assay yielded many products, and we selected three separated, clear bands for quantification. The sum of the three bands yielded in the presence of CT RNA was set as ‘1’, to which the relative DNA cleavage efficiency in the presence of all other target RNAs were calculated.
Cyclic oligoadenylates (cOA) synthesis assay
The cOA synthesis assay is conducted in a 10-µl reaction mixture containing 20 mM Mes (pH 6.0), 5 mM MnCl2, 5 mM DTT, ca. 1 nM α[Citation32]P-ATP (PerkinElmer, Waltham, MA, USA.), 100 µM ATP, about 5 nM wild type or mutated Cmr-α-RNP, and 200 nM different target RNAs. The mixture was incubated at 70°C for 20 min, followed by supplementation of 10 µl 2× RNA loading dye and heated at 95°C for 2 min. After cooling on ice for 5 min, the samples were loaded onto a 24% polyacrylamide gel, and the synthesized cOA was detected by exposing the gel to a phosphor screen and scanning the screen with a Typhoon FLA 7000 (GE Healthcare, Waukesha, WI, USA). The amount of cOA yielded in the presence of CT RNA was set as ‘1’, to which the relative cOA synthesis efficiency in the presence of all other target RNAs were calculated.
Supplemental Material
Download MS Word (2.9 MB)Acknowledgments
The research was supported by National Science Foundation of China (Grant No. 31771380), the Independent Research Fund Denmark—Natural Sciences (DFF-4181-00274) and Huazhong Agricultural University Scientific & Technological Self-innovation Foundation. Tong Guo is the recipient of a PhD studentship from the China Scholarship Council.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Materials:
Supplemental Materials data for this article can be accessed here.
Additional information
Funding
References
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712.
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845.
- McGinn J, Marraffini LA. Molecular mechanisms of CRISPR-Cas spacer acquisition. Nature Rev Microbiol. 2019;17:7–12.
- Brouns SJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964.
- Carte J, Wang R, Li H, et al. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496.
- Mohanraju P, Makarova KS, Zetsche B, et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147.
- Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78.
- Chen JS, Ma E, Harrington LB, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439.
- Gootenberg JS, Abudayyeh OO, Kellner MJ, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444.
- Li SY, Cheng QX, Liu JK, et al. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018;28:491–493.
- Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573.
- Hale CR, Zhao P, Olson S, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956.
- Elmore JR, Sheppard NF, Ramia N, et al. Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR-Cas system. Genes Dev. 2016;30:447–459.
- Estrella MA, Kuo FT, Bailey S. RNA-activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Genes Dev. 2016;30:460–470.
- Kazlauskiene M, Tamulaitis G, Kostiuk G, et al. Spatiotemporal control of type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol Cell. 2016;62:295–306.
- Han W, Li Y, Deng L, et al. A type III-B CRISPR-Cas effector complex mediating massive target DNA destruction. Nucleic Acids Res. 2017;45:1983–1993.
- Niewoehner O, Garcia-Doval C, Rostol JT, et al. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017;548:543–548.
- Kazlauskiene M, Kostiuk G, Venclovas C, et al. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017;357:605–609.
- Rouillon C, Athukoralage JS, Graham S, et al. Control of cyclic oligoadenylate synthesis in a type III CRISPR system. eLife. 2018;7:e36734.
- Han W, Stella S, Zhang Y, et al. A Type III-B Cmr effector complex catalyzes the synthesis of cyclic oligoadenylate second messengers by cooperative substrate binding. Nucleic Acids Res. 2018;46:10319–10330.
- Staals RH, Agari Y, Maki-Yonekura S, et al. Structure and activity of the RNA-targeting type III-B CRISPR-Cas complex of thermus thermophilus. Mol Cell. 2013;52:135–145.
- Tamulaitis G, Kazlauskiene M, Manakova E, et al. Programmable RNA shredding by the type III-A CRISPR-Cas system of streptococcus thermophilus. Mol Cell. 2014;56:506–517.
- Ramia NF, Spilman M, Tang L, et al. Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR-Cas complex. Cell Rep. 2014;9:1610–1617.
- Benda C, Ebert J, Scheltema RA, et al. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol Cell. 2014;56:43–54.
- Zhu X, Ye K. Cmr4 is the slicer in the RNA-targeting Cmr CRISPR complex. Nucleic Acids Res. 2015;43:1257–1267.
- Tamulaitis G, Venclovas C, Siksnys V. Type III CRISPR-Cas immunity: major differences brushed aside. Trends Microbiol. 2017;25:49–61.
- Deveau H, Barrangou R, Garneau JE, et al. Phage response to CRISPR-encoded resistance in streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400.
- Gasiunas G, Barrangou R, Horvath P, et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–86.
- Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821.
- Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771.
- Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571.
- Osawa T, Inanaga H, Sato C, et al. Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol Cell. 2015;58:418–430.
- Taylor DW, Zhu Y, Staals RH, et al. Structural biology. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science. 2015;348:581–585.
- You L, Ma J, Wang J, et al. Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference. Cell. 2019;176:239–53 e16.
- Jia N, Mo CY, Wang C, et al. Type III-A CRISPR-Cas Csm complexes: assembly, periodic RNA cleavage, DNase activity regulation, and autoimmunity. Mol Cell. 2018;73:264–73 e5.
- Golovanov IB, Zhenodarova SM, Ivanitskii GR. Structure-property correlation and prediction of melting temperature of RNA duplexes. Doklady biological sciences: proceedings of the Academy of Sciences of the USSR. Bio sci Sect. 2001;380:504–507.
- Meeske AJ, Marraffini LA. RNA guide complementarity prevents self-targeting in type VI CRISPR systems. Mol Cell. 2018;71:791–801 e3.
- Lasa I, Toledo-Arana A, Dobin A, et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci U S A. 2011;108:20172–20177.
- Lillestol RK, Shah SA, Brugger K, et al. CRISPR families of the crenarchaeal genus sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009;72:259–272.
- Pyenson NC, Gayvert K, Varble A, et al. Broad targeting specificity during bacterial type III CRISPR-Cas immunity constrains viral escape. Cell Host Microbe. 2017;22:343–53 e3.
- Silas S, Lucas-Elio P, Jackson SA, et al. Type III CRISPR-Cas systems can provide redundancy to counteract viral escape from type I systems. eLife. 2017;6:e27601.
- Gleditzsch D, Pausch P, Muller-Esparza H, et al. PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. Rna Biol. 2018;16:1–14.
- Hale CR, Cocozaki A, Li H, et al. Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex. Genes Dev. 2014;28:2432–2443.
- Li Y, Zhang Y, Lin J, et al. Cmr1 enables efficient RNA and DNA interference of a III-B CRISPR-Cas system by binding to target RNA and crRNA. Nucleic Acids Res. 2017;45:11305–11314.
- Mogila I, Kazlauskiene M, Valinskyte S, et al. Genetic dissection of the type III-A CRISPR-Cas system csm complex reveals roles of individual subunits. Cell Rep. 2019;26:2753–65 e4.
- Peng N, Deng L, Mei Y, et al. A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon sulfolobus islandicus. Appl Environ Microbiol. 2012;78:5630–5637.
- Warrens AN, Jones MD, Lechler RI. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene. 1997;186:29–35.
- Peng N, Han W, Li Y, et al. Genetic technologies for extremely thermophilic microorganisms of sulfolobus, the only genetically tractable genus of crenarchaea. Sci China Life Sci. 2017;60:370–385.
- Zhang J, White MF. Expression and purification of the CMR (Type III-B) complex in sulfolobus solfataricus. Methods Mol Biol. 2015;1311:185–194.
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–4.
