ABSTRACT
La-Related Protein 1 (LARP1) is an RNA-binding protein that regulates the stability and translation of mRNAs encoding the translation machinery, including ribosomal proteins and translation factors. These mRNAs are characterized by a 5ʹ-terminal oligopyrimidine (TOP) motif that coordinates their temporal and stoichiometric expression. While LARP1 represses TOP mRNA translation via the C-terminal DM15 region, the role of the N-terminal La-Module in the recognition and translational regulation of TOP mRNAs remains elusive. Herein we show that the LARP1 La-Module also binds TOP motifs, although in a cap-independent manner. We also demonstrate that it recognizes poly(A) RNA. Further, our data reveal that the LARP1 La-Module can simultaneously engage TOP motifs and poly(A) RNA. These results evoke an intriguing molecular mechanism whereby LARP1 could regulate translation and stabilization of TOP transcripts.
Introduction
La-related Proteins (LARPs) are a diverse family of RNA-binding proteins that are conserved throughout eukaryotic evolution and function in nearly all stages of RNA metabolism [Citation1–3]. Each LARP subfamily has evolved to recognize and regulate the metabolism of particular RNAs. The target RNAs of these subfamilies range from non-coding RNAs to messenger and viral RNAs.
LARPs are characterized by an N-terminal La-Motif (LAM) and are classified into subfamilies based on sequence conservation of the LAM: LARP1, LARP2, Genuine La (LARP3), LARP4, LARP5, LARP6, and LARP7 [Citation1]. The LAM adopts a winged helix-turn-helix fold [Citation4] and is followed by an RNA recognition motif (RRM). The RRM has a canonical βαββαβ structure or an RRM-like (RRML) fold, predicted to fold similarly to an RRM, but lacking the consensus RNP motifs [Citation1,Citation5,Citation6]. Together, the LAM and RRM comprise the ‘La-Module.’ In addition to the La-Module, most LARPs have a C-terminal RNA-binding domain that enhances RNA recognition and biological function [Citation1–3].
Phylogenetic analyses reveal that the LAM and RRM co-evolved in LARPs, suggesting the importance of the entirety of the La-Module in RNA recognition [Citation1]. Consistent with this, initial structural and biochemical studies in Genuine La and LARP7 show that the canonical RNA-binding surfaces of the LAM and RRM do not engage RNA [Citation3,Citation7–9]. Instead, the LAM and RRM function synergistically; RNAs make intimate contacts with a conserved patch of hydrophobic residues in the LAM, and are supported by contacts with the edge of the RRM β2 strand [Citation7–9]. Similarly, in LARP6 the LAM and RRM synergistically bind RNA, but also require participation of the interdomain linker [Citation10]. However, in LARP4 the La-Module plays a minor role in RNA recognition [Citation11]. Instead, intrinsically disordered regions N-terminal to the La-Module drive RNA-binding [Citation11]. Thus, while the main RNA-binding unit is conserved, additional unstructured features and conformational plasticity within the La-Module coordinate RNA recognition.
Although all LARPs share a La-Module, sequence and structural variations give rise to distinct RNA binding specificities and roles in RNA metabolism [Citation1,Citation3,Citation12]. In Genuine La, the prototypical LARP, the La-Module binds the UUUOH 3ʹ termini of premature tRNAs to promote their folding and maturation [Citation7,Citation8,Citation13]. The LARP7 La-Module also recognizes UUUOH, however it binds the 3ʹ terminus of 7SK non-coding RNA to facilitate transcription regulation [Citation9,Citation14]. The LARP6 La-Module evolved to engage a stem-loop within the 5ʹUTR of collagen mRNAs to increase their translation [Citation15,Citation16]. The La-Modules of LARP4 and LARP5, the most closely related to LARP1, bind poly(A) and AU-rich regions of mRNAs, respectively, and also stimulate mRNA stability and translation [Citation11,Citation17–19]. Thus, despite seemingly convergent features, La-Modules display adaptable and specified RNA target selection.
LARP1 regulates the stability and translation of mRNAs that encode components of the translation machinery, such as ribosomal proteins and translation factors [Citation20–26]. These transcripts, known as TOP mRNAs, are characterized by a terminal oligopyrimidine (TOP) motif in the 5ʹUTR, immediately after the 5ʹcap [Citation27]. The TOP motif, comprised of 4–14 pyrimidines followed by a GC-rich region [Citation27], allows for the coordinated translation of TOP mRNAs downstream of mTORC1 [Citation28]. Under conditions of mTORC1 inhibition, the LARP1 C-terminal DM15 region binds the 5ʹ cap and TOP motif [Citation23,Citation25]. This obstructs the formation of the translation initiation complex and thereby represses TOP mRNA translation during metabolically unfavourable conditions [Citation23,Citation25].
However, thus far, the role of the LARP1 La-Module in recognizing TOP mRNAs and regulating their translation is unknown. A few lines of evidence led us to hypothesize that the La-Module binds the poly(A) tails of TOP mRNAs. First, LARP1 associates with Poly(A)-Binding Protein (PABP) via a putative PABP-interacting motif 2 (PAM2) located between the LAM and RRM [Citation21,Citation24]. Second, LARP1 co-sediments with PABP through polysome gradients with TOP mRNAs [Citation21,Citation24,Citation29]. Finally, LARP1 from human cell extracts immunoprecipitates with poly(A) RNA, but not with poly (U), poly(C), or poly(G) [Citation26].
Here we show that the LARP1 La-Module directly engages poly(A) RNA. Unexpectedly, we find that the La-Module also binds some TOP motifs, in a cap-independent manner, with similar affinity to poly(A) RNA. We also present evidence that the LARP1 La-Module can simultaneously bind poly(A) and the TOP motif RNA. Integration of these data with the established roles of the DM15 region allow us to assemble a model for the role of LARP1 in TOP mRNA recognition that reconciles many of the observations of LARP1 published hitherto. We establish a direct role for LARP1 at both the 5ʹ and 3ʹ ends of TOP mRNAs, with the La-Module recognizing distinguishing features at either end of TOP transcripts.
Results
LARP1 La-Module binds poly(A) RNA
To identify the RNA targets of the LARP1 La-Module we analysed in vitro binding assays by native gel electrophoresis. Because LARP1 associates with poly(A) RNA [Citation26], and also with PABP via a PAM2 located within the La-Module [Citation21,Citation24], we hypothesized that the La-Module recognizes the poly(A) tails of TOP mRNAs. To test this, we determined the relative affinity of purified recombinant human LARP1 La-Module for homopolymeric RNA sequences using electrophoretic mobility shift assays (EMSAs) (, Supp. ). EMSAs conducted with a 25-nucleotide poly(A) RNA in the presence of non-specific competitors demonstrated that the La-Module directly binds this sequence with an apparent Kd of 40 ± 1 nM ( A,B). The shifted poly(A) oligonucleotide largely remained in the well, possibly due to the formation of multimeric protein-RNA complexes (); as the recombinant protein was observed to be folded (Supp. ), and isothermal titration calorimetry (ITC) experiments confirmed the specific binding of the La-Module to poly(A) RNA (Supp. ), non-specific aggregation in the well is unlikely. Further, substitution of tRNA or salmon sperm DNA for poly(dI-dC) as a non-specific competitor allowed the La-Module-poly(A) complex to enter the gel (Supp. ).
Figure 1. LARP1 La-Module binds poly(A) RNA. (A) EMSA analysis of binding assays of WT La-Module with poly(A) 25-mer. (B) Quantification of three independent EMSAs of WT La-Module with poly(A) RNA. Bars are standard deviation. (C-F) EMSAs analysing WT La-Module with (C) poly(U) 20-mer, (D) poly(G) 19-mer (E) poly(C) 20-mer, and (F) poly(A) 25-mer 3ʹPO4.
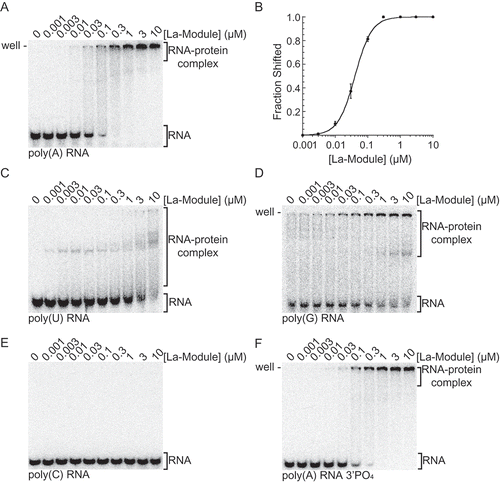
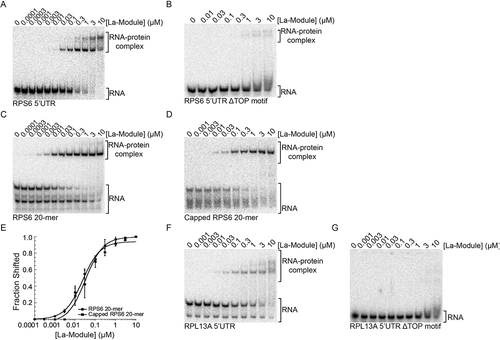
Figure 2. LARP1 La-Module binds TOP mRNA 5ʹUTRs in a TOP motif-dependent manner. EMSAs of binding assays of WT La-Module with (A) RPS6 5ʹUTR, (B) RPS6 5ʹUTR lacking the polypyrimidine region of the TOP motif, (C) RPS6 20-mer, and (D) Capped RPS6 20-mer. (E) Quantification of three independent EMSAs of WT La-Module with indicated RNAs. Bars are standard deviation. EMSAs of binding assays of the LARP1 La-Module with (F) RPL13A 5ʹUTR and (G) RPL13A lacking the polypyrimidine region of the TOP motif
We also determined the affinities of the La-Module for each of the other homopolymeric oligonucleotides. Importantly, the LARP1 La-Module bound poly(U) RNA, the cognate binding target of Genuine La and LARP7, with poor affinity (). Similarly, the La-Module had low affinity for poly(G) RNA, and did not bind poly(C) RNA ( ,E); ITC experiments confirmed that the La-Module does not recognize poly(C) RNA (Supp. ).
Because the chemical moiety at the 3ʹ end of the RNA is required for binding by Genuine La and LARP7 [Citation7–9], we then tested the binding of the LARP1 La-Module with a poly(A) 25-mer modified with a 3ʹPO4 group. While it did not abolish binding, assays conducted with this modified poly(A) RNA showed a two-fold decrease in apparent affinity of 85 ± 5 nM (), suggesting a small role for the role of the 3ʹ chemical moiety in LARP1 La-Module recognition.
LARP1 La-Module binds various TOP motifs
Given that the La-Module binds poly(A) RNA, we predicted that it would not bind the 5ʹUTRs of TOP transcripts. Surprisingly, the LARP1 La-Module shifted an oligonucleotide representing the complete 42-nucleotide 5ʹUTR of RPS6 mRNA (). Furthermore, deletion of the first 10 nucleotides corresponding to the TOP motif abrogated binding (). To validate the interaction between the La-Module and RPS6 TOP motif, we tested a 20-mer RNA representing the first 20 nucleotides of the RPS6 5ʹUTR (RPS6 20-mer), containing the 10-nucleotide pyrimidine tract followed by the GC-rich region. The La-Module bound the RPS6 20-mer with an apparent affinity of 31 ± 3 nM, comparable to the 40 ± 1 nM affinity observed for poly(A) RNA (, E). Interestingly, unlike the DM15 region [Citation25], the affinity of the La-Module for the RPS6 20-mer did not increase upon capping of the oligonucleotide, showing an apparent affinity of 32.8 ± 5 nM (, E). The La-Module also shifts an oligonucleotide representing the RPL13A 5ʹUTR, and accordingly, deletion of the TOP motif pyrimidine tract also abrogates binding (, G). The La-Module did not bind oligonucleotides corresponding to the 5ʹUTR of RPS18 and PABPC1 mRNA (Supp. , B), suggesting that, like the DM15 region [Citation30], the La-Module does not bind all TOP sequences. We next tested whether the LARP1 La-Module recognizes TOP mRNA 3ʹUTRs; it did not shift an oligonucleotide representing the 3ʹUTR of RPS6 mRNA (Supp. ).
LARP1 La-Module simultaneously engages poly(A) RNA and the RPS6 TOP motif
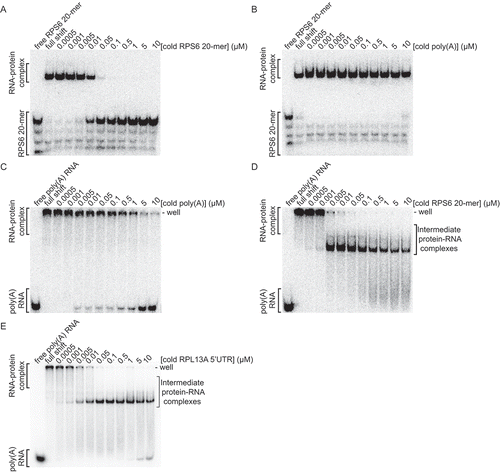
Because the LARP1 La-Module bound the poly(A) and RPS6 20-mer RNAs with similar affinities, we utilized competition assays to delineate specificity (). We first pre-bound the La-Module to radiolabeled RPS6 20-mer and competed with a titration of cold RPS6 20-mer or poly(A) RNA. Cold RPS6 20-mer at 50 nM was sufficient to displace the pre-bound RPS6 20-mer, whereas cold poly(A) RNA did not compete for the radiolabeled RNA even at 10 μM ( A,B). We next pre-bound the La-Module to poly(A) RNA and competed with a titration of cold poly(A) or RPS6 20-mer RNA. Cold poly(A) RNA weakly displaced pre-bound poly(A) RNA, only achieving maximal displacement at 10 μM (). However, when pre-bound poly(A) RNA was competed with cold RPS6 20-mer, beginning at 5 nM cold competitor, a complex formed that migrated further into the gel, between the shifted and free poly(A) RNA (; intermediate complex). Similarly, an intermediate complex formed when pre-bound poly(A) RNA was competed with cold RPL13A 5ʹUTR ().
Figure 3. An intermediate complex forms upon the addition of cold RPS6 5ʹTOP motif or RPL13A 5ʹUTR to the La-Module-poly(A) RNA complex. Competition assays conducted in the presence of poly(dI-dC) and analysed by native gel of: the La-Module-RPS6 20-mer RNA complex with cold (A) RPS6 20-mer RNA, and (B) poly(A) 25-mer RNA. Competition assays conducted in the presence of poly(dI-dC) and analysed by native gel of: the La-Module-poly(A) RNA complex with: cold (C) poly(A) 25-mer RNA, (D) RPS6 20-mer RNA, and (E) RPL13A 5ʹUTR
These results led us to ask: in the absence of tRNA, why does the La-Module-poly(A) RNA complex get stuck in the well, but moves into the gel in the presence of cold RPS6 20-mer competitor? The simplest explanation is that the register of binding to poly(A) RNA is undefined because of its homopolymeric nature, thereby allowing for the assembly of multimeric complexes. We hypothesized that addition of a non-adenylate competitor could sequester molecules of La-Module away from the multimeric complex, thus allowing bound poly(A) RNA to enter the gel. To test this, we performed the competition assay with cold poly(C) RNA. However, the bound poly(A) remained within the well such that the intermediate complex was not recapitulated ().
Figure 4. Formation of an intermediate complex is not an artefact of aggregation or RNA base pairing, and is dependent upon an intact TOP motif. Competition assays conducted in the presence of poly(dI-dC) and analysed by native gel of the La-Module-poly(A) RNA complex with: cold (A) poly(C) RNA, (B) poly(G) RNA, (C) RPS6 5ʹUTR lacking the pyrimidine stretch of the TOP motif, and (D) RPL13A 5ʹUTR lacking the pyrimidine stretch of the TOP motif
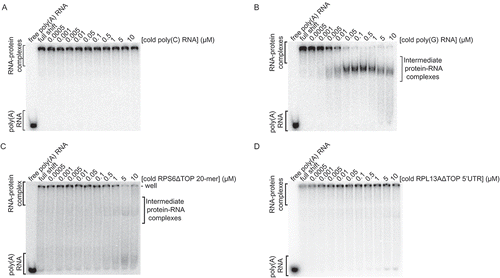
Given that the competitions were analysed by native gel, we hypothesized that the intermediate complex has increased negative charge, and therefore migrated faster towards the positive electrode. This could occur if the La-Module simultaneously binds radiolabeled poly(A) and unlabelled RPS6 20-mer RNA. Alternatively, the intermediate complex could be an artefact of uridines within the RPS6 20-mer base pairing with the poly(A) RNA. To exclude this possibility, we competed La-Module pre-bound to poly(A) RNA with cold poly(G) RNA (); if the intermediate complex is an artefact of RNA duplex formation, we would not expect to reproduce the intermediate complex. However, we observed the formation of an intermediate complex that migrated between the shifted and free poly(A) RNA (). Additionally, we competed the La-Module-poly(A) RNA complex with cold RPS6 20-mer and RPL13A 5ʹUTR with TOP motif deletions. Using the TOP motif deletion RNA also did not reproduce the intermediate complex (, D). To further validate these results, we performed the competitions in the presence of tRNA as a non-specific competitor. La-Module pre-bound to RPS6 20-mer RNA was not competed by the addition of cold poly(A) RNA (Supp. ). However, once again, when we pre-bound the La-Module to poly(A) RNA and competed with cold RPS6 20-mer, a distinct RNP complex formed (Supp. ). Thus, we hypothesized that the intermediate complex might be a ternary complex of the La-Module, RPS6 20-mer, and poly(A) RNA.
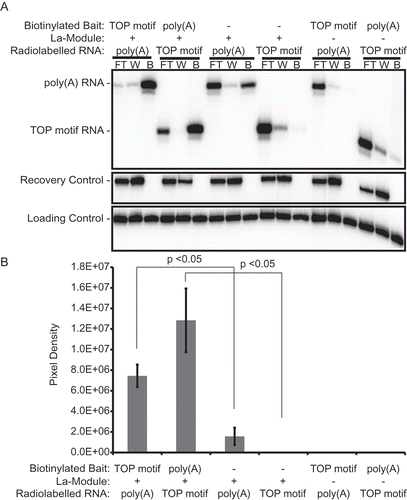
To test this hypothesis, we conducted biotin pull-down assays. We pulled down the LARP1 La-Module using either biotinylated poly(A) or RPS6 20-mer RNA, prior to introducing radiolabeled RPS6 20-mer or poly(A) RNA (). Enrichment of radiolabeled signal relative to the controls would suggest a ternary complex of the biotinylated bait, La-Module, and radiolabeled RNA. Using biotinylated poly(A) RNA as bait, we observed enrichment of the radiolabeled RPS6 20-mer relative to controls when either bait or La-Module was omitted (, B). Similarly, biotinylated RPS6 20-mer bait enriched radiolabeled poly(A) RNA relative to the controls (, B). Thus, both biotinylated bait RNA and La-Module were required to capture the radiolabeled RNA.
Figure 5. The La-Module simultaneously binds to poly(A) RNA and RPS6 TOP motif in biotin pull-down experiments. (A) Denaturing gel analysing the flow through (FT), wash (W), and pull-down streptavidin beads (B) of the indicated binding experiments. Poly(A), poly(A) 25-mer; TOP motif, RPS6 20-mer. Ethanol precipitation recovery control, loading control, RPS6 20-mer and poly(A) RNA size markers are indicated. (B) Quantification of biotin pull-down assays. Bars indicate standard error
Discussion
In this paper we identify the RNA binding partners of the LARP1 La-Module to elucidate its contribution to TOP mRNA recognition. We found that the LARP1 La-Module binds poly(A) RNA (, Supp. ). Surprisingly, the LARP1 La-Module also binds the 5ʹUTRs of some TOP mRNAs (). Deletion of the TOP motif abrogated La-Module binding to TOP mRNA 5ʹUTRs, suggesting that the La-Module requires the polypyrimidine portion of TOP sequences (). Indeed, the La-Module binds the RPS6 TOP motif with comparable affinity to poly(A) RNA, and does so in a cap-independent manner ().
The affinity of the LARP1 La-Module for poly(A) RNA may indicate a role at poly(A) tails. This is consistent with the association of LARP1 with PABP and poly(A) RNA [Citation21,Citation24,Citation26]. The PAM2 located within the interdomain linker between the LAM and RRM could bind PABP [Citation24], while the LAM and RRM bind poly(A) tails of TOP transcripts or other mRNAs.
The coordinated translation of TOP mRNAs is mediated by a characteristic 5ʹTOP motif followed by a GC-rich region within the 5ʹUTRs of these transcripts [Citation27]. Our data also implicate the LARP1 La-Module at the 5ʹUTRs of TOP mRNAs via recognition of the TOP motif region. The La-Module binds the 5ʹUTRs of RPS6 and RPL13A mRNAs, but not those of RPS18 or PABPC1 (, Supp. ). These data are consistent with differential levels of TOP mRNA repression by LARP1 [Citation24], and the specificity of the DM15 region for only certain TOP mRNAs [Citation30]. While the DM15 region binds the 5ʹ cap and first four nucleotides of TOP mRNAs [Citation25], the La-Module binds in a cap-independent manner (), suggesting that the La-Module recognizes sequences downstream of the DM15 binding site. Consistent with this, the La-Module binds the RPS6 20-mer, but not an RPS6 15-mer that shortens the GC-rich region (data not shown), suggesting a contribution of that sequence to La-Module binding; in addition, although it is weak, the La-Module binds to poly(G) RNA in the presence of non-specific competitor (). These observations are consistent with data showing that, at least in some eukaryotic cell lines, both the 5ʹTOP motif and the GC-rich region immediately downstream are necessary for TOP mRNA translation regulation [Citation27,Citation31], and the mapping of LARP1 footprints to 3ʹ ends of TOP 5ʹUTRs [Citation32]. Binding of the La-Module to TOP motifs may enhance TOP mRNA recognition by LARP1. As the C-terminal DM15 region binds the 5ʹ cap and TOP motif to repress TOP mRNA translation [Citation23,Citation25], the N-terminal La-Module might bind cooperatively to the TOP motif to strengthen the interaction and promote stringent specificity for recognizing select TOP mRNAs.
More notably, the LARP1 La-Module can simultaneously bind both poly(A) and TOP motif RNA. We observed the formation of an intermediate complex upon competition of La-Module-poly(A) complex with TOP motif RNA (). Biotin pull-down experiments indicated that this intermediate complex corresponds to the LARP1 La-Module simultaneously engaging both poly(A) RNA and TOP motif RNA ().
The unprecedented ability of the LARP1 La-Module to bind two distinct RNAs at the same time not only expands the known binding repertoire of LARPs, but also might explain the biological functions of LARP1. Importantly, the targets of the LARP1 La-Module correspond to the 5ʹ and 3ʹ ends of TOP mRNAs – TOP motifs and poly(A) tails (). The simultaneous binding of poly(A) tails and the TOP motif would imply that the La-Module brings the 5ʹ and 3ʹ ends of bound transcripts into close proximity.
While the mRNA circularization model [Citation33,Citation34] has recently been reassessed [Citation35], other studies have provided evidence for its role in promoting oncogenic translation and tumorigenesis [Citation36]. Indeed, overexpression of LARP1 in epithelial malignancies, such as cervical and ovarian cancers, is correlated with tumour progression and poor clinical outcome [Citation37,Citation38]. Consistent with this, LARP1 increases the stability of several oncogenic transcripts [Citation37,Citation38] and enhances protein synthesis [Citation21,Citation29].
The translational repression of transcripts in P-bodies and stress granules is also associated with 5ʹ and 3ʹ UTR proximity. Thus, binding of the LARP1 La-Module to both ends of a single transcript may also explain its localization to P-bodies and stress granules [Citation24,Citation39] (), where RNAs are translationally repressed in condensed conformations [Citation40]. This is consistent with the necessity of LARP1 for recruiting and anchoring TOP mRNAs to these granules [Citation41].
Figure 6. Model of potential roles for the LARP1 La-Module in TOP mRNA translation and localization to P-bodies and stress granules. (A) The LARP1 La-Module could bind the poly(A) tails and TOP motifs of different TOP transcripts to form multimeric complexes. This might contribute to the phase separation characteristic of P-bodies and stress granules, while repressing translation and minimizing transcript degradation. (B) The LARP1 La-Module could bind the poly(A) tail and TOP motif of the same TOP transcript to enhance translation (left); recognition of the 5ʹ cap by the DM15 region of LARP1 could then repress the translation of bound transcripts (right)
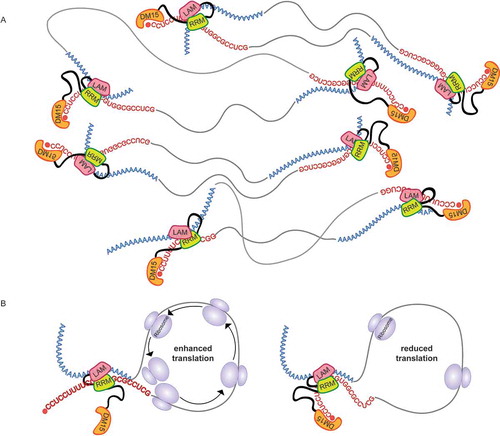
The role of the LARP1 La-Module in TOP mRNA recognition may also shed light on seemingly incongruous data regarding the function of LARP1 in TOP mRNA translation regulation. If the La-Module binds to both ends of TOP mRNAs, it might increase TOP mRNA translation (). However, upon the binding of the DM15 region to the 5ʹ cap, the translation initiation complex would be occluded, resulting in translation inhibition (). This idea perhaps explains the observations of LARP1 playing opposing stimulatory [Citation21,Citation29] and repressive [Citation23–25] roles in TOP translation regulation. Since our data do not directly address translation levels, it is also possible that the simultaneous interaction of the LARP1 La-Module stabilizes TOP transcripts.
Other La-Modules have been shown to play a role in stimulating translation and stabilizing transcripts. Cytoplasmic La regulates both cap-dependent [Citation42,Citation43] and cap-independent [Citation44–47] translation in a transcript-dependent manner. Cytoplasmic La also binds binding to poly(A) tails of messenger RNAs and facilitates entry into polysomes [Citation48]. LARPs 4 and 5 associate with PABP, RACK1, polysomes, and increase mRNA translation [Citation18,Citation19]. The LARP6 La-Module recognizes a stem-loop motif within the 5ʹUTR of collagen I- and III- encoding mRNAs to promote their translation [Citation15,Citation16]. Thus, there is precedent for proposing a stimulatory role for the LARP1 La-Module.
Herein we demonstrate that the LARP1 La-Module binds both poly(A) RNA and TOP motifs with comparable affinities. While we observed that the LARP1 La-Module can bind poly(A) and TOP motif RNA at the same time, we do not know the stoichiometry of the ternary complex: does the La-Module dimerize, with each protein molecule binding one RNA? Or does one protein bind both RNAs using different binding surfaces? In most LARPs, the LAM and RRM bind synergistically to RNAs without using their canonical RNA-binding surfaces [Citation2,Citation3]. It might be possible that the canonical binding surface of the LAM or RRM can bind a second RNA; indeed, Genuine La has recently been shown to bind poly(A) RNA via the canonical binding surface of the LAM [Citation48]. Thus, much remains to be uncovered about the atypical binding modes of LARPs. Nonetheless, dual RNA recognition by the LARP1 La-Module offers a tantalizing explanation for the mechanisms by which LARP1 regulates the translation and stability of TOP transcripts.
Materials and methods
Protein cloning, expression, and purification
The La-Module region of the LARP1 coding sequence (amino acids 310–540 from 1019-amino acid isoform LARP1a) (Integrated DNA Technologies) was PCR amplified and inserted into a pET28a vector (Novagen Inc) using NdeI and BamHI sites. The resulting construct expresses the La-Module with an N-terminal 6XHis tag followed by a Tobacco Etch Virus (TEV) protease cleavage site. The 6XHis-La-Module fusion protein was expressed in E. coli BL21(DE3) and cultured at 37°C for two hours, shifting to 17.5°C for 18 hours. Cells were harvested, frozen in liquid nitrogen, and stored at −80°C.
Cells were resuspended in lysis buffer [50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 10 mM imidazole, 10% v/v glycerol, one cOmplete EDTA-free Protease Inhibitor™ tablet (Roche)]. Cells were lysed using homogenization and lysate was cleared via centrifugation. The 6XHis-La-Module was purified in batch using nickel agarose affinity chromatography (ThermoScientific) and eluted with 50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 300 mM imidazole, 10% v/v glycerol. The 6XHis tag was removed by cleavage with 0.5 mg TEV per 10 mL eluate overnight at 4°C in dialysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 0.5 mM EDTA, 0.5 mM DTT). Nucleic acid and protein contaminants were removed using HiTrap Heparin followed by tandem HiTrap S and HiTrap QP (GE Healthcare Lifesciences) chromatography with an NaCl gradient (150 mM-1M), with La-Module eluting from the Heparin and Q columns. Fractions containing the La-Module were collected, concentrated, dialysed into storage buffer (50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 25% glycerol, 4 mM DTT), frozen in liquid nitrogen, and stored at −80°C. Samples for ITC were exchanged in 50 mM sodium phosphate pH 7.6 via 10K MWCO Centriprep concentrator and assessed for folding via circular dichroism spectroscopy (Supp. ) prior to use.
RNAs
RNA oligonucleotides (Sigma-Aldrich) have the following sequences:
Poly(A) RNA: 5ʹ- AAAAAAAAAAAAAAAAAAAAAAAAAOH
Poly(G) RNA: 5ʹ- GGGGGGGGGGGGGGGGGGGOH
Poly(C) RNA: 5ʹ- CCCCCCCCCCCCCCCCCCCCOH
Poly(U) RNA: 5ʹ- UUUUUUUUUUUUUUUUUUUUOH
Poly(A) RNA-PO4: 5ʹ- AAAAAAAAAAAAAAAAAAAAAAAAAPO4
RPS6 TOP 20-mer: 5ʹ- CCUCUUUUCCGUGGCGCUC
RPS6 5ʹUTR: 5ʹ- CCUCUUUUCCGUGGCGCCUCGGAGGCGUUCAGCUGCUUCAAG
RPS6 5ʹUTR ΔTOP: 5ʹ- GUGGCGCCUCGGAGGCGUUCAGCUGCUUCAAG
RPS18 5ʹUTR: 5ʹ-CUCUCUUCCACAGGAGGCCUACACGCCGCCGCUUGUGCUGCAGCC
PABPC1 42-mer: 5ʹ- CCUUCUCCCCGGCGGUUAGUGCUGAGAGUGCGGAGUGUGUG
RPL13A 5ʹUTR: 5ʹ- CCUUUUCCAAGCGGCUGCCGAAG
RPL13A 5ʹUTR ΔTOP: 5ʹ- AAGCGGCUGCCGAAG
RPS6 3ʹUTR: 5ʹ- AAGAUUUUUUGAGUAACAAAU
Loading Control: 5ʹ- CCAGUCAUGCUAGCCAUAUGCCUGGUCCGCCUGUUGC
Recovery Control: 5ʹUCCUGAAUGCUACGUUAAUCGGUAUCCAGCAGUUCUUUCAGUUU
CG
Electrophoretic mobility shift assays
RNA oligonucleotides were 5ʹ-end labelled with [γ-32P]-ATP (Perkin Elmer) using T4-polynucleotide kinase (New England Biolabs) and gel purified. Capped RPS6 20-mer was prepared as previously described[Citation25]. La-Module 5X stocks were prepared in dilution buffer (50 mM Tris-HCl pH, 7.5, 100 mM NaCl, 25% glycerol, 4 mM DTT). 10 μL binding reactions contained final concentrations of 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 7.5% v/v glycerol, 1mM DTT, 1 μM BSA (Thermo Fisher Scientific), ≤ 2 nM radiolabeled RNA, and either: 10 U/mL poly(dI-dC) (Sigma-Aldrich), 5 μM Yeast tRNA (Ambion) or 3 μM salmon sperm DNA (Thermo Fisher Scientific). La-Module was titrated at 0, 0.01, 0.03, 0.1, 0.3, 1, 3, 10 μM. Reactions were incubated on ice for 30 min then run on 7% polyacrylamide (29:1) native 0.5X TBE gels at 125 V for 45 min at 4°C. Exposed phosphor screens (GE Healthcare Lifesciences) were imaged on a Typhoon FLA plate reader (GE Healthcare Lifesciences) and quantitated using Imagequant TL (GE Healthcare Lifesciences). Dissociation constants were determined by plotting (KaleidaGraph) the fraction of shifted RNA versus the concentration of protein after band intensities were corrected for background (ImageQuant)TL.
Electrophoretic mobility shift competition assays
La-Module was pre-bound to radiolabeled RNA on ice in 8 μL reactions as described above. 2 μL 5X cold competitor RNA was titrated to yield final concentrations of 0, 0.0005, 0.001, 0.005, 0.01, 0.05, 0.1, 0.5 1, 5, 10 μM. Competition reactions were incubated on ice for an additional 30 min prior to loading and run as described above. Gels were dried, exposed overnight, and analysed as described for EMSAs.
Biotin pull-downs
3ʹ Biotinylated RNA oligonucleotides (IDT) were pre-bound to streptavidin magnetic beads and 1 μM La-Module was pulled down as per kit instructions (Pierce # 20,164), with the exception that the La-Module was incubated in 1X binding buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 7.5% glycerol, 1 mM DTT) in the presence of 1 μM BSA and 10 U/mL poly(dI-dC) (Sigma-Aldrich) as non-specific competitors. Beads were washed twice with 1X binding buffer to remove excess La-Module prior to the addition of ≤ 25 nM 5ʹ radiolabelled RNA in 100 μL 1X binding buffer in the presence of non-specific competitors as described above. Reactions were incubated for an additional 30 min at 4°C, then washed with 100 μL 1X binding buffer to remove unbound radiolabeled RNA. Flow through and washes were ethanol precipitated and resuspended in 10 μL 47.5% formamide, 0.1% bromophenol blue. Beads were resuspended in 2X formamide to a final volume of 10 μL. All samples were loaded onto 10% polyacrylamide (29:1) 1X TBE 7M urea sequencing gels and run at 90 W for 45 min at room temperature. Gels were dried, exposed overnight, and analysed as described for EMSAs above.
Isothermal titration calorimetry
Purified La-Module was titrated with poly(A) or poly(C) RNA in a TA instruments nanoITC to confirm selectivity. Representative traces (Supp. ) used 300 μM polyC RNA titrated into 110 μM La-Module or 200 μM polyA RNA into 85 μM La-Module with both titrant and titrand in 50 mM sodium phosphate, pH 7.6, at 25°C. Initial injection was 0.18 μL and subsequent 33 injections were 1.49 μL each into an active cell volume of 200 μL (overfilled to 300 μL). Stirring speed was 300 rpm with a 100 second injection interval and 1 second data collection interval.
Circular dichroism spectroscopy
La-Module folding was assessed in a JASCO J1500 circular dichroism spectrophotometer using 20 uM La-Module in 50 mM sodium phosphate, pH 7.6. Spectra were collected in a 0.5 mm cuvette from 280 to 180 nm at 25°C with 0.5 nm steps at a rate of 50 nm/min. Eight repetitions were averaged and smoothed with a 5 nm Savitzky-Golay filter[Citation49]. Ellipticity was normalized to mean residue molar ellipticity (Supp. ).
Supplemental Material
Download Zip (11 MB)Acknowledgments
We thank members of the LARP society, Dr. Corlett Wolfe Wood, and Dr. Sara Keubbing for critical discussion and feedback, and Sarah Waldron for technical help.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Bousquet-Antonelli C, Deragon JM. A comprehensive analysis of the La-motif protein superfamily. RNA. 2009;15:750–764.
- Maraia RJ, Mattijssen S, Cruz-Gallardo I, et al. The La and related RNA-binding proteins (LARPs): structures, functions, and evolving perspectives. Wiley Interdiscip Rev RNA. 2017;8.
- Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim Biophys Acta. 2010;1799:365–378.
- Dong G, Chakshusmathi G, Wolin SL, et al. Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J. 2004;23:1000–1007.
- Alfano C, Babon J, Kelly G, et al. Resonance assignment and secondary structure of an N-terminal fragment of the human La protein. J Biomol NMR. 2003;27:93–94.
- Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18:290–298.
- Teplova M, Yuan YR, Phan AT, et al. Structural basis for recognition and sequestration of UUU(OH) 3ʹ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell. 2006;21:75–85.
- Alfano C, Sanfelice D, Babon J, et al. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol. 2004;11:323–329.
- Uchikawa E, Natchiar KS, Han X, et al. Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7. Nucleic Acids Res. 2015;43:3373–3388.
- Martino L, Pennell S, Kelly G, et al. Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module. Nucleic Acids Res. 2015;43:645–660.
- Isabel Cruz-Gallardo LM, Geoff Kelly R, Atkinson A, et al. LARP4A recognizes polyA RNA via a novel binding mechanism mediated by disordered regions and involving the PAM2w motif, revealing. Nucleic Acids Res. 2019;47(8):4272–4291.
- Maraia RJ, Mattijssen S, Cruz-Gallardo I, et al. The La and related RNA-binding proteins (LARPs): structures, functions, and evolving perspectives. WIREs RNA. 2017;8:e1430.
- Chakshusmathi G, Kim SD, Rubinson DA, et al. A La protein requirement for efficient pre-tRNA folding. EMBO J. 2003;22:6562–6572.
- Krueger BJ, Jeronimo C, Roy BB, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229.
- Cai L, Fritz D, Stefanovic L, et al. Binding of LARP6 to the conserved 5ʹ stem-loop regulates translation of mRNAs encoding type I collagen. J Mol Biol. 2010;395:309–326.
- Stefanovic L, Longo L, Zhang Y, et al. Characterization of binding of LARP6 to the 5ʹ stem-loop of collagen mRNAs: implications for synthesis of type I collagen. RNA Biol. 2014;11:1386–1401.
- Schäffler K, Schulz K, Hirmer A, et al. A stimulatory role for the La-related protein 4B in translation. RNA. 2010;16:1488–1499.
- Yang R, Gaidamakov SA, Xie J, et al. La-related protein 4 binds poly(A), interacts with the poly(A)-binding protein MLLE domain via a variant PAM2w motif, and can promote mRNA stability. Mol Cell Biol. 2011;31:542–556.
- Küspert M, Murakawa Y, Schäffler K, et al. LARP4B is an AU-rich sequence associated factor that promotes mRNA accumulation and translation. RNA. 2015;21:1294–1305.
- Fonseca BD, Lahr RM, Damgaard CK, et al. LARP1 on TOP of ribosome production. WIREs RNA. 2018; 9.
- Tcherkezian J, Cargnello M, Romeo Y, et al. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5ʹTOP mRNA translation. Genes Dev. 2014;28:357–371.
- Sungki Hong MAF, Han T, Kamath A, et al. LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. eLife. 2017;6:e25237.
- Philippe L, Vasseur J, Debart F, et al. La-related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap-binding domain and controlled by an adjacent regulatory region. Nucleic Acids Res. 2018;46:1457–1469.
- Fonseca BD, Zakaria C, Jia JJ, et al. La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1). J Biol Chem. 2015;290:15996–16020.
- Lahr RM, Fonseca BD, Ciotti CE, et al. La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. eLife. 2017;6:e24146.
- Aoki K, Adachi S, Homoto M, et al. LARP1 specifically recognizes the 3ʹ terminus of poly(A) mRNA. FEBS Lett. 2013;587:2173–2178.
- Meyuhas O, Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta. 2014;1849(7):801–811.
- Thoreen CC, Chantranupong L, Keys HR, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113.
- Burrows C, Abd Latip N, Lam SJ, et al. The RNA binding protein Larp1 regulates cell division, apoptosis and cell migration. Nucleic Acids Res. 2010;38:5542–5553.
- Lahr RM, Mack SM, Heroux A, et al. The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5ʹTOP sequence. Nucleic Acids Res. 2015;43:8077–8088.
- Avni D, Shama S, Gabrizio Loreni MO. Vertebrate mRNAs with a 5ʹ-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterizationof the translational cis-regulatory element. Mol Cell Biol. 1994;14:3822–3833.
- Hong S, Freeberg MA, Han T, et al. LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. eLife. 2017;6:e25237.
- Wells SE, Hillner PE, Vale RD, et al. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140.
- Afonina ZA, Myasnikov AG, Shirokov VA, et al. Formation of circular polyribosomes on eukaryotic mRNA without cap-structure and poly(A)-tail: a cryo electron tomography study. Nucleic Acids Res. 2014;42:9461–9469.
- Vicens Q, Kieft JS, Rissland OS. Revisiting the closed-loop model and the nature of mRNA 5′–3′ communication. Mol Cell. 2018;72:805–812.
- Choe J, Lin S, Zhang W, et al. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–560.
- Mura M, Hopkins TG, Michael T, et al. LARP1 post-transcriptionally regulates mTOR and contributes to cancer progression. Oncogene. 2014;34:5025–5036.
- Hopkins TG, Mura M, Abd Latip N, et al. The RNA-binding protein LARP1 regulates survival and tumorigenesis in ovarian cancer. London, UK: Imperial College London; 2015.
- Nykamp K, Lee MH, Kimble J. C. elegans La-related protein, LARP-1, localizes to germline P bodies and attenuates Ras-MAPK signaling during oogenesis. RNA. 2008;14:1378–1389.
- Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol. 2012;4:a012286-a012286.
- Wilbertz JH, Voigt F, Horvathova I, et al. Single-molecule imaging of mRNA localization and regulation during the integrated stress response. Mol Cell. 2019;73:946–958.
- Cardinali B, Carissimi C, Gravina P, et al. La protein is associated with terminal oligopyrimidine mRNAs in actively translating polysomes. J Biol Chem. 2003;278:35145–35151.
- Crosio C, Boyl PP, Loreni F, et al. La protein has a positive effect on the translation of TOP mRNAs in vivo. Nucleic Acids Res. 2000;28:2927–2934.
- Ali N, Pruijn GJM, Kenan‖ DJ, et al. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J Biol Chem. 2000;275:27531–27540.
- Meerovitch K, Svitkin YV, Lee HS, et al. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807.
- Izumi RE, Das S, Barat B, et al. A peptide from autoantigen la blocks poliovirus and hepatitis C virus cap-independent translation and reveals a single tyrosine critical for La RNA binding and translation stimulation. J Virol. 2004;78:3763–3776.
- Kim YK, Back SH, Rho J, et al. La autoantigen enhances translation of BiP mRNA. Nucleic Acids Res. 2001;29:5009–5016.
- Vinayak J, Marrella SA, Hussain RH, et al. Human La binds mRNAs through contacts to the poly(A) tail. Nucleic Acids Res. 2018;4:4228–4240.
- Savitzky A, Golay MJE. Smoothing + Differentiation of Data by Simplified Least Squares Procedures. Anal Chem. 1964;36:1627–1639.
